Introduction
Glioblastoma is one of the most common malignant
primary tumors and develops in brain (1). Glioblastoma is highly aggressive and
the survival rate of patients with glioblastoma was very low
(2,3). Every year, large amounts of patients
died from glioblastoma (4,5). Surgical resection, radiotherapy and
chemotherapy are the main methods for glioblastoma therapy. In
spite of some advances in tumor therapy, treatments for
glioblastomas still remain challenging. The tumor recurrence rate
is still very high because of infiltrative growth of glioblastoma
(6,7). Up to now, many efforts have been made
to understand the knowledge of glioblastoma biology. To develop
gene therapies toward targeted sites, which show an exciting
prospect, it is necessary to uncover the underlying molecular
mechanisms that regulate glioblastoma.
MicroRNAs (miRNAs) belong to noncoding RNAs, which
exert all kind of biological functions (8–10).
miRNAs can bind to the 3′UTR of target mRNAs to promote their
degradation and regulate gene expression with a
post-transcriptional manner (11).
Increasing evidences show that miRNAs participate in various
cellular processes such as development, proliferation and tumor
metastasis (12–15). Therefore, miRNAs may serve as
effective therapeutic targets and diagnostic biomarkers. Previous
study also showed that miRNAs possess important roles in
glioblastomas. For example, MicroRNA-141-3p promotes glioblastoma
cell growth by directly targeting p53 (16). However, most important miRNAs in
glioblastoma remains to be revealed. As for miR-500a-5p, Zhao et
al (17) reported that
miR-500a overexpression enhances hepatocarcinoma metastasis by
repressing PTEN expression. Degli et al (18), showed that miR-500a-5p regulates
breast cancer progression and predicts cancer survival.
Additionally, Guo et al (19), indicated that miR-500a promotes
migration and invasion in hepatocellular carcinoma by activating
the Wnt/β-catenin signaling pathway. The role of miR-500a-5p in
glioblastoma remains elusive and required to be defined.
In the present study, we aimed to investigate the
function and mechanism of miR-500a-5p in glioblastoma. By reverse
transcription-quantitative polymerase chain reaction (RT-qPCR), we
showed that miR-500a-5p was upregulated in glioblastoma tissues and
cells. CCK8 and Transwell assays indicated that miR-500a-5p
overexpression promoted glioblastoma cell proliferation, migration
and invasion in vitro. Moreover, xenograft experiments
illustrated that miR-500a-5p inhibition delayed tumor growth in
vivo. In mechanism, we found that miR-500a-5p directly targeted
CHD5. Summarily, our study highlighted the essential role of
miR-500a-5p on glioblastoma development and progression via
miR-500a-5p/CHD5 axis.
Materials and methods
Patient samples
All 60 tissue samples were collected from Harbin
Medical University (Heilongjiang, China). The present study
received ethical approval from Harbin Medical University. The
clinical characters of these samples were listed in Table I. Written informed consent
approving this study was obtained from each patient. Human
glioblastoma cell lines U-87MG (www.atcc.org/Products/All/HTB-14.aspx) and U251 were
from the American Tissue Culture Collection (ATCC, Manassas, VA,
USA). The U-87MG cell line is known to be cross-contaminated with
another cell line of unknown origin, but it is likely to be another
glioblastoma cell line (20).
U-87MG cells were grown in Minimum Essential Medium (HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). U251 cells were maintained in Dulbecco's High Glucose
Modified Eagle Medium (HyClone; GE Healthcare Life Sciences) with
10% FBS. Normal human astrocytes (NHA) obtained from Lonza
(www.lonza.com/products-services/bio-research/primary-cells/human-cells-and-media/neural-cells-and-media/nha-normal-human-astrocytes.aspx)
were cultured in the provided astrocyte growth media and 5% FBS.
Cells were incubated in a humidified atmosphere with 5%
CO2 at 37°C.
 | Table I.Clinical parameters of
microRNA-500a-5p in glioblastoma samples. |
Table I.
Clinical parameters of
microRNA-500a-5p in glioblastoma samples.
| Characteristics | Low expression | High expression | P-value |
|---|
| Age (years) |
|
| 1.00 |
| ≤60 | 19 | 18 |
|
|
>60 | 11 | 12 |
|
| Gender |
|
| 0.43 |
| Male | 20 | 16 |
|
|
Female | 10 | 14 |
|
| Grade |
|
| 0.02 |
| II | 14 | 6 |
|
|
III | 10 | 8 |
|
| IV | 6 | 16 |
|
| Tumor size
(cm) |
|
| 0.01 |
| ≤4 | 17 | 6 |
|
|
>4 | 13 | 24 |
|
Construction and infection
NC mimic, miR-500a-5p mimic and its corresponding
miR-500a-5p inhibitor were purchased from Guangzhou RiboBio Co.,
Ltd., (Guangzhou, China). Cell transfection was performed with
Lipofectamine 2000 Reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to manufacturer's protocol. After incubating, cells
were harvested then experiments were performed. Loss of CHD5
expression was achieved using small interfering RNA (siRNA) of
CHD5. To establish a stably CHD5-silenced cell line, a GV113
plasmid containing CHD5 lentivirus short hairpin RNAs
(TGGTTAAGGGCAGTGATAG) were separately transduced into U-87MG
cells.
CCK8 assay
Cell proliferation was detected by Cell Counting Kit
(7 sea biotech, Shanghai, China). Cells were grown in 96-well plate
with 1×104 per well and incubated in 37°C with 5%
CO2 until cell confluent rate reached 70%. After
transfected with plasmid for 48 h, cells were still incubated for
24, 48 and 72 h. 10 µl CCK8 solution was seed into each well. The
absorbance at 450 nm was measured with SUNRISE Microplate Reader
(Tecan Group, Ltd., Mannedorf, Switzerland).
Apoptosis assay
Cell apoptosis was measured by flow cytometry
following the instructions of the Annexin V-FITC/PI apoptosis
detection kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China).
Cultured cells were harvested and washed twice in PBS, re-suspended
in binding buffer, and then incubated with Annexin V-FITC and
propidium iodide (PI) for 15 min at room temperature. Afterwards,
flow cytometry was performed to determine rate of apoptosis on a
FACSAria flow cytometer (BD Biosciences, Franklin Lakes, NJ,
USA).
In vitro migration and invasion
assays
Twenty-four-well Transwell chambers with 8-µm pore
size polycarbonate (Corning Incorporated, Corning, NY, USA) were
used for cell migration and invasion assays. For invasion assays,
the top side of the membrane was coated with Matrigel (BD
Biosciences), and then 1×105 cells (in each well) in
serum-free DMEM or RPMI 1640 medium were seeded on the chambers.
DMEM or RPMI 1640 containing 10% FBS was added to the wells under
the chamber. For migration analysis, 5×104 cells (in
each well) in serum-free DMEM or RPMI 1640 medium were seeded on
the chambers without Matrigel. After 24 h of incubation, cotton
swabs were used to remove the cells inside the upper chamber, while
the cells on the other side of the membrane surface were fixed and
stained with 0.5% crystal violet solution. Five random fields were
counted in each well.
Bioinformatics analysis
The potential targets of miR-500a-5p were predicted
using the TargetScan (www.targetscan.org), microRNA (www.microrna.org/) and miRDB (www.mirdb.org). The common genes of these algorithms
were selected for further analysis.
RT-qPCR
The total RNA of the tissue samples and cells was
isolated using TRIzol Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), in accordance with manufacturer's instructions.
Then, 1 µg of total RNA was reverse-transcribed in a volume of 20
µl using random and oligo dT primers under standard conditions, in
accordance with the instructions of the PrimeScript RT kit (Takara
Biotechnology Co., Ltd., Dalian, China). For RT-qPCR assays, we
used SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) to
determine the expression level of CHD5, in accordance with
manufacturer's instructions. The thermocycling conditions were as
follows: 95°C for 30 sec, followed by 40 cycles at 95°C for 15 sec
and 60°C for 32 sec and dissociation at 95°C for 60 sec, 55°C for
30 sec and 95°C for 30 sec. The expression data of CHD5 were
normalized to the expression of glyceraldehyde-3-phosphate
dehydrogenase (GAPDH). The primers used were as follows: Forward
primer, 5′-CTGACTTCAACAGCGACACC-3′ and reverse primer,
5′-TCTGACTTCAACAGCGACACC-3′. The relative fold change was
calculated using the 2−ΔΔCq method (21).
In vivo assay
The protocol was described previously (22). Four-week-old athymic BALB/c nude
mice were maintained under specific pathogen-free conditions. The
mice were manipulated in accordance with the protocols approved by
Harbin Medical University. The U-87MG cells were harvested and
washed with PBS. Then, 1×107 cells were subcutaneously
injected into the ventral side of each mouse for tumor formation
assays. Six mice were used for each group. The tumor volumes were
examined every 7 days and calculated. The protocol was approved by
the Animal Care Ethics Committee of Harbin Medical University.
Details in Sukru's cohort
(GSE90598)
In this dataset, 16 fresh-frozen glioblastoma
multiforme samples, 7 healthy brain tissues, a NHA cell line and
human fetal astrocyte cell line were analyzed by using miRNA and
whole transcriptome microarray chips.
Luciferase reporter assay
U-87MG cells were seeded into a 24-well plate. Cells
were co-transfected with wild-type, mutated CHD5 reporter plasmid
or pMIR vector, and miR-500a-5p mimics or miR-500a-5p inhibitor.
Luciferase assays were conducted 24 h after transfection using the
Dual Luciferase Reporter Assay System (Promega Corporation,
Madison, WI, USA).
Statistical snalysis
Statistical analysis was performed using SPSS v18.0
software (SPSS, Inc., Chicago, IL, USA). Each experiment was
repeated at least three times. All data are expressed as the mean ±
standard deviation. The Kaplan-Meier method was used to calculate
the survival curve, and log-rank test to determine statistical
significance. Student's t-test and one-way analysis of variance
followed by Tukey's post hoc test were used to analyze 2 or
multiple groups, respectively, for statistical significance. A
paired Student's t-test was used to compare differences between
human glioblastoma and normal adjacent tissue samples. Diagnostic
potential of miR-500a-5p expression was assessed by receiver
operating characteristic (ROC) analysis. The respective area under
the curve (AUC) was analyzed by the Hanley and McNeil method. For
analysis of correlation between miR-500a-5p levels and clinical
features, chi-square tests were used. Pearson correlation
coefficient analysis was used to determine the correlations.
P<0.05 was considered to indicate a statistically significant
difference.
Results
MiR-500a-5p is significantly
upregulated in glioblastoma tissues
To explore the function miR-500a-5p, we firstly
examined its expression in glioblastoma tissues by RT-qPCR. We
found that miR-500a-5p was significantly upregulated in
glioblastoma tissues compared with adjacent normal tissues
(Fig. 1A). Similarly, the high
expression of miR-500a-5p was verified in glioblastoma cell lines
(Fig. 1B). Then we divided these
samples into two groups according to miR-500a-5p expression levels
(mean value was the cut-off). Kaplan-Meier analysis was used to
evaluate survival. The results showed that patients with higher
miR-500a-5p expression possessed poorer survival (Fig. 1C). Furthermore, ROC curves were
performed to evaluate the sensitivity and specificity of
miR-500a-5p expression in predicting glioblastoma tissues from
normal tissues. Notably, miR-500a-5p displayed predictive, with an
AUC of 0.753 (P = 0.046; Fig. 1D).
These results implied that miR-500a-5p might provide imperative
clinical significance in glioblastoma diagnosis.
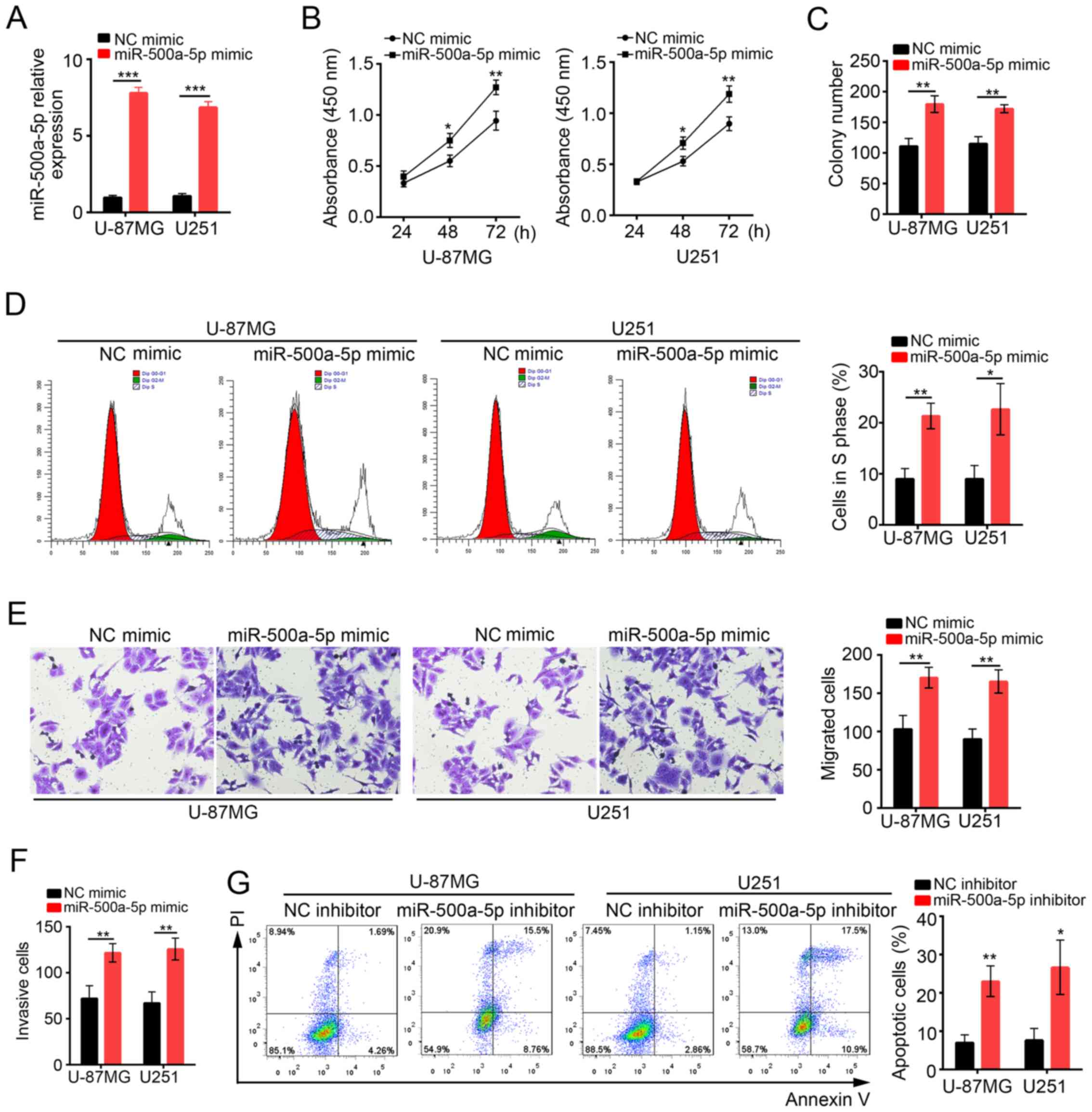
MiR-500a-5p enhances glioblastoma cell
proliferation, migration and invasion in vitro
To determine the functions of miR-500a-5p in
glioblastoma, we overexpressed miR-500a-5p in U-87MG and U251 cells
by transduction with NC mimic or miR-500a-5p mimic. At 48 h after
transduction with miR-500a-5p, miR-500a-5p expression was
significantly increased compared with NC mimic group (Fig. 2A). Then CCK8 assay was conducted to
analyze the proliferation of U-87MG and U251 cells at 24, 48 and 72
h post transfection (Fig. 2B).
Overexpression of miR-500a-5p observably promoted cell
proliferation in both U-87MG MG and U251 cells. Similarly, colony
formation assay showed that cells transfected with miR-500a-5p
formed more clones (Fig. 2C). In
consistence, we found that more U-87MG and U251 cells transfected
with miR-500a-5p entered into S phase than control (Fig. 2D). To evaluate the effect of
miR-500a-5p on cell migration and invasion, we performed transwell
assays. We found that overexpression of miR-500a-5p promoted cell
migration and invasion (Fig. 2E and
F). In addition, we inhibited miR-500a-5p in U-87MG and U251
cells to evaluate the effect on cell apoptosis. Flow Cytometry by
Annexin V/PI staining showed that apoptotic ratio was significantly
higher in miR-500a-5p inhibitor-transfected cells than control
(Fig. 2G). In collection,
miR-500a-5p promoted cell proliferation, migration and invasion in
glioblastoma, but inhibited cell apoptosis.
MiR-500a-5p delayed tumor growth in
vivo
To further determine the effects of miR-500a-5p on
glioblastoma cells in vivo, we examined tumor growth in nude
mice. U-87MG cells transfected with NC inhibitor or miR-500a-5p
inhibitor were subcutaneously inoculated into the armpits of the
recipient nude mice. Every other 7 day, we measured the tumor
volumes. We found that miR-500a-5p knockdown significantly
inhibited tumor growth in vivo (Fig. 3A). At the endpoint of the
experiments, the tumor weights were measured. The tumors in
miR-500a-5p knockdown group were significantly lighter than that in
control group (Fig. 3B).
MiR-500a-5p specifically targeted CHD5
3′-UTR in glioblastoma
To explore the molecular mechanism through which
miR-500a-5p promoted glioblastoma progression, three computational
algorithms including TargetScan, miRanda and PicTar were used in
combination to search for potential targets of miR-500a-5p. Among
the candidates, the chromatin remodeler and tumor suppressor CHD5
was predicted to be a target of miR-500a-5p. The predicted
interaction between miR-500a-5p and 3′UTR of CHD5 was illustrated
(Fig. 4A). In order to verify this
prediction, we cloned 3′UTR-wt and 3′UTR-mut into pMIR-REPORT
vector. As expected, dual luciferase assay demonstrated that
miR-500a-5p overexpression remarkably inhibited the luciferase
activity while miR-500a-5p knockdown increased the luciferase
activity (Fig. 4B). In contrast,
the effect of miR-500a-5p on luciferase activity observed in
pMIR-3′-UTR-wt was absent in pMIR-3′-UTR-mut (Fig. 4B). Moreover, we found that
overexpression of miR-500a-5p decreased the mRNA and protein levels
of CHD5 in U-87MG and U251 cells while miR-500a-5p knockdown got
the inverse result (Fig. 4C and
D). In addition, by RT-qPCR we found that the expression of
miR-500a-5p was inversely correlated with that of CHD5 in
glioblastoma tissues (Fig. 4E).
Furthermore, the dataset (GSE90598) in GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90598)
showed that the expression of CHD5 was significantly downregulated
in glioblastoma tissues compared with normal tissues (Fig. 4F). Similarly, RT-qPCR also showed
that the mRNA levels of CHD5 were lower in glioblastoma samples
than normal tissues (Fig. 4G).
These results indicated that CHD5 mRNA 3′-UTR is a specific
functional target of miR-500a-5p in glioblastoma cells.
MiR-500a-5p regulated glioblastoma
cell proliferation, migration and invasion by targeting CHD5 in
vitro and in vivo
To investigate whether the regulation of cell
proliferation, migration and invasion of glioblastoma cells by
miR-500a-5p is executed via a CHD5-dependent manner, we
co-transfected U-87MG and U251 cells with miR-500a-5p inhibitor and
CHD5 siRNA. Compared with cells transfected with miR-500a-5p
inhibitor, the cells transfected with both miR-500a-5p inhibitor
and CHD5 siRNA exhibited a lower expression on CHD5 protein level
(Fig. 5A). CCK8 and colony
formation assays showed that CHD5 knockdown reversed the inhibitory
effects by miR-500a-5p inhibition on cell proliferation potential
in U-87MG and U251 cells (Fig. 5B and
C). In addition, transwell assays showed that CHD5 knockdown
reversed the inhibitory effects by miR-500a-5p inhibition on cell
migration and invasion potential in U-87MG and U251 cells (Fig. 5D and E). Furthermore, CHD5
knockdown decreased the cell apoptosis induced by miR-500a-5p
inhibition (Fig. 5F). What's more,
CHD5 knockdown also increased tumor weights to the control level
in vivo (Fig. 5G). Besides,
WB assay with formed tumor tissues showed that CHD5 knockdown
reversed the inhibitory effects by miR-500a-5p inhibition on cell
proliferation, migration and invasion in vivo (Fig. 5H). Accumulating studies showed that
Wnt/β-catenin signaling is indispensable for cancer development. We
then performed WB assays with formed tumor tissues and found that
miR-500a-5p inhibition downregulated Wnt/β-catenin signaling while
CHD5 knockdown upregulated it (Fig.
5I), which indicated that miR-500a-5p/CHD5 regulated
glioblastoma development and progression through Wnt/β-catenin
signaling at least in part.
 | Figure 5.miR-500a-5p regulates glioblastoma
cell proliferation, migration and invasion by targeting CHD5 in
vitro and in vivo. (A) Western blot assay showed the
protein levels of CHD5 in U-87MG and U251 cells transfected with
miR-363-3p inhibitor. (B) Cell Counting Kit-8 proliferation assay
demonstrated the effects of miR-500a-5p and/or CHD5 knockdown on
U-87MG and U251 cell proliferation. (C) Colony formation assay
revealed the effects of miR-500a-5p and/or CHD5 knockdown on U-87MG
and U251 cell proliferation. (D and E) Migration and invasion
assays demonstrated the effects of miR-500a-5p and/or CHD5
knockdown on the migration and invasion of U-87MG and U251 cells
(magnification, ×200). (F) Fluorescence activated cell sorter assay
by staining with Annexin V/PI showed the effects of miR-500a-5p
and/or CHD5 knockdown on U-87MG and U251 cell apoptosis. (G) Tumor
weights of U-87MG xenograft tumors were measured at the end of
experiments. (H) Western blot assay revealed the effects of
miR-500a-5p and/or CHD5 knockdown on tumor proliferation and
migration in vivo. (I) Western blot assay indicated that
miR-500a-5p activated Wnt/β-catenin signaling by targeting CHD5 in
glioblastoma. All data are representative of three independent
experiments and expressed as the mean ± standard deviation.
*P<0.05, **P<0.01 and ***P<0.001 vs. the NC group. miR,
microRNA; CHD5, chromodomain helicase DNA binding protein 5; PI,
propidium iodide; sh-, short hairpin RNA; TCF1, transcription
factor 7; BCL2, B-cell lymphoma-2; PCNA, proliferating cell nuclear
antigen; TWIST, Twist family basic helix-loop-helix transcription
factor; MMP9, matrix metalloproteinase; SNAI1, Snail family
transcription repressor 1; NC, negative control. |
Discussion
Glioblastoma was one of the most common clinical
primary brain tumors. However, the mechanism that regulates
glioblastoma development and progression remains largely unknown.
Previous studies demonstrated that miRNAs exerted pivot functions
in all kinds of cancers including pancreatic cancer, glioblastoma,
breast carcinoma, hepatocellular carcinoma and so on (23–26).
In glioblastoma, some miRNAs are reported to exert important
functions. For instance, microRNA-101 inhibits proliferation,
migration and invasion of human glioblastoma by targeting SOX9
(27). Nevertheless, the functions
of most of miRNAs are unknown in glioblastoma. Therefore, there is
an urgent need to define the molecular mechanism that regulates the
genesis of glioblastoma, in order to develop effective
therapeutics. Previous research shows that miR-500a increases
cancer stem cells properties by STAT3 pathway in human
hepatocellular carcinoma (28).
Besides, MicroRNA-500a also enhances migration and invasion in
hepatocellular carcinoma by activating the Wnt/β-catenin signaling
pathway (19). However, the
functions of miR-500a-5p remain to be elucidated in glioblastoma.
In our study, we showed that the expression of miR-500a-5p was
significantly upregulated in glioblastoma tissues compared to
normal tissues, which indicated that miR-500a-5p may act as on
oncogene in glioblastoma.
Aberrant miRNAs expression is closely related to
various types of tumors (29). In
the present study, we demonstrated the highly expression of
miR-500a-5p in glioblastoma tissues and cell lines. In addition, by
analysis with Kaplan-Meier survival curve and ROC, we showed that
the expression of miR-500a-5p in glioblastoma can serve as a new
biomarker for the diagnosis and prognosis of patients with
glioblastoma. To further determine the roles of miR-500a-5p in
glioblastoma, we overexpressed miR-500a-5p in U-87MG and U251
cells. By CCK8, colony formation and transwell assays, we showed
that overexpression of miR-500a-5p promoted cell proliferation,
migration and invasion in U-87MG and U251 cells in vitro.
Besides, we also knocked down miR-500a-5p by transfection with
miR-500a-5p inhibitor. By FACS with Annexin V/PI staining, we found
that miR-500a-5p increased the apoptosis of U-87MG and U251 cells.
To further explore the physiological function of miR-500a-5p, we
conducted xenograft experiments, which indicated that miR-500a-5p
inhibition significantly inhibited glioblastoma growth in
vivo.
Up to now, the targets of miR-500a-5p have not been
identified. On the basis of bioinformatics analysis, we predicted
CHD5 as a target of miR-500a-5p in glioblastoma. CHD5 is a
chromatin remodeler and serves as a tumor suppressor in various
tumors. For example, CHD5 is a potential tumor suppressor in
non-small cell lung cancer (NSCLC) (30). In glioblastoma, a report showed
that CHD5 is downregulated in human glioblastoma (31). Nevertheless, how the expression of
CHD5 is regulated remains unknown in glioblastoma. By luciferase
reporter assay, we showed that the miR-500a-5p directly bond to the
3′-UTR of CHD5 mRNA. RT-qPCR and WB results showed that miR-500a-5p
inhibited the mRNA and protein level of CHD5 in glioblastoma cells.
Besides, we showed that miR-500a-5p activated Wnt/β-catenin
signaling while CHD5 inhibited this signaling in glioblastoma. Our
results indicated that miR-500a-5p promoted cell proliferation,
migration and invasion by targeting CHD5 and subsequently
activating Wnt/β-catenin signaling. However, how CHD5 mediates
activation of Wnt/β-catenin pathway needs further investigation.
And the roles of CHD5-mediated activation of Wnt/β-catenin
signaling on glioblastoma progression still require to be
defined.
In summary, the present study provides new insights
into the mechanism of glioblastoma progression, and suggests that
miR-500a-5p might potentially serve as therapeutic target for
glioblastoma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZL and JM conceived and designed the present study,
analyzed and interpreted the results, and wrote the manuscript. DS
and XQ performed the experiments. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
For the use of human samples, the protocol for the
present study was approved by the Institutional Ethics Committee of
Harbin Medical University (Heilongjiang, China) and all enrolled
patients signed a written informed consent document. In addition,
all procedures involving animals conformed to the national
guidelines of and were approved by the Animal Care Ethics Committee
of Harbin Medical University.
Patient consent for publication
All patients recruited to the present study provided
written informed consent for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ilkanizadeh S, Lau J, Huang M, Foster DJ,
Wong R, Frantz A, Wang S, Weiss WA and Persson AI: Glial
progenitors as targets for transformation in glioma. Adv Cancer
Res. 121:1–65. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang K, Sun X, Zhou X, Han L, Chen L, Shi
Z, Zhang A, Ye M, Wang Q, Liu C, et al: Long non-coding RNA HOTAIR
promotes glioblastoma cell cycle progression in an EZH2 dependent
manner. Oncotarget. 6:537–546. 2015.PubMed/NCBI
|
|
3
|
Sadetzki S, Zach L, Chetrit A, Nass D,
Hoffmann C, Ram Z, Zaaroor M, Umansky F, Rappaport ZH, Cohen A, et
al: Epidemiology of gliomas in Israel: A nationwide study.
Neuroepidemiology. 31:264–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee JH, Jung TY, Jung S, Kim IY, Jang WY,
Moon KS and Jeong EH: Performance status during and after
radiotherapy plus concomitant and adjuvant temozolomide in elderly
patients with glioblastoma multiforme. J Clin Neurosci. 20:503–508.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liang BC, Thornton AF Jr, Sandler HM and
Greenberg HS: Malignant astrocytomas: Focal tumor recurrence after
focal external beam radiation therapy. J Neurosurg. 75:559–563.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Héricé C, Khalil R, Moftah M, Boraud T,
Guthrie M and Garenne A: Decision making under uncertainty in a
spiking neural network model of the basal ganglia. J Integra
Neurosci. 15:515–538. 2016. View Article : Google Scholar
|
|
8
|
Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B,
Du Y, Gao G, Tian Y, He L and Fan Z: LncBRM initiates YAP1
signalling activation to drive self-renewal of liver cancer stem
cells. Nat Commun. 7:136082016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu
P, Du Y, Wu J, Qin X, Chen R, et al: Long noncoding RNA lncKdm2b is
required for ILC3 maintenance by initiation of Zfp292 expression.
Nat Immunol. 18:499–508. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smolle MA, Leithner A, Posch F, Szkandera
J, Liegl-Atzwanger B and Pichler M: MicroRNAs in different
histologies of soft tissue sarcoma: A comprehensive review. Int J
Mol Sci. 18:pii: E1960. 2017. View Article : Google Scholar
|
|
11
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Townley-Tilson WH, Callis TE and Wang D:
MicroRNAs 1, 133, and 206: Critical factors of skeletal and cardiac
muscle development, function, and disease. Int J Biochem Cell Biol.
42:1252–1255. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Xu D, Li N, Li Y, He Y, Hu X, Lyu
L and He L: Downregulation of microRNA-31 inhibits proliferation
and induces apoptosis by targeting HIF1AN in human keloid.
Oncotarget. 8:74623–74634. 2017.PubMed/NCBI
|
|
14
|
Zhou C, Jiang CQ, Zong Z, Lin JC and Lao
LF: miR-146a promotes growth of osteosarcoma cells by targeting
ZNRF3/GSK-3β/β-catenin signaling pathway. Oncotarget.
8:74276–74286. 2017.PubMed/NCBI
|
|
15
|
Qu Y, Liu H, Lv X, Liu Y, Wang X, Zhang M,
Zhang X, Li Y, Lou Q, Li S and Li H: MicroRNA-16-5p overexpression
suppresses proliferation and invasion as well as triggers apoptosis
by targeting VEGFA expression in breast carcinoma. Oncotarget.
8:72400–72410. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou X, Wu W, Zeng A, Nie E, Jin X, Yu T,
Zhi T, Jiang K, Wang Y, Zhang J and You Y: MicroRNA-141-3p promotes
glioma cell growth and temozolomide resistance by directly
targeting p53. Oncotarget. 8:71080–71094. 2017.PubMed/NCBI
|
|
17
|
Zhao Y and Wang Y and Wang Y: Up-regulated
miR-500a enhances hepatocarcinoma metastasis by repressing PTEN
expression. Biosci Rep. 37:2017. View Article : Google Scholar
|
|
18
|
Esposti Degli D, Aushev VN, Lee E, Cros
MP, Zhu J, Herceg Z, Chen J and Hernandez-Vargas H: miR-500a-5p
regulates oxidative stress response genes in breast cancer and
predicts cancer survival. Sci Rep. 7:159662017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo Y, Chen L, Sun C and Yu C:
MicroRNA-500a promotes migration and invasion in hepatocellular
carcinoma by activating the Wnt/β-catenin signaling pathway. Biomed
Pharmacother. 91:13–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re32016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gaur AB, Holbeck SL, Colburn NH and Israel
MA: Downregulation of Pdcd4 by mir-21 facilitates glioblastoma
proliferation in vivo. Neuro Oncol. 13:580–590. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu YF, Hannafon BN, Zhao YD, Postier RG
and Ding WQ: Plasma exosome miR-196a and miR-1246 are potential
indicators of localized pancreatic cancer. Oncotarget.
8:77028–77040. 2017.PubMed/NCBI
|
|
24
|
Rizzo S, Cangemi A, Galvano A, Fanale D,
Buscemi S, Ciaccio M, Russo A, Castorina S and Bazan V: Analysis of
miRNA expression profile induced by short term starvation in breast
cancer cells treated with doxorubicin. Oncotarget. 8:71924–71932.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li W, Yang W, Liu Y, Chen S, Chin S, Qi X,
Zhao Y, Liu H, Wang J, Mei X, et al: MicroRNA-378 enhances
inhibitory effect of curcumin on glioblastoma. Oncotarget.
8:73938–73946. 2017.PubMed/NCBI
|
|
26
|
Zhu W, Qian J, Ma L, Ma P, Yang F and Shu
Y: MiR-346 suppresses cell proliferation through SMYD3 dependent
approach in hepatocellular carcinoma. Oncotarget. 8:65218–65229.
2017.PubMed/NCBI
|
|
27
|
Liu N, Zhang L, Wang Z, Cheng Y, Zhang P,
Wang X, Wen W, Yang H, Liu H, Jin W, et al: MicroRNA-101 inhibits
proliferation, migration and invasion of human glioblastoma by
targeting SOX9. Oncotarget. 8:19244–19254. 2017.PubMed/NCBI
|
|
28
|
Jiang C, Long J, Liu B, Xu M, Wang W, Xie
X, Wang X and Kuang M: miR-500a-3p promotes cancer stem cells
properties via STAT3 pathway in human hepatocellular carcinoma. J
Exp Clin Cancer Res. 36:992017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mei Z, He Y, Feng J, Shi J, Du Y, Qian L,
Huang Q and Jie Z: MicroRNA-141 promotes the proliferation of
non-small cell lung cancer cells by regulating expression of PHLPP1
and PHLPP2. FEBS Lett. 588:3055–3061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Baykara O, Tansarikaya M, Bulut P,
Demirkaya A and Buyru N: CHD5 is a potential tumor suppressor in
non small cell lung cancer (NSCLC). Gene. 618:65–68. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang L, He S, Tu Y, Ji P, Zong J, Zhang J,
Feng F, Zhao J, Gao G and Zhang Y: Downregulation of chromatin
remodeling factor CHD5 is associated with a poor prognosis in human
glioma. J Clin Neurosci. 20:958–963. 2013. View Article : Google Scholar : PubMed/NCBI
|