Introduction
Breast cancer is the leading cause of
cancer-associated mortality in women (1,2). The
mortality rate is >450,000 individuals per year (3). The World Health Organization's
previous data suggested the number of breast cancer cases has been
gradually increasing worldwide since 2008 (4). Despite the availability of advanced
therapeutic modalities, current options to manage breast cancer
remain inadequate (5,6). Using drugs to treat or manage the
progression of breast cancer is the recommended option, however,
its efficacy has been seriously compromised due to the development
of resistance (7,8). Therefore, it is vital to identify
drugs that are able to target novel sites and act in a specific
manner. Previous studies reported that epidermal growth factor
receptor-tyrosine kinase (EGFR-TK) was aberrantly overexpressed in
breast cancer (9,10). As a result, EGFR-TK has attracted
interest as a potential target from a number of studies across the
globe searching for novel therapy agents (11–13).
Various studies have reported that EGFR and Wnt signaling cascades
are associated with and conserved in neoplasia. EGFR and Wnt
signaling pathways are considered to serve key roles in embryonic
development and cell proliferation. It is well established that
dysregulation of these two pathways frequently leads to
tumorigenesis with a poor prognosis. EGFR mutations are associated
with a better prognosis in non-small cell lung cancer patients with
unmethylated Wnt antagonists compared with patients with
methylations, which points to a functional cooperation. Thus, it is
hypothesized that activation of β-catenin by inactivation of
E-cadherin activates EGFR, and thus effectively creates a positive
feedback loop and potential targets for anticancer agents.
Dueto their excellent pharmacological activity,
1,3,5-triazine derivatives have received considerable attention
from medicinal chemists (14).
These derivatives have been demonstrated to exert strong
antibacterial (15,16), antiviral (17), antifungal (18), antimalarial (19,20),
antidiabetic (21) and cystic
fibrosis transmembrane conductance regulator-modulatory effect
(22). Previously, these
derivatives have been demonstrated to exert potent anticancer
activity and it has been reported that substitution of various
chemical groups on 1,3,5-triazine exhibited pronounced effect on
the tumor cell proliferation (13,23).
Therefore, in the present study, the EGFR-TK inhibitory activity of
certain previously reported 1,3,5-triazines as anti-breast cancer
agents were investigated. Furthermore, the effect of the compounds
were evaluated on multiple breast cancer cell lines, specifically,
the highly metastatic MDA-MB-231, human epidermal growth factor
receptor 2 (HER2)-positive BT-474 and estrogen receptor
(ER)-positive MCF7 cell lines. The effect of the compounds on
β-catenin was also assessed.
Materials and methods
Synthesis of compounds
The synthesis of the compounds was described in a
previous study and the purity of the compounds was assessed on the
basis of standard melting point (21).
Molinspiration
Physicochemical parameters have a vital role in the
modulation of the bioactivity of chemical entities. Molinspiration
(www.molinpiration.com), web-based
software was used to obtain various parameters, including
octanol/water partition coefficient (MiLogP), topological polar
surface area (TPSA) and drug likeness. The MiLogP is calculated
using the method developed by Molinspiration by summing the
fragment-based contributions and correction factors. The method is
robust and is able to process practically all organic and most
organometallic molecules. TPSA is calculated based on the
methodology published by Ertl et al (24) as a sum of the fragment-based
contributions (25) in which O-
and N-centered polar fragments are considered and the surface areas
that are occupied by oxygen and nitrogen atoms and by hydrogen
atoms attached to them are calculated. TPSA has been demonstrated
to be a useful factor for characterizing drug absorption, including
intestinal absorption, bioavailability, Caco-2 permeability and
blood brain barrier penetration. Therefore, TPSA is associated with
the hydrogen bonding potential of a compound. The compounds were
evaluated on the basis of these parameters.
Docking study
Docking calculations were performed using Docking
Server (26). Gasteiger partial
charges were added to the ligand atoms. Non-polar hydrogen atoms
were merged and rotatable bonds were defined. Docking calculations
were carried out on compounds (ligands) using the EGFR kinase
domain model (Protein code: 1M17.pdb; rcsb.org/3d-view/1M17). Essential hydrogen atoms,
Kollman united atom type charges and solvation parameters were
added with the aid of AutoDock tools (27). Affinity (grid) maps of 60×60×60 Å
grid points and 0.375 Å spacing were generated using the Autogrid
program. AutoDock parameter set- and distance-dependent dielectric
functions were used in the calculation of van der Waals and the
electrostatic forces, respectively. Docking simulations were
performed using the Lamarckian genetic algorithm and the Solis-Wets
local search method (28). Initial
position, orientation and torsions of the ligand molecules were set
randomly. All rotatable torsions were released during docking. Each
docking experiment was derived from 10 different runs that were set
to terminate following a maximum of 250,000 energy evaluations. The
population size was set to 150. During the search, a translational
step of 0.2 Å and quaternion, and torsion steps of 5 were
applied.
Cell lines
Three breast cancer cell lines, specifically, the
highly metastatic MDA-MB-231, HER2-positive BT-474 and ER-positive
MCF7 cell lines were obtained from the American Type Culture
Collection (Manassas, VA, USA). The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM), RPMI (Sigma-Aldrich,
Merck KGaA, Darmstadt, Germany) and α-minimal essential medium
(Sigma-Aldrich, Merck KGaA), for the MDA-MB-231, HER2-positive
BT-474 and ER-positive MCF7 cells, respectively. Cells were
cultured at 37°C with 5% CO2 and 100% humidity. The
medium was supplemented with 10% fetal bovine serum (FBS; HyClone;
GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml penicillin
and 100 µg/ml streptomycin.
MTT assay
MTT (Sigma-Aldrich; Merck KGaA) assay was used to
evaluate the effect of compounds (1a-1d) on cell proliferation
capacity. Cells were cultured in a 96-well plate at a density of
7×103 cells/well and in a volume of 200 µl. Stock
solutions of the compounds were prepared in dimethyl sulfoxide
(DMSO). The cells were then treated with compound 1d (0, 10, 25 and
50 nM). The final concentration of the solvent in the medium was
0.5%. At appropriate time intervals, the medium was removed and
replaced with 200 µl 0.5 mg/ml MTT in the growth medium and then
the plates were transferred to a 37°C incubator for 3 h. Then the
medium was removed and the purple formazan crystals were dissolved
in DMSO (200 µl/well). Absorbance was determined on an ELISA plate
reader (Biotek Instruments, Inc., Winooski, VT, USA) with a test
wavelength of 570 nm and a reference wavelength of 630 nm to obtain
the sample signal optical density (OD; 570–630 nm).
EGFR-TK inhibitory activity
Kinase activity was determined using Kinase-Glo Plus
luminescence kinase assay kit (Promega Corporation, Madison, WI,
USA) by calculating the amount of adenosine triphosphate (ATP)
remaining in the kinase reactionsolution. The luminescent signal is
correlated with the residual amount present and it was inversely
associated with kinase activity. The tested compounds were diluted
to 100 mM in 10% DMSO, then 5 ml the dilution was added to a 50 ml
reaction. All of the enzymatic reactions were performed at 30°C for
40 min using a 50 ml reaction volume containing 10 mM
MgCl2, 40 mM Tris, (pH 7.4), 0.1 mg/ml bovine serum
albumin, 0.2 mg/ml poly (Glu, Tyr) substrate, 10 mM ATP and EGFR.
The plate was incubated for 5 min at room temperature then 50 ml
Kinase-GloPlus Luminescence kinase assay regent was added to each
reaction. ADP-Glo assay kit (Promega Corporation) is a protein
kinase assays used to determine IC50 values in which adenosine
diphosphate (ADP) generation was measured which leads to an
increase in the luminescence signal. The reaction mixture was
incubated in a 96-well plate at 30°C for 30 min, following the
incubation period 25 ml ADP-Glo reagent was added to terminate the
assay. The 96-well plate agitated for 30 min at ambient temperature
and incubated, then 50 ml kinase detection reagentwas added. The
96-well plate was read using the ADP-Glo Luminescence reader. All
the assay components were added to the blank control except the
substrate. The blank control value was used to correct the activity
for each protein kinase target is determined.
Apoptosis assay
Cells at a density of 1×105 cells/well
were cultured in 96-well plates in medium supplemented with 10% FBS
for 24 h and was followed by treatment with compound 1d (0, 10, 25
or 50 nM). After 48 h, cells were collected by 300–350 × g for 5
min at room temperature, rinsed with PBS, fixed in 4%
paraformaldehyde for 30 min at room temperature and then rinsed
again with PBS to remove the fixing agent. The fixed cells were
resuspended in PBS that already containing 5 µg/ml Hoechst 33258
and incubated at room temperature for 15 min in the dark room. The
cells were examined to record the percentages of apoptotic cells by
determining the nuclear condensation and chromatin fragmentation
via fluorescence microscopy. A total of 250 nuclei from random
microscopic fields were examined for the quantification of the
apoptotic rate.
Western blot analysis
Total proteins were extracted using a RIPA buffer
(Beyotime Institute of Biotechnology, Haimen, China). A
bicinchoninic protein assay kit was used to quantify protein
concentrations. Then, the protein samples were separated by 12%
SDS-PAGE and transferred to polyvinylodene difluoride membranes.
The membranes were blocked with 5% skim milk powder overnight at
4°C. Primary antibodies against β-catenin (~90 kDa) and β-actin
were purchased from Santa Cruz Biotechnology, Inc.,
(anti-β-catenin; cat. no. sc-7963; 1:200; anti-β-actin, cat. no.
sc-130301; 1:10,000) and incubated overnight at 4°C. The secondary
antibody, goat anti-mouse immunoglobulin (Ig) G-horseradish
peroxidase, was purchased from Santa Cruz Biotechnology, Inc.,
(cat. no. sc-2005; 1:5,000) and incubated at 37°C for 1 h. Finally,
the bands were visualized using an enhanced chemiluminescence kit
(Solarbio, Beijing, China) and analyzed by Image pro plus (version
9.3; Media Cybernetics, Inc., Rockville, MD, USA).
Statistical analysis
All experiments were done in triplicate or more.
One-way analysis of variance was used to estimate overall
significance followed by post-hoc Tukey's tests corrected for
multiple comparisons. Data are presented as the mean ± standard
deviation of the mean. P<0.05 was considered to indicate a
statistically significant difference. Statistical analyses were
performed with GraphPad Prism 5.0 (GraphPad Software, Inc., San
Diego, CA, USA).
Results
Molecular properties of
1,3,5-triazines
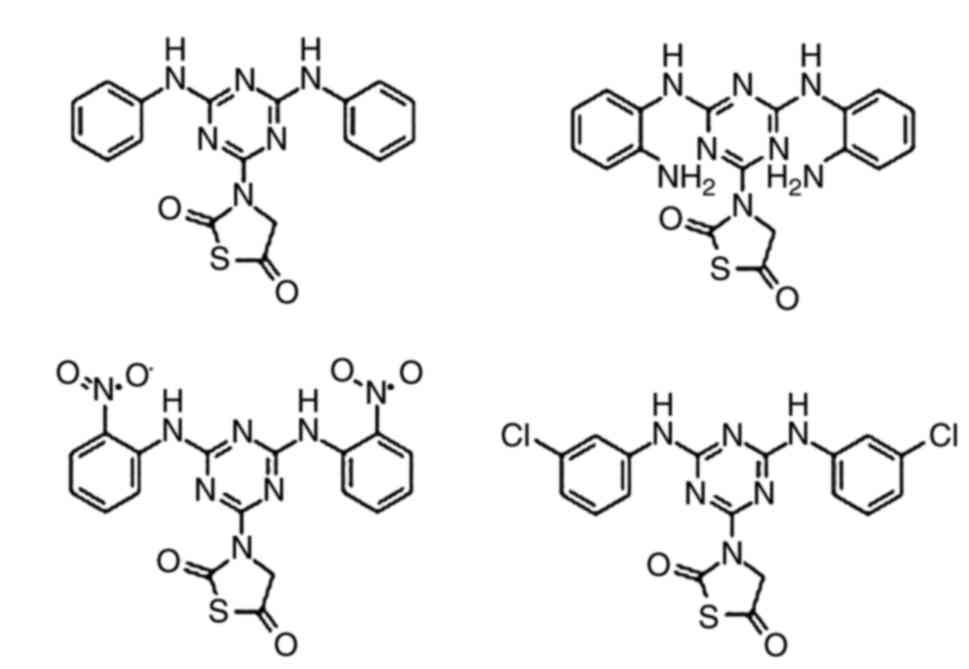
In order to find a probable candidate for an
anti-breast cancer agent as shown in Fig. 1, four previously reported
1,3,5-triazine compounds were selected for the present study. The
molecular properties of the compounds are presented in Table I, where the molecules were assessed
on the parameters of the Lipinski's rule of five (29). The molecules demonstrated no
violations of the rules and proved to be effective drug candidate
to be act as anticancer agent.
 | Table I.Molecular property calculation of
target compounds. |
Table I.
Molecular property calculation of
target compounds.
| Compound | Mutagenic | Tumorigenic | Irritant | Reproductive
effective | ClogP | Solubility | MW | TPSA | Drug likeness | Drug score |
|---|
| 1A | +++ | + | +++ | + | 3.51 | −5.31 | 378.0 | 125.4 | 2.11 | 0.2 |
| 1B | +++ | + | +++ | + | 2.16 | −5.46 | 408.0 | 177.4 | 1.63 | 0.2 |
| 1C | +++ | + | +++ | + | 1.67 | −6.23 | 468.0 | 217.0 | −3.25 | 0.09 |
| 1D | +++ | + | +++ | + | 4.73 | −6.78 | 446.0 | 125.4 |
3.78 | 0.13 |
Docking analysis of 1,3,5-triazine at
the EGFR-TK
As the drug likeness profile of the 1,3,5-triazine
derivatives was good, docking analysis with EGFR-TK protein domain
was performed. The results have been presented in Table II. It has been demonstrated that
all compounds exhibited good inhibition constants against the
target protein ranging from 0.44 nM to 3.10 µM and engaging the key
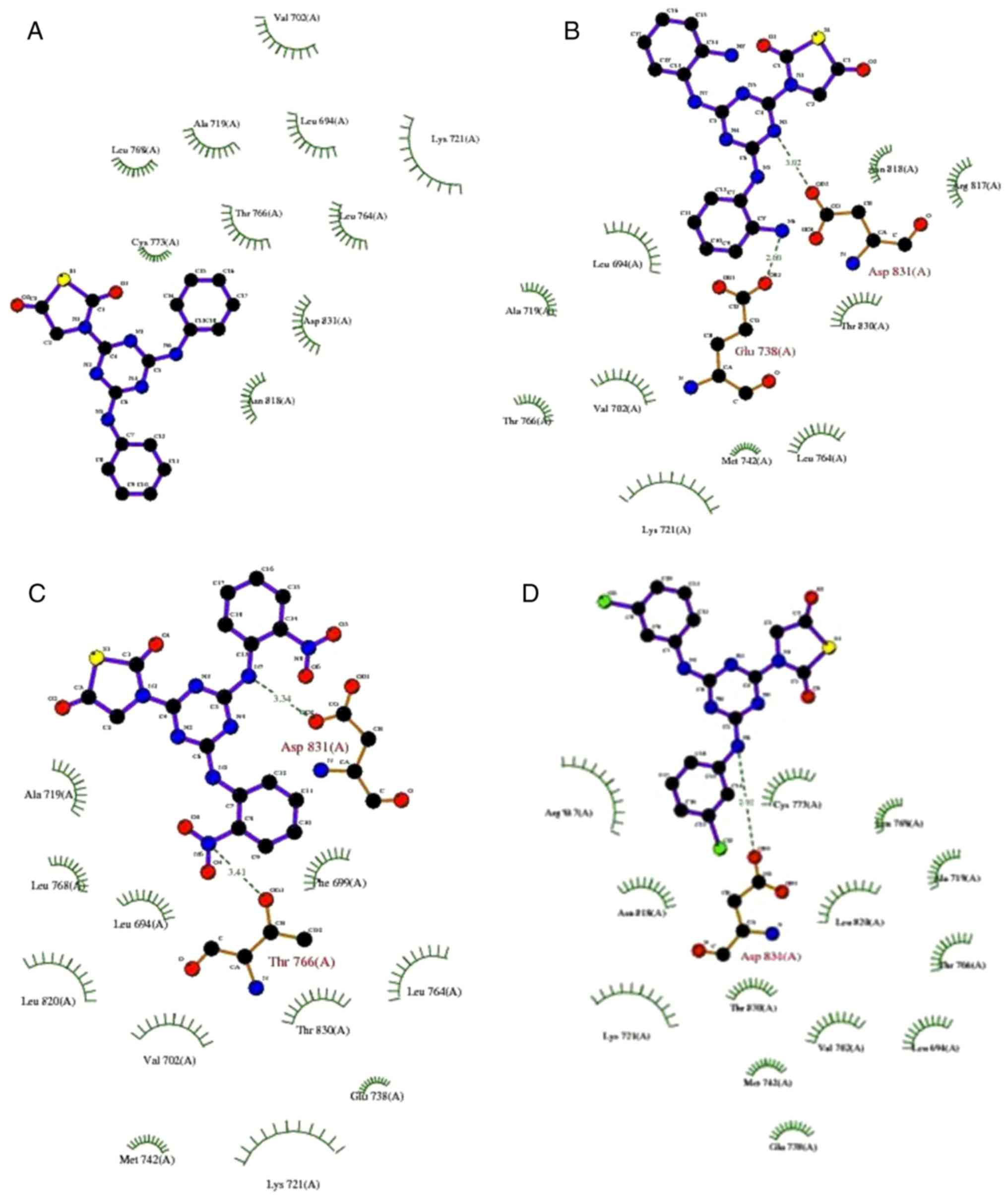
active site as shown in Fig. 2.
The molecules also demonstrated considerable interaction with key
catalytic residues, including Asn818, Lys721, Leu694, Val702, Met
742 and other residues as exhibited in Table II. Particularly, in the case of
compound 1a, as presented in Fig.
3, it had an inhibitory constant (Ki) of 3.10 µM with polar
interaction with Lys721 and Asn818. It also had interaction with
Leu694, Val702 and Leu678. Compound 1b exhibited marked improvement
in the Ki value as a result of improved binding contacts with the
target protein, including additional residues Arg817, Met442 and
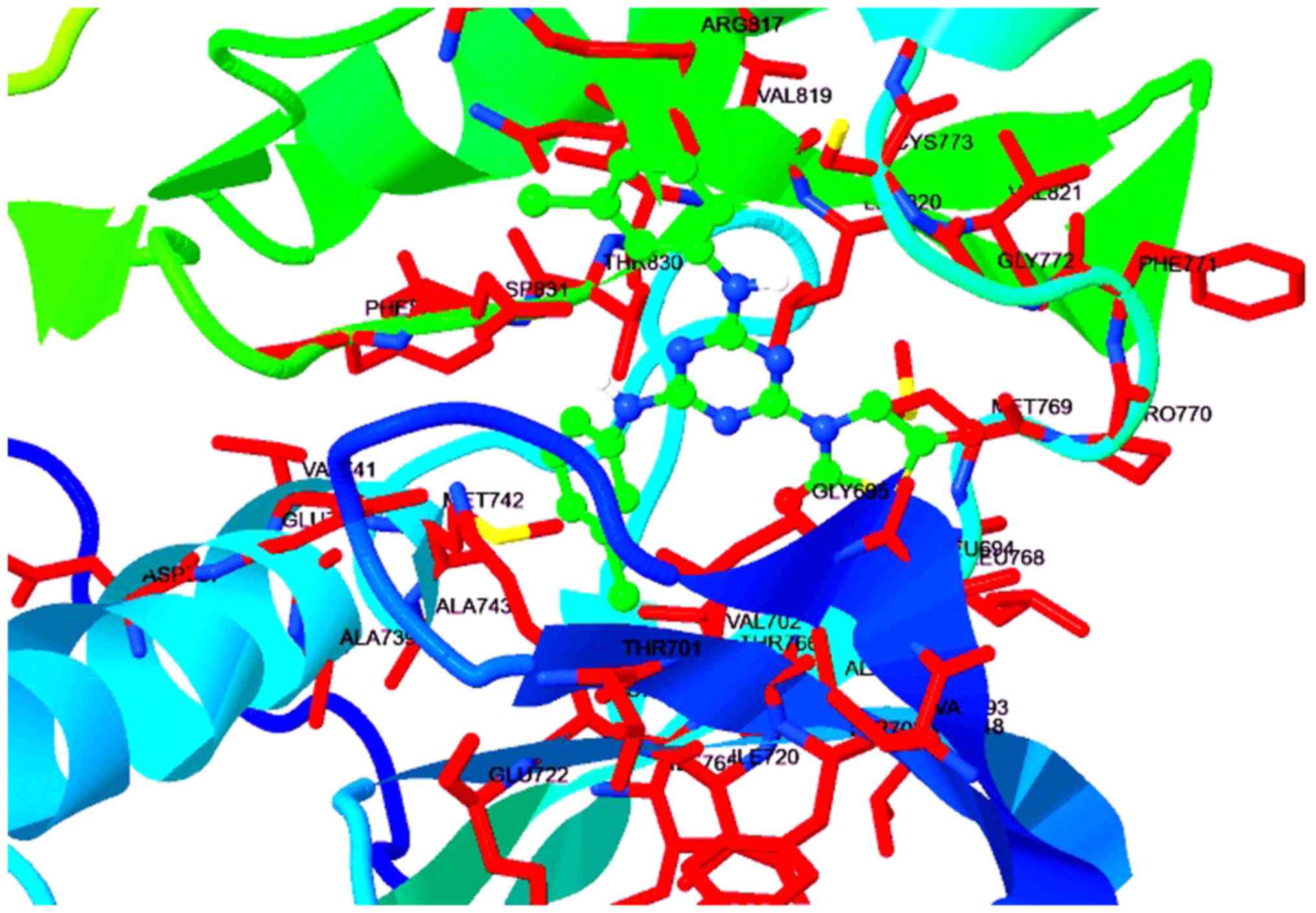
halogen binding with Glu738, Asp831, and Lys721, Fig. 4. Fig.
5 demonstrated that compound 1c exhibited 1.80 µM via
interacting with Thr766, Leu694 and Met742. Compound 1d had the
most potent activity, where the compound exhibited Ki value of 0.44
nM with improved contacts in the 3D protein structure (Fig. 6). The docking interactions were
further demonstrated to be in agreement with the 2D H-plot, as
shown in Fig. 2.
 | Table II.Scoring and interacting residue of
the compounds against the epidermal growth factor receptor tyrosine
kinase domain. |
Table II.
Scoring and interacting residue of
the compounds against the epidermal growth factor receptor tyrosine
kinase domain.
| Code | Est. free energy of
binding (kcal/mol) | Est. inhibition
constant (Ki) | vdW + Hbond +
desolvenergy | Electrostatic
energy | Total
intermolecular energy | Polar | Hydrophobic | Halogen bond |
|---|
| 1a | −7.51 | 3.10 µM | −8.26 | −0.06 | −8.32 | Lys721, Asn818 | Leu694, Val702,
Leu678 | – |
| 1b | −7.91 | 1.60 µM | −8.51 | −0.14 | −8.64 | Lys721, Arg817,
Asn818 | Leu694, Val702,
Met442 | Glu738, Asp831,
Lys721 |
| 1c | −7.84 | 1.80 µM | −8.93 | −0.06 | −8.99 | Thr766 | Leu694, Met742,
Leu764 | Thr766, Asp831 |
| 1d | −8.66 | 0.44 nM | −9.32 | −0.06 | −9.37 | Asp831 | Leu694, Met742,
Leu820 | Asp831 |
In vitro EGFR-TK inhibitory
activity
As a result of the excellent drug likeness profile
and considerable docking affinity of the 1,3,5-triazine
derivatives, these derivatives were synthesized and tested for
inhibitory activity against EGFR-TK. The results are presented in
Table III.
 | Table III.Epidermal growth factor
receptor-tyrosine kinase inhibitory activity of target
molecules. |
Table III.
Epidermal growth factor
receptor-tyrosine kinase inhibitory activity of target
molecules.
| Compound | IC50 (in
µM) |
|---|
| 1a | 112.45±21.52 |
| 1b | 34.05±4.11 |
| 1c | 8.45±0.65 |
| 1d | 2.54±0.22 |
| Dacomitinib | 0.06±0.01 |
It has been demonstrated that synthesized
derivatives exhibit excellent inhibitory activity compared with
dacomitinib as a standard in the present study. For instance,
compound 1d was demonstrated to exhibit the most potent activity
among all the tested analogues. The lowest activity against EGFR
was reported by compound 1a. The other compounds demonstrated mild
to moderate activity. These results further confirmed the EGFR-TK
inhibitory effect of 1,3,5-triazines as determined in docking
experiments.
In vitro anticancer activity
Subsequent to determining the inhibitory activity,
compound 1d was further analyzed for anticancer activity in breast
cancer cell lines, specifically, the highly metastatic MDA-MB231,
HER2-positive BT-474 and ER-positive MCF7 cell lines. The compound
was synthesized as described elsewhere and characterized using the
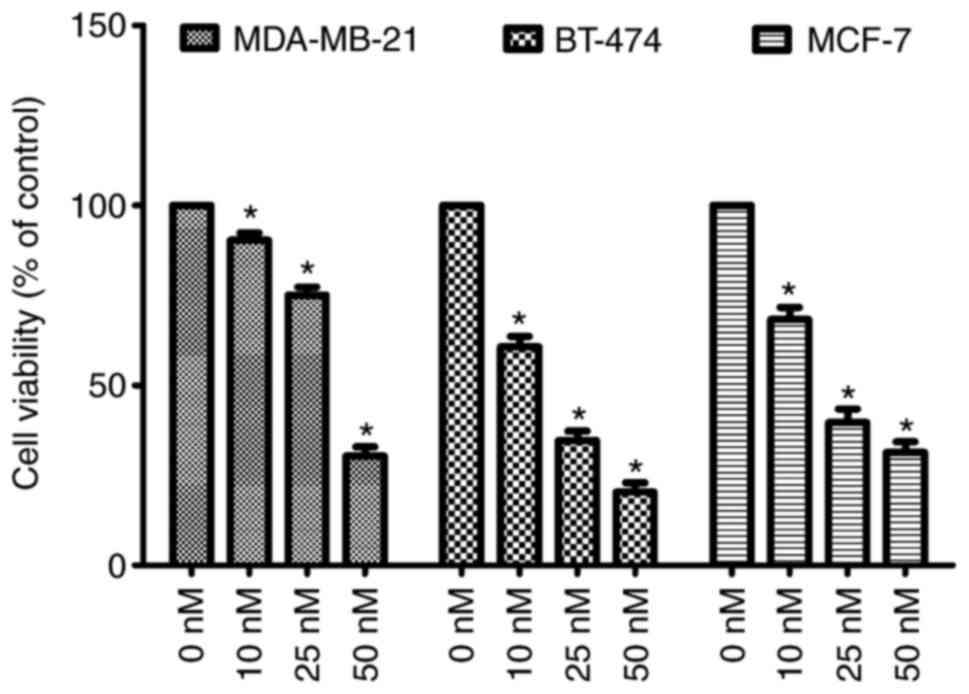
melting point as compared with the reported value (21). As presented in Fig. 7, it has been demonstrated that the
cellular viability of all the cells were decreased in a significant
manner (P<0.05). Out of the tested cells, compound 1d causes
inhibition of the BT-474 and least activity against MCF-7 at 50 nM.
The inhibitory activity of compound 1d was revealed to be
concentration dependent with maximum at 50 nM and minimum at 10 nM.
Whereas 25 nM demonstrated considerable effects on the viability of
cells.
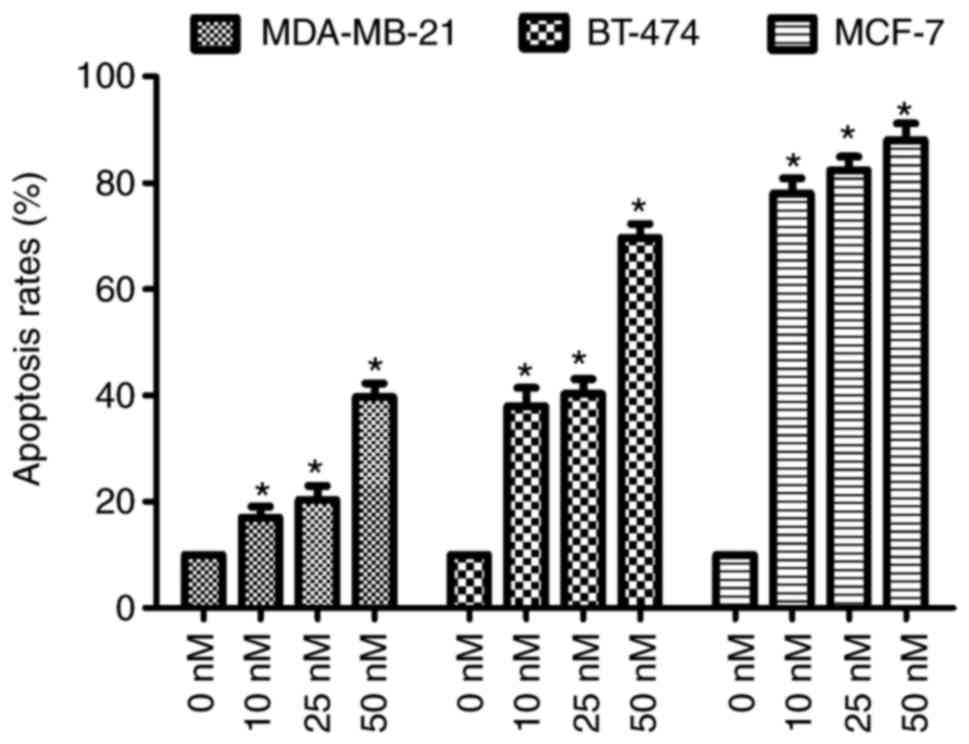
The effect of compound 1d was further evaluated by
measuring the apoptosis of the all three tested breast cancer
cells. The cells were treated with the vehicle (0 nM) and different
concentrations (10, 25 or 50 nM) of the compound 1d for 48 h. The
apoptotic rates were determined via the staining of all three cells
treated with compound 1d or the control cells with Hoechst 33258.
As shown in Fig. 8, compound 1d
increased in apoptosis in all three cells. At the
highestconcentration of compound 1d (50 nM), the level of apoptosis
in all the cells were demonstrated to be increased significantly
(P<0.05; Fig. 8).
The results suggested that compound 1d causes
significant increases in the apoptotic rates. The effect of
compound 1d on the degradation of β-catenin was also evaluated. The
total protein was isolated and the expression levels of β-catenin
were determined by immunoblot analysis using β-actin as a loading
control. As shown in Fig. 9, it
has compound 1d decreased the protein expression of β-catenin in
MDA-MB-231 compared with the control. These results indicated that
compound 1d caused a significant decline in the protein expression
of β-catenin expression in the MDA-MB-231 cells (P<0.05). Thus,
compound 1d may exertan anticancer effect via inhibition of the
β-catenin signaling pathway.
Discussion
The burden of cancer and themultifactorial etiology
has attracted thousands of studies across the world to investigate
this disease (2). Cancer can be
induced by the aberrant expression of receptors and growth factors
(3). It can also be modulated via
various oncogenic activators and inactivation of tumor suppression
genes (5). Various intracellular
pathways have also been demonstrated to be deregulated in cancer.
Consequently, the advancement of novel clinical techniques advances
the understanding of the underlying mechanisms behind the
generation and progression of cancer (6). This leads to the development of novel
treatments targeted to specific molecules and pathways, which are
considered to be critical for the development of cancer. Among the
well-known targets, EGFR a transmembrane growth factor receptor TK
was demonstrated to be frequently overexpressed in epithelial
tumors. Particularly in breast cancer, EGFR has a major role in
promoting cellular proliferation and tumor growth (9). It is considered to be a
proto-oncogene, which encodes a 170 kDa transmembrane protein that
results in the dimerization and autophosphorylation of the receptor
in presence of the EGFR ligand. EGFR activation leads to the
recruitment of downstream signaling molecules, which have critical
roles in cellular proliferation, survival and migration. EGFR has
been reported to be overexpressed in the majority of the triple
receptor-negative breast tumors (30). The canonical Wnt signaling pathway
(β-catenin dependent) regulates numerous genes that are responsible
for diverse cellular functions including morphogenesis,
differentiation and proliferation. Therefore, aberrant activation
of the pathway is considered to be involved in the pathogenesis of
multiple types of human cancers, and particularly in breast cancer.
A recent study reported that Wnt/β-catenin signaling activation is
preferentially found in a subgroup of invasive breast cancers of
triple negative breast cancer and is associated with a poor
clinical outcome (12). Thus, the
selective inhibition of EGFR-TK and β-catenin offers a number of
advantages. 1,3,5-Triazinesare well known for their anticancer
activity and are an important group of agentsamong novel drugs with
potential for cancer therapy. Recently certain 1,3,5-triazines have
been demonstrated to exhibit anti-EGFR-TK inhibitory activity
(31). As a result, in the present
study the putative EGFR-TK inhibitory activity of specific
1,3,5-triazine derivatives was investigated (21). As presented in Table I, it has been found compounds
demonstrated a considerable druglikness score, as indicated by
Lipinski's rule of five. Lipinski's rule of five, also known as the
Pfizer's rule of five, or simply the rule of five is used to
evaluate drug-likeness or to determine if a chemical compound with
a certain pharmacological or biological activity has properties
that would make it a suitable candidate as anorally active drug in
humans. The rule was formulated by based on the observation that
the majority of orally administered drugs are relatively small and
moderately lipophilic molecules (29). The rule describes the molecular
properties important for drug pharmacokinetics in the human body,
including their absorption, distribution, metabolism and excretion.
However, the rule does not predict if a compound is
pharmacologically active. Thus, the compounds were investigated to
determine if they have sufficient efficacy to act as drug
molecules. The molecules were analyzed for docking with EGFR-TKs.
In previous studies, it has been demonstrated that potent EGFR
inhibitors were able to interact with the EGFR protein and the
docking results revealed that three amino acids Leu694, Lys721 and
Asp831 located in the binding pocket of the protein had a vital
role in binding with compounds (10–13).
The compounds in the present study interacted with these residues
and similarly exhibited EGFR-TK inhibitory activity. The compounds
also significantly inhibited EGFR-TK in an enzymatic inhibitory
assay (Table III). Thus, it was
hypothesized that these derivatives may have potential to inhibit
EGFR in abreast cancer cell system assay. As confirmed by the in
vitro analysis, the most active compound exhibited potent
anticancer activity in the all tested cell lines in a
dose-dependent manner, with the highest in the activity at 50 nM.
The results of the study were in accordance with a previous study
where 1,3,5-triazine attenuated EGFR-TK with anti-breast cancer
activity (30). The activity
against apoptosis on different cells was investigated. Apoptosis is
evaded by cancer cells, resulting in malignant cells that will not
die spontaneously. The mechanism of apoptosis is multifaceted and
involves a number of signaling pathways. Defects can occur at any
point along these pathways, leading to malignant transformation of
the affected cells, tumor metastasis and resistance to anticancer
drugs. The tested compound exhibited potent induction of apoptosis.
The effect of compound 1d on β-catenin was also investigated via
western blot analysis. Compound 1d reduced the expression of
β-catenin in the MDA-MB-321 cells at various doses, with prominent
activity at 50 nM dose. However the present study lacks ITC data
for each compound and the effect on non-cancerous cells to further
understand the spectrum of activity.
In conclusion, a series of 1,3,5-triazines were
investigated as inhibitors of β-catenin and EGFR-TK inhibitors, and
acted as potent anti-breast cancer agents. Currently, the more data
is being acquired on the inhibitory mechanism of these compounds
via ITC experiments and effect on non-cancerous cells, which will
be reported in due course. Thus, derivatives may act as potent
anti-breast cancer agents.
Acknowledgements
The authors would like to thank Shanghai Jiao Tong
University School of Medicine, China for providing laboratory
facilities.
Funding
The present study was supported by Shanghai Jiaotong
University Medical Crossover fund (grant no. YG2016MS63).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
JH designed the study. WY and YZ performed the
experiments. WY, YZ and JH analyzed the data and wrote the
paper.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schneider KA: All about breast
cancerCounseling About Cancer: Strategies for Genetic Counseling.
Wiley-Blackwell; Hoboken, NJ: pp. 151–185. 2011, View Article : Google Scholar
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boyd NF, Martin LJ, Bronskill M, Yaffe MJ,
Duric N and Minkin S: Breast tissue composition and susceptibility
to breast cancer. J Natl Cancer Inst. 102:1224–1237. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ripperger T, Gadzicki D, Meindl A and
Schlegelberger B: Breast cancer susceptibility: Current knowledge
and implications for genetic counselling. Eur J Hum Genet.
17:722–731. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Osborne CK and Schiff R: Mechanisms of
endocrine resistance in breast cancer. Annu Rev Med. 62:233–247.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pohlmann PR, Mayer IA and Mernaugh R:
Resistance to trastuzumab in breast cancer. Clin Cancer Res.
15:7479–7491. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mueller KL, Yang ZQ, Haddad R, Ethier SP
and Boerner JL: EGFR/Met association regulates EGFR TKI resistance
in breast cancer. J Mol Signal. 5:82010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ferrer-Soler L, Vazquez-Martin A, Brunet
J, Menendez JA, De Llorens R and Colomer R: An update of the
mechanisms of resistance to EGFR-tyrosine kinase inhibitors in
breast cancer: Gefitinib (Iressa)-induced changes in the expression
and nucleo-cytoplasmic trafficking of HER-ligands (Review). Int J
Mol Med. 20:3–10. 2007.PubMed/NCBI
|
|
11
|
Mowafy S, Farag NA and Abouzid KAM:
Design, synthesis and in vitro anti-proliferative activity of
4,6-quinazolinediamines as potent EGFR-TK inhibitors. Eur J Med
Chem. 61:132–145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lo HW, Hsu SC and Hung MC: EGFR signaling
pathway in breast cancers: From traditional signal transduction to
direct nuclear translocalization. Breast Cancer Res Treat.
95:211–218. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh UP and Bhat HR: PUB081 Discovery of
1,3,5-triazine based novel EGFR-tyrosine kinase inhibitor against
human lung carcinoma. J Thor Oncol. 12(Suppl): S1494–S1495. 2017.
View Article : Google Scholar
|
|
14
|
Giacomelli G, Porcheddu A and De Luca L:
[1,3,5]-triazine: A versatile heterocycle in current applications
of organic chemistry. Curr Org Chem. 8:1497–1519. 2004. View Article : Google Scholar
|
|
15
|
Singh B, Bhat HR, Kumawat MK and Singh UP:
Structure-guided discovery of 1,3,5-triazine-pyrazole conjugates as
antibacterial and antibiofilm agent against pathogens causing human
diseases with favorable metabolic fate. Bioorg Med Chem Lett.
24:3321–3325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dubey V, Pathak M, Bhat HR and Singh UP:
Design, facile synthesis, and antibacterial activity of hybrid
1,3,4-thiadiazole-1,3,5-triazine derivatives tethered via -S-
bridge. Chem Biol Drug Des. 80:598–604. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Maarouf AR, Farahat A, Selim KB and Eisa
HM: Synthesis and antiviral activity of benzimidazolyl- and
triazolyl-1,3,5-triazines. Med Chem Res. 21:703–710. 2012.
View Article : Google Scholar
|
|
18
|
Singh UP, Bhat HR and Gahtori P:
Antifungal activity, SAR and physicochemical correlation of some
thiazole-1,3,5-triazine derivatives. J Mycol Med. 22:134–141. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bhat HR, Singh UP, Yadav PS, Kumar V,
Gahtori P, Das A, Chetia D, Prakash A and Mahanta J: Synthesis,
characterization and antimalarialactivity of
hybrid4-aminoquinoline-1,3,5-triazinederivatives. Arabian J Chem.
9(Suppl): S625–S631. 2016. View Article : Google Scholar
|
|
20
|
Bhat HR, Singh UP, Gahtori P, Ghosh SK,
GogoiKabita, Prakash A and Singh RK:
4-Aminoquinoline-1,3,5-triazine: Design, synthesis, in vitro
antimalarial activity and docking studies. New J Chem.
37:2654–2662. 2013. View Article : Google Scholar
|
|
21
|
Srivastava JK, Dubey P, Singh S, Bhat HR,
Kumawat MK and Singh UP: Discovery of novel
1,3,5-triazine-thiazolidine-2,4-diones as dipeptidyl peptidase-4
(DPP-4) inhibitor targeting S1 pocket for the treatment of type 2
diabetes along with antibacterial activity. RSC Adv. 5:14095–14102.
2015. View Article : Google Scholar
|
|
22
|
Srivastava JK, Awatade NT, Bhat HR, Kmit
A, Mendes K, Ramos M, Amaral MD and Singh UP: Pharmacological
evaluation of hybrid thiazolidin-4-one-1,3,5-triazines for NF-κB,
biofilm and CFTR activity. RSC Adv. 5:88710–88718. 2015. View Article : Google Scholar
|
|
23
|
Nie Z, Perretta C, Erickson P, Margosiak
S, Lu J, Averill A, Almassy R and Chu S: Structure-based design and
synthesis of novel macrocyclicpyrazolo[1,5-a] [1,3,5]triazine
compounds as potent inhibitors of protein kinase CK2 and their
anticancer activities. Bioorg Med Chem Lett. 18:619–23. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ertl P, Rohde B and Selzer P: Fast
calculation of molecular polar surface area as a sum of
fragment-based contributions and its application to the prediction
of drug transport properties. J Med Chem. 43:3714–3717. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pervez A, Meshram J, Tiwari V, Sheik J,
Dongre R, Youssoufi MH and Ben Hadda T: Pharmacophores modeling in
terms of prediction of theoretical physico-chemical properties and
verification by experimental correlations of novel coumarin
derivatives produced via Betti's protocol. Eur J Med Chem.
45:4370–4378. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bikadi Z and Hazai E: Application of the
PM6 semi-empirical method to modeling proteins enhances docking
accuracy of AutoDock. J Cheminform. 1:152009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Morris GM, Goodsell DS, Halliday RS, Huey
R, Hart WE, Belew RK and Olson AJ: Automated docking using a
Lamarckian genetic algorithm and an empirical binding free energy
function. J Comput Chem. 19:1639–1662. 1998. View Article : Google Scholar
|
|
28
|
Solis FJ and Wets RJB: Minimization by
random search techniques. Math Operat Res. 6:19–30. 1981.
View Article : Google Scholar
|
|
29
|
Lipinski CA, Lombardo F, Dominy BW and
Feeney PJ: Experimental and computational approaches to estimate
solubility and permeability in drug discovery and development
settings. Adv Drug Deliv Rev. 46:3–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ,
Kim YJ, Kim JH, Kang E, Kim SW, Kim IA and Park SY: High EGFR gene
copy number predicts poor outcome in triple-negative breast cancer.
Mod Pathol. 27:1212–1222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Srivastava JK, Pillai GG, Bhat HR, Verma A
and Singh UP: Design and discovery of novel
monastrol-1,3,5-triazines as potent anti-breast cancer agent via
attenuating epidermal growth factor receptor tyrosine kinase. Sci
Rep. 7:58512017. View Article : Google Scholar : PubMed/NCBI
|