Introduction
Lung cancer is one of the most prevalent cancers and
has the highest mortality rate worldwide (1). Currently, lung cancer has become a
severe public health problem in humans (2). Of all lung cancer patients, non-small
cell lung cancer (NSCLC) accounts for 80% cases and demonstrates a
poor prognosis (3). The high
potential of proliferation, invasion and metastasis remains a major
obstacle for the treatment of NSCLC patients (4). Currently, it is very difficult to
impede the development and progression of NSCLC, particularly at an
advanced stage. Therefore, in order to identify effective
biomarkers for diagnosis and therapeutic targets, it is necessary
to investigate the mechanisms of NSCLC progression.
MicroRNAs (miRs) are a class of short noncoding RNAs
(~20–22 nucleotides) and that have been demonstrated to regulate
gene expression by binding to the complementary site in the
3′-untranslated region (UTR) of target mRNAs (5,6). An
increasing number of studies indicate that miRs are involved in the
regulation of a number of biological processes, including cell
survival, division, migration and tumor angiogenesis (7,8). Due
to their extensive biological functions, abnormal expression of
miRs also results in occurrence of human malignances including
NSCLC (8). For example, Yang et
al (9) reported that
miR-769-5p is downregulated in NSCLC and suppresses tumor cell
proliferation, migration and invasion. Li et al (10) indicated that miR-9-5p promotes cell
growth and metastasis in NSCLC through the repression of tumor
growth factor b receptor 2. In addition, Wang et al
(11) demonstrated that miR-124
inhibits growth and enhances radiation-induced apoptosis in NSCLC
by inhibiting signal transducer and activator of transcription 3.
In addition, several miRs have been demonstrated to be promising
indicators for NSCLC diagnosis or prognosis (12,13).
Therefore, identifying the function and mechanism of miRs in NSCLC
is necessary.
Up to now, the knowledge regarding miR-3120-5p is
limited. Only a recent report indicated that miR-3120-5p promotes
colon cancer stem cell stemness and invasiveness (14). The function of miR-3120-5p in NSCLC
remains unclear. In the present study, it was demonstrated that
miR-3120-5p was significantly upregulated in NSCLC tissues and
serves as a diagnostic biomarker. Furthermore, it was demonstrated
that overexpression of miR-3120-5p remarkably enhanced the
proliferation and invasion of NSCLC cells through directly
targeting Krueppel-like factor 4 (KLF4). In conclusion, the results
of the present study provided a novel mechanism that the
miR-3120-5p/KLF4 axis regulates NSCLC progression and miR-3120-5p
may be an indicator for NSCLC diagnosis.
Materials and methods
Clinical specimens
A total of 39 NSCLC tissues (28 males and 11
females) and adjacent normal tissues were obtained from patients
who underwent surgery at The Second Affiliated Hospital of Harbin
Medical University (Harbin, China) between January 2013 and
December 2016. All patients were diagnosed as NSCLC by two
professional pathologists. All patients had no preoperative
adjuvant therapy. All tissue samples were immediately frozen in
liquid nitrogen and stored at −80°C until RNA analysis. These
samples were divided into two groups (miR-3120-5p high expression
group and low expression group) using the median value of
miR-3120-5p expression as the cutoff. Association of the expression
of miR-3120-5p with clinicopathological features is listed in
Table I. Written informed consent
was obtained from all of the participants and the study was
approved by the Institutional Review Board of The Second Affiliated
Hospital of Harbin Medical University.
 | Table I.Association of the expression of
miR-3120-5p with clinicopathological features. |
Table I.
Association of the expression of
miR-3120-5p with clinicopathological features.
|
| miR-3120-5p |
|
|---|
|
|
|
|
|---|
| Clinicopathological
parameters | High (n=19) | Low (n=20) | P-value |
|---|
| Age (years) |
|
| 0.731 |
| ≤50 | 13 | 15 |
|
|
>50 | 6 | 5 |
|
| Tumor size (cm) |
|
| 0.054 |
| ≤4 | 5 | 12 |
|
|
>4 | 14 | 8 |
|
| Lymph node
metastasis |
|
| 0.003 |
|
Negative | 3 | 13 |
|
|
Positive | 16 | 7 |
|
| Stage |
|
| 0.025 |
| I–II | 5 | 13 |
|
| III | 14 | 7 |
|
Cell culture and transfection
Four human NSCLC cell lines (A549, H1299, H1970 and
H460) and one normal bronchial epithelial cell line BEAS-2B were
purchased from the Institute of Biochemistry and Cell Biology of
the Chinese Academy of Sciences (Shanghai, China). Cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and supplemented with 10% fetal bovine
serum (FBS; HyClone, GE Healthcare Life Sciences Logan, UT, USA),
100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C in an environment containing 5%
CO2.
The miR-3120-5p mimic (5′-CCUGUCUGUGCCUGCUGUACA-3′),
mimic scrambled control (miR-NC; 5′-UCACAACCUCCUAGAAAGAGUAGA-3′),
miR-3120-5p inhibitor (5′-UGUACAGCAGGCACAGACAGG-3′) were chemically
synthesized by GenePharma Co., Ltd. (Shanghai, China). KLF4 coding
sequence was constructed into pcDNA3.1 vector (Addgene, Inc.,
Cambridge, MA, USA) to obtain pcDNA3.1-KLF4 plasmid. A549 and H460
cells in logarithmic phase were transfected with 100 nM
miRs/controls or 1 µg pcDNA3.1-KLF4 using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Further experiments were carried out at 48
h post-transfection.
Cell viability assay
Cell viability was determined using a Cell Counting
Kit (CCK-8; Beyotime Institute of Biotechnology, Jiangsu, China)
according to the manufacturer's protocol. Briefly, the transfected
cells (2×104 cells/ml) were seeded into a 96-well plate
and then 10 µl CCK-8 solution was added into each well of the plate
and the plates were cultured for 24, 48 and 72 h. The absorbance of
each well at a wavelength of 450 nm was measured using a
multidetection microplate reader (BMG Labtech, GmbH, Ortenberg,
Germany).
Transwell invasion assay
Cell invasion was detected using Transwell chambers
(8-µm pore size; BD Biosciences, Franklin Lakes, NJ, USA) coated
with Matrigel (BD, Franklin Lakes, NJ, USA). The transfected cells
(5×104) suspended in 150 µl serum free RPMI-1640 medium
were added into the upper chamber and the bottom compartment of the
chamber were filled up with 500 µl RPMI-1640 medium with 20% (v/v)
FBS as a chemoattractant. Following incubation for 48 h, the
non-invading cells inside the upper chamber were scraped off by a
cotton swab. The invaded cells on the lower chamber were then fixed
with 4% formaldehyde at 25°C for 30 min and stained with 0.1%
crystal violet at 25°C for 30 min. The cells were counted from five
independent fields under a light microscope using a ×200
magnification.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cultured cells using
the TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol and cDNA
was synthesized from total RNA by a PrimerScript RT Reagent kit
(Takara Bio Inc. Tokyo, Japan). miR from total RNA was reverse
transcribed using the Prime-Script miRNA cDNA Synthesis kit (Takara
Bio Inc.). The RT reaction was performed at 37°C for 1 h and
terminated at 85°C for 5 min. qPCR was performed with the SYBR
green Premix Ex Taq II (Takara Bio Inc.) on an Applied Biosystems
Step One Plus Real-Time PCR System (Applied Biosystems, Thermo
Fisher Scientific, Inc.). The thermocycling conditions were as
follows: Initial denaturation at 95°C for 10 min; followed by 40
cycles of denaturation at 95°C for 15 sec and elongation at 60°C
for 1 min. GAPDH was used as the endogenous control for detection
of mRNA expression level, while U6 was used as endogenous control
for miR expression analysis. Relative expression fold was
calculated according to the 2−ΔΔCq method (15). Primer details were as follows: U6
forward, 5′-AACGAGACGACGACAGAC-3′ and reverse,
5′-GCAAATTCGTGAAGCGTTCCATA-3′; GAPDH forward,
5′-ATGTTGCAACCGGGAAGGAA-3′ and 5′-AGGAAAAGCATCACCCGGAG-3′;
miR-3120-5p forward, 5′-AACGAGACGACGACAGAC-3′ and reverse,
5′-CCTGTCTGTGCCTGCTGTACA-3′; KLF4 forward,
5′-ATGCTCACCCCACCTTCTTC-3′ and reverse,
5′-TTCTCACCTGTGTGGGTTCG-3′.
Flow cytometry analysis (FACS)
FACS was used to analyze the cell cycle
distributions. The A549 and H460 cells were fixed with 75% ethanol
for 4 h at 4°C, washed with PBS, and stained with propidium iodide
(PI) supplemented with RNaseA overnight at 4°C. Following staining,
the cells were analyzed by flow cytometry using a BD FACScan flow
cytometer (BD Biosciences) coupled with BD Cell Quest Pro™ software
(version 2; BD Biosciences).
Western blotting
A549 and H460 cells were lysed in cold
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.), and the protein concentration was determined using a
Bicinchoninic Acid Protein Assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Proteins (40 µg/lane) were separated via 10%
SDS-PAGE and transferred to a polyvinylidene difluoride membrane
(Thermo Fisher Scientific, Inc.). Following this, the membrane was
blocked in 5% non-fat milk in PBS containing 0.1% Tween-20
(Sigma-Aldrich; Merck KGaA) at room temperature for 3 h.
Subsequently, the PVDF membrane was incubated with anti-PCNA
(1:1,000; cat. no. ab29; Abcam, Cambridge, MA, USA),
anti-N-cadherin (1:1,000; cat. no. ab18203; Abcam), anti-E-cadherin
(1:1,000; cat. no. ab1416; Abcam), anti-KLF4 (1:1,000; cat. no.
ab106629; Abcam) and anti-GAPDH (1:1,000; cat. no. ab9485; Abcam)
primary antibodies at room temperature for 2 h. Following washing
with PBS for 10 min, the PVDF membrane was incubated with
horseradish peroxidase-tagged goat anti-rabbit secondary antibodies
(1:5,000; cat. no. ab7090; Abcam) at room temperature for 1 h.
Membranes were washed with PBS for 10 min, and the protein bands
were visualized using an Enhanced Chemiluminescence Western
Blotting kit (Pierce; Thermo Fisher Scientific, Inc.), in
accordance with the manufacturer's protocol. Protein densitometry
was performed using ImageJ software (version 1.41; National
Institutes of Health, Bethesda, MD, USA).
Luciferase reporter assay
The potential target binding sites of miR-3120-5p
were predicted using the TargetScan tool (http://www.targetscan.org/vert_71/). The wild-type
3′UTR KLF4 and mutant type 3-′UTR KLF4 were inserted into pmiR-GLO
vector (Promega Corporation, Madison, WI, USA). A549 and H460 cells
were co-transfected with miR-3120-5p mimic, inhibitor or miR-NC as
well as wild-type 3′UTR KLF4 or mutant 3′UTR KLF44 (100 ng) using
Lipofectamine 2000 reagent according to the manufacturers'
protocol. Luciferase activity was detected and analyzed by
Dual-luciferase reporter system according to the manufacturers'
protocol (Promega Corporation) 24 h post-transfection. Relative
activity was normalized to Renilla luciferase activity.
Statistical analysis
Each experiment was repeated at least three times.
Data were analyzed using SPSS software version 20.0 (IBM Corp.,
Armonk, NY, USA) and expressed as the mean ± standard deviation.
The Kaplan-Meier method was used to calculate the survival curve
and log-rank test to determine statistical significance. The
differences between groups were analyzed using Two-tail Student's
t-test or analysis of variance followed by Tukey's post hoc test.
Association of the expression of miR-3120-5p with
clinicopathological features was analyzed using the χ2
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-3120-5p is upregulated in NSCLC
tissues and serves as a prognostic biomarker
The expression patterns of miR-3120-5p in NSCLC
tissues and adjacent normal tissues were first analyzed by RT-qPCR.
As presented in Fig. 1A, it was
demonstrated that miR-3120-5p was significantly upregulated in
NSCLC tissues compared with adjacent normal tissues (P<0.0001).
To further check the association between miR-3120-5p expression and
clinical characteristics, these samples were divided into two
subgroups based on tumor metastasis. RT-qPCR analysis indicated
that the NSCLC tissues with metastasis (n=23) displayed
significantly increased levels of miR-3120-5p compared with
non-metastatic tissues (n=16) (P<0.05; Fig. 1B). In addition, it was observed
that increased expression of miR-3120-5p was associated with
advanced clinical stage (stage III, n=21; Fig. 1C). Consistently, the relative
miR-3120-5p expression was examined in four NSCLC cell lines
including A549, H1975, H1299 and H460 cells and a normal lung
epithelial cell line BEAS-2B. The results indicated that
miR-3120-5p expression was higher in the NSCLC cell lines compared
with in the BEAS-2B cells (Fig.
1D). To determine whether miR-3120-5p could serve as a
biomarker for NSCLC diagnosis, receiver operating characteristic
analysis was performed. The results indicated that miR-3120-5p
displayed a predictive signature, with an area under curve of 0.848
(P<0.01; Fig. 1E). Taken
together, the aforementioned data suggested that miR-3120-5p was
upregulated in NSCLC tissues and served as a predictor for NSCLC
diagnosis.
miR-3120-5p promotes NSCLC cell
proliferation and invasion
To investigate the function of miR-3120-5p in NSCLC,
A549 and H460 cells were chosen for experiments, as miR-3120-5p
expression was the highest in these two cell lines (Fig. 1D). A549 and H460 cells were
transfected with miR-3120-5p mimic, inhibitor or negative controls
(miR-NC). RT-qPCR analysis indicated that miR-3120-5p was
significantly upregulated or downregulated when the mimic or
inhibitor was used, respectively (P<0.05; Fig. 2A). Then, CCK-8 and colony formation
assays were performed to measure the effects of miR-3120-5p on
NSCLC cell proliferation. It was demonstrated that overexpression
of miR-3120-5p significantly promoted (P<0.05) the proliferation
and colony formation whereas inhibition of miR-3120-5p suppressed
cellular proliferation in A549 and H460 cells (Fig. 2B and C). It was demonstrated
miR-3120-5p expression is associated with cancer metastasis. A
Transwell assay was then used to determine the effect of
miR-3120-5p on cell invasion. As presented, overexpression of
miR-3120-5p significantly enhanced the invasion of A549 and H460
cells and inhibition resulted in the opposite effect (P<0.05;
Fig. 2D).
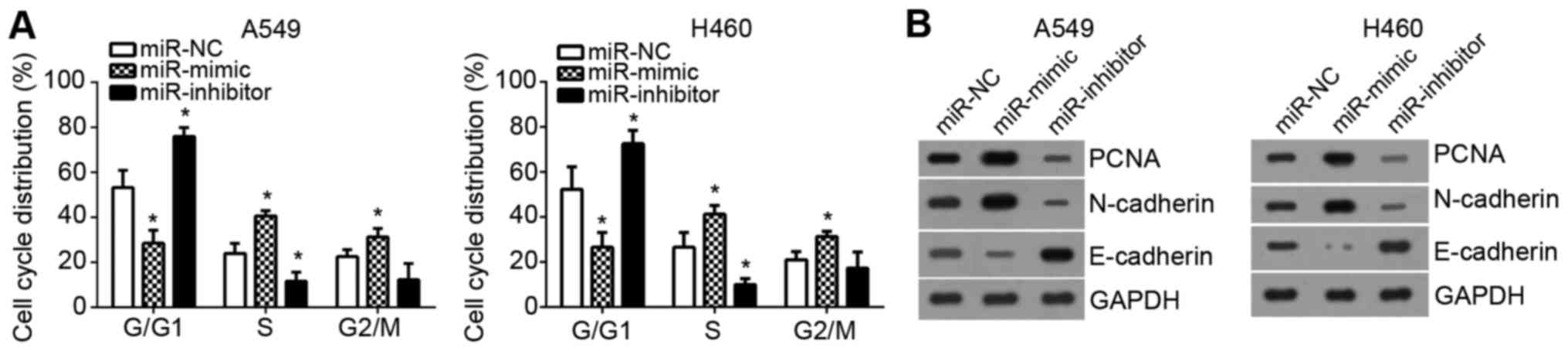
Effects of miR-3120-5p expression on
cell cycle and epithelial mesenchymal transition (EMT) markers of
NSCLC cells
Subsequently, the effects of miR-3120-5p on cell
cycle were examined. By FACS, it was demonstrated that
overexpression of miR-3120-5p significantly reduced the cells in
G0/G1 phase, however increased the cell
percentage in S phase and G2/M phase (P<0.05;
Fig. 3A). Furthermore, the
expression levels of proliferation-associated protein proliferating
cell nuclear antigen (PCNA) and EMT-associated markers [epithelial
(E)-cadherin and neural (N)-cadherin] was measured by western
blotting. It was demonstrated that PCNA and N-cadherin levels were
markedly upregulated following miR-3120-5p overexpression while
E-cadherin expression was downregulated and the opposite effects
were observed following miR-3120-5p inhibition (Fig. 3B). Therefore, these data indicated
that miR-3120-5p promoted cell cycle progression and NSCLC
malignant behaviors.
KLF4 is a target of miR-3120-5p in
NSCLC cells
To further investigate the downstream mechanism of
miR-3120-5p in NSCLC, the target genes of miR-3120-5p were searched
for by TargetScan tool. Among all potential targets, KLF4 ranked
top and has been demonstrated to suppress NSCLC progression.
Therefore, KLF4 was chosen for the following investigation. First,
a wild-type-KLF4-3′-UTR or mutant-KLF4-3′-UTR luciferase reporter
vector was constructed (Fig. 4A).
Using a luciferase reporter assay, it was demonstrated that
overexpression of miR-3120-5p significantly inhibited the
luciferase activity in A549 and H460 cells, and inhibition resulted
in the opposite effects (P<0.05; Fig. 3B and C), which indicated
miR-3120-5p directly targeted KLF4. Furthermore, RT-qPCR analysis
and western blotting also demonstrated that miR-3120-5p
overexpression significantly inhibited the mRNA and protein levels
of KLF4 in A549 and H460 cells (P<0.05; Fig. 4D and E). Taken together, the
present study proved KLF4 was a target gene of miR-3120-5p in NSCLC
cells.
miR-3120-5p enhances proliferation and
invasion by regulating KLF4 in NSCLC cells
To further determine whether KLF4 is involved in the
regulation of miR-3120-5p-mediated promotion of NSCLC progression,
a series of experiments were performed. Through RT-qPCR analysis
and western blotting, it was demonstrated that the expression of
KLF4 was significantly downregulated in NSCLC tissues compared with
adjacent normal tissues (P<0.05; Fig. 5A and B). Similarly, KLF4 was also
downregulated in NSCLC cell lines compared with normal cell line
BEAS-2B (Fig. 5C). Then, KLF4 was
overexpressed by transfection with pcDNA3.1-KLF4 plasmid in A549
and H460 cells (Fig. 5D). The
CCK-8 assays demonstrated that miR-3120-5p overexpression enhanced
cell proliferation and overexpression of KLF4 inhibited cell
proliferation, compared with the control groups (Fig. 5E). However, cotransfection with
miR-3120-5p mimic plus pcDNA3.1-KLF4 reversed the effects of
miR-3120-5p mimic on cell proliferation in A549 and H460 cells
(Fig. 5E). Consistently, the
cellular invasion was also enhanced by miR-3120-5p mimic and
inhibited by KLF4 overexpression (Fig.
5F). Restoration of KLF4 also abolished the effect of
miR-3120-5p on NSCLC cell invasion (Fig. 5F). Taken together, these results
suggested that miR-3120-5p promoted cell proliferation and invasion
by regulating KLF4 expression in NSCLC cells.
 | Figure 5.miR-3120-5p enhances proliferation and
invasion by regulating KLF4 in NSCLC cells. (A) Expression of KLF4
was examined in NSCLC tissues and adjacent normal tissues using
RT-qPCR analysis. ***P<0.001 vs. normal group. (B) Protein
levels of KLF4 in NSCLC tissues and paired adjacent normal tissues
were measured by western blotting. (C) Expression of KLF4 was
examined in NSCLC cell lines (A549, H1299, H1970 and H460) and one
normal bronchial epithelial cell line BEAS-2B using RT-qPCR
analysis. *P<0.05, **P<0.01 vs. BEAS-2B cells. (D) KLF4 was
upregulated in A549 and H460 cells by transfecting with
pcDNA3.1-KLF4. (E) Cell Counting Kit-8 assay was used to assess
cell growth following transfection with miR-NC, miR-3120-5p mimic,
miR-3120-5p mimic plus pcDNA3.1-KLF4 or pcDNA3.1-KLF4 in A549 and
H460 cells. *P<0.05 vs. control group. (F) Cell invasion was
measured by Transwell assays in A549 and H460 cells transfected
with miR-NC, miR-3120-5p mimic, miR-3120-5p mimic plus
pcDNA3.1-KLF4 or pcDNA3.1-KLF4. *P<0.05 vs. control group. miR,
microRNA; NC, negative control; NSCLC, non-small cell lung cancer;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; KLF4, Krueppel-like factor 4; N, normal tissues; T, NSCLC
tissues; oe, overexpression; Vec, vector. |
Discussion
As the most common and prevalent type of cancer
around the world, NSCLC gives rise to a large number of
cancer-associated mortalities every year (1). There are still no effective
therapeutic strategies for NSCLC intervention. Therefore, it is
crucial to determine the underlying molecular mechanisms of NSCLC
progression. miRs have been widely reported to participate in the
progression of human cancer, including NSCLC (16). A number of studies indicate that
miRs may be promising therapeutic targets for cancer treatment
(17). Therefore, identifying the
regulatory mechanism of miRs in tumorigenesis will contribute to
the development of effective therapeutic methods. The present study
investigated the expression pattern of miR-3120-5p in NSCLC
tissues, investigated its effects on NSCLC cells and demonstrated
its functional mechanism.
An increasing number of studies suggest that miRs
could regulate the proliferation, migration, invasion and apoptosis
of cancer cells (18). Quite a
number of miRs were observed to be dysregulated in cancer,
including in NSCLC (19). For
example, miR-520e is downregulated in NSCLC tissues and suppresses
cancer growth by targeting the Zbtb7a-mediated Wnt signaling
pathway (20). Upregulation of
miR-146a increases the sensitivity of NSCLC to cisplatin by
downregulating cyclin J (21).
Downregulation of miR-218 contributes to EMT and tumor metastasis
in lung cancer by targeting Slug/ZEB2 signaling (22). As for miR-3120-5p, a recent study
indicated that it regulates colon cancer stem cell stemness and
invasiveness (14). Nevertheless,
whether miR-3120-5p promotes NSCLC progression remains unknown. In
the present study, it was demonstrated that miR-3120-5p was
upregulated in NSCLC tissues. Furthermore, high miR-3120-5p
expression was associated with NSCLC metastasis and tumor advanced
stages. Furthermore, miR-3120-5p was identified as a diagnostic
biomarker for NSCLC patients. Through functional experiments, it
was demonstrated that miR-3120-5p promoted the proliferation and
invasion of NSCLC cells.
KLF4 belongs to the Krueppel family of transcription
factors (23) and is thought to
control the G1-to-S transition of the cell cycle. KLF4 has been
demonstrated to serve the role of oncogene or tumor suppressor in
several types of cancer (24,25).
A previous study demonstrated that KLF4 serves as a tumor
suppressor in lung cancer (24).
Enhanced expression of KLF4 in lung cancer cells induces G1/S cell
cycle arrest and inhibits tumor growth in vitro and in
vivo (26). For example, a
study indicated that KLF4 inhibits invasion of lung cancer cells
via suppression of secreted protein acidic and rich in cysteine
(27). Another study demonstrated
that KLF4 is significantly downregulated in lung adenocarcinoma
compared with adjacent normal tissues (28). However, how KLF4 expression is
regulated in lung cancer requires further investigation. In the
present study, it was demonstrated that KLF4 was a direct target of
miR-3120-5p in NSCLC cells through bioinformatics analysis. The
results of the present study indicated that overexpression of
miR-3120-5p suppressed the expression of KLF4 in A549 and H460
cells. Furthermore, it was also demonstrated that KLF4 was
downregulated in NSCLC tissues compared with adjacent normal
tissues. Furthermore, through CCK-8 and Transwell assays, it was
demonstrated that overexpression of KLF reversed the effects of
miR-3120-5p on NSCLC cell proliferation and invasion.
In conclusion, the results of the present study
demonstrated that miR-3120-5p promoted the proliferation and
invasion of NSCLC cells through regulating KLF4 expression.
Furthermore, these results implied miR-3120-5p could serve as a
diagnostic marker for NSCLC patients and may serve as a potential
therapeutic target for NSCLC intervention.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HX initiated, designed this work, analyzed,
interpreted the results and wrote the manuscript. QW performed the
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
For the use of human samples, the protocol for the
present study was approved by the Institutional Ethics Committee of
The Second Affiliated Hospital of Harbin Medical University and all
enrolled patients signed a written informed consent document.
Patient consent for publication
All patients within this study provide consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen KD, Huang KT, Lin CC, Weng WT, Hsu
LW, Goto S, Nakano T, Lai CY, Kung CP, Chiu KW, et al: MicroRNA-27b
enhances the hepatic regenerative properties of adipose-derived
mesenchymal stem cells. Mol Ther Nucleic Acids. 5:e2852016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karlsen TA, de Souza GA, Odegaard B,
Engebretsen L and Brinchmann JE: microRNA-140 inhibits inflammation
and stimulates chondrogenesis in a model of interleukin 1β-induced
Osteoarthritis. Mol Ther Nucleic Acids. 5:e3732016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lemjabbar-Alaoui H, Hassan OU, Yang YW and
Buchanan P: Lung cancer: Biology and treatment options. Biochim.
Biophys Acta. 1856:189–210. 2015.
|
|
5
|
Huang K, Dong X, Sui C, Hu D, Xiong T,
Liao S and Zhang H: MiR-223 suppresses endometrial carcinoma cells
proliferation by targeting IGF-1R. Am J Transl Res. 6:841–849.
2014.PubMed/NCBI
|
|
6
|
Huang X, Huang M, Kong L and Li Y: miR-372
suppresses tumour proliferation and invasion by targeting IGF2BP1
in renal cell carcinoma. Cell Prolif. 48:593–599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
8
|
Kim G, An HJ, Lee MJ, Song JY, Jeong JY,
Lee JH and Jeong HC: Hsa-miR-1246 and hsa-miR-1290 are associated
with stemness and invasiveness of non-small cell lung cancer. Lung
Cancer. 91:15–22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Z, He J, Gao P, Niu Y, Zhang J, Wang
L, Liu M, Wei X, Liu C, Zhang C, et al: miR-769-5p suppressed cell
proliferation, migration and invasion by targeting TGFBR1 in
non-small cell lung carcinoma. Oncotarget. 8:113558–113570. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li G, Wu F, Yang H, Deng X and Yuan Y:
MiR-9-5p promotes cell growth and metastasis in non-small cell lung
cancer through the repression of TGFBR2. Biomed Pharmacother.
96:1170–1178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang M, Meng B and Liu Y, Yu J, Chen Q and
Liu Y: MiR-124 inhibits growth and enhances radiation-induced
apoptosis in non-small cell lung cancer by inhibiting STAT3. Cell
Physiol Biochem. 44:2017–2028. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu G, Fu D, Jia C, Chai L, Han Y, Liu J,
Wu T, Xie R, Chang Z, Yang H, et al: Reduced miR-105-1 levels are
associated with poor survival of patients with non-small cell lung
cancer. Oncol Lett. 14:7842–7848. 2017.PubMed/NCBI
|
|
13
|
Zhou Q, Huang SX, Zhang F, Li SJ, Liu C,
Xi YY, Wang L, Wang X, He QQ, Sun CC and Li DJ: MicroRNAs: A novel
potential biomarker for diagnosis and therapy in patients with
non-small cell lung cancer. Cell Prolif. 50:2017. View Article : Google Scholar :
|
|
14
|
Hongdan L and Feng L: miR-3120-5p promotes
colon cancer stem cell stemness and invasiveness through targeting
Axin2. Biochem Biophys Res Commun. 496:302–308. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cai Y, Ruan J, Yao X, Zhao L and Wang B:
MicroRNA-187 modulates epithelial-mesenchymal transition by
targeting PTRF in non-small cell lung cancer. Oncol Rep.
37:2787–2794. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng R, Lu C, Zhang G, Zhang G and Zhao
G: Overexpression of miR-203 increases the sensitivity of NSCLC
A549/H460 cell lines to cisplatin by targeting Dickkopf-1. Oncol
Rep. 37:2129–2136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu G, Shao G, Pan Q, Sun L, Zheng D, Li M,
Li N, Shi H and Ni Y: MicroRNA-9 regulates non-small cell lung
cancer cell invasion and migration by targeting eukaryotic
translation initiation factor 5A2. Am J Transl Res. 9:478–488.
2017.PubMed/NCBI
|
|
19
|
Chen S, Jiang S, Hu F, Xu Y, Wang T and
Mei Q: Foxk2 inhibits non-small cell lung cancer
epithelial-mesenchymal transition and proliferation through the
repression of different key target genes. Oncol Rep. 37:2335–2347.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhijun Z and Jingkang H: MicroRNA-520e
suppresses non-small-cell lung cancer cell growth by targeting
Zbtb7a-mediated Wnt signaling pathway. Biochem. Biophys Res Commun.
486:49–56. 2017. View Article : Google Scholar
|
|
21
|
Shi L, Xu Z, Wu G, Chen X, Huang Y, Wang
Y, Jiang W and Ke B: Up-regulation of miR-146a increases the
sensitivity of non-small cell lung cancer to DDP by downregulating
cyclin J. BMC Cancer. 17:1382017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi ZM, Wang L, Shen H, Jiang CF, Ge X, Li
DM, Wen YY, Sun HR, Pan MH, Li W, et al: Downregulation of miR-218
contributes to epithelial-mesenchymal transition and tumor
metastasis in lung cancer by targeting Slug/ZEB2 signaling.
Oncogene. 36:2577–2588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ye B, Liu B, Hao L, Zhu X, Yang L, Wang S,
Xia P, Du Y, Meng S, Huang G, et al: Klf4 glutamylation is required
for cell reprogramming and early embryonic development in mice. Nat
Commun. 9:12612018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li S, Huang L, Gu J, Wu J, Ou W, Feng J,
Liu B, Xu X and Zhou Y: Restoration of KLF4 Inhibits Invasion and
Metastases of Lung Adenocarcinoma through Suppressing MMP2. J
Cancer. 8:3480–3489. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu LP, Wu J, Shang A, Yang M, Li LL, Yu J,
Xu LR, Wang CB, Wang WW, Zhu JJ and Lu WY: miR-124 inhibits
progression of hepatocarcinoma by targeting KLF4 and promises a
novel diagnostic marker. Artif Cells Nanomed Biotechnol. Dec
18–2017.(Epub ahead of print). View Article : Google Scholar
|
|
26
|
Hu W, Hofstetter WL, Li H, Zhou Y, He Y,
Pataer A, Wang L, Xie K, Swisher SG and Fang B: Putative
tumor-suppressive function of Kruppel-like factor 4 in primary lung
carcinoma. Clin Cancer Res. 15:5688–5695. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou Y, Hofstetter WL, He Y, Hu W, Pataer
A, Wang L, Wang J, Zhou Y, Yu L, Fang B and Swisher SG: KLF4
inhibition of lung cancer cell invasion by suppression of SPARC
expression. Cancer Biol Ther. 9:507–513. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fadous-Khalife MC, Aloulou N, Jalbout M,
Hadchity J, Aftimos G, Paris F and Hadchity E: Kruppel-like factor
4: A new potential biomarker of lung cancer. Mol Clin Oncol.
5:35–40. 2016. View Article : Google Scholar : PubMed/NCBI
|