Introduction
Osteosarcoma (OS) arises from primitive bone-forming
mesenchymal cells and is the most common type of primary bone
neoplasm in children and adolescents (1). It is characterized by rapid
progression and high metastatic potential and is associated with
high mortality rates among adolescents and young adults (2,3).
Despite recent developments in combined treatment including
surgery, chemotherapy, and radiotherapy, the overall therapeutic
outcomes remain unsatisfactory with a median survival time of only
23 months (4). The unfavorable
prognosis of patients with OS is largely due to the high rates of
recurrence and local or distant metastasis of this malignancy
(5). Genetic alterations,
environmental ionizing radiation, and lesions are closely
associated with OS development; nevertheless, the detailed
mechanisms underlying OS pathogenesis remain largely unknown
(6). Therefore, fully
understanding the occurrence and development of OS is necessary to
develop alternative therapeutic strategies that can improve the
clinical management of this disease.
MicroRNA (miRNA/miR) are a group of noncoding RNA
molecules 22–25 nucleotides in length (7). MiRNA have been identified as critical
gene regulators (8). Given that
~30% of human protein-coding genes may be regulated by miRNA
(9), they may modulate thousands
of genes and serve key roles in a wide range of biological
processes, including cell proliferation, cycle, development,
differentiation, and metabolism (10). Numerous studies have demonstrated
that miRNA expression levels are dysregulated in almost all types
of human malignancies, including OS (11), breast (12), lung (13), nasopharyngeal (14), and cervical cancers (15). Dysregulated miRNA have been
implicated as oncogenes or tumor suppressors in OS formation and
progression. For example, the upregulation of miR-93a promotes OS
cell proliferation through the negative regulation of
cyclin-dependent kinase inhibitor 1A (16). MiR-203 overexpression restricts
cell proliferation and invasion, inducing apoptosis in OS through
blockage of runt-related transcription factor 2 (17). Considering their important role in
OS, the in-depth investigation of the molecular mechanisms
underlying the roles of miRNA in cancer is important to facilitate
the identification of novel therapeutic agents or targets for
OS.
MiR-944 is frequently dysregulated in human cancers,
such as endometrial (18),
cervical (19) and breast cancer
(20). However, the expression
pattern, detailed functions and underlying mechanisms of miR-944 in
OS remain largely elusive. Hence, the present study aimed to detect
the expression pattern, role, and governing mechanisms of miR-944
in OS.
Materials and methods
Tissue collection
The present study was approved by the Ethics
Committee of Jining No. 1 People's Hospital, and was performed
according to the principles of the Declaration of Helsinki. Written
informed consent was also provided from all patients enrolled in
the present study. A total of 38 cancerous and adjacent tissues
were obtained from OS patients (22 males, 16 females; age range,
16–53 years) in Jining No. 1 People's Hospital between March 2015
and February 2017. None of the patients had received radiotherapy
or chemotherapy prior to surgical resection. All cancerous and
adjacent tissues (~0.5–1 cm thick) were snap-frozen and were kept
in liquid nitrogen until further use.
Cell culture
Four human OS cell lines (MG-63, SAOS-2, HOS, and
U2OS) and the normal human osteoblast hFOB1.19 were acquired from
American Type Culture Collection (Manassas, VA, USA). All cells
were cultured in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin
and 100 U/ml streptomycin (all from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and grown at 37°C in a
humidified atmosphere containing 5% CO2. Cells were
harvested using 0.25% trypsin-0.02% EDTA solution (Gibco; Thermo
Fisher Scientific, Inc.) once cell density reached ~80–90%
confluency.
Transfection of oligonucleotides and
plasmids
For transfection, cells were seeded into 24-well
plates at a density of 5×105 cells/well. MiR-944 mimics
(Shanghai GenePharma Co., Ltd., Shanghai, China) were introduced
into cells to increase miR-944 expression. MiRNA mimic negative
control (miR-NC) was used as an internal control. The miR-944
mimics sequence was 5′-AAAUUAUUGUACAUCGGAUGAG-3′ and the miR-NC
sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. In order to increase VEGF
expression, cells were transfected with the VEGF-overexpressing
plasmid pcDNA3.1-VEGF (Integrated Biotech Solutions, Shanghai,
China). An empty pcDNA3.1 plasmid was used as control. Cells were
transfected with miR-944 mimics (100 pmol), miR-NC (100 pmol),
pcDNA3.1-VEGF (4.0 µg) or empty pcDNA3.1 plasmid (4.0 µg) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's protocol.
Transfected cells were incubated at 37°C in a humidified atmosphere
containing 5% CO2. Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) was performed to detect miR-944
expression at 48 h post-transfection. At 72 h post-transfection,
western blot analysis was used to determine VEGF protein
expression. MTT and Transwell invasion assays were conducted at 24
and 48 h post-transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or all cultured
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. For
analysis of miR-944 expression levels, complementary DNA (cDNA) was
synthesized from total RNA with a TaqMan MicroRNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The temperature protocol for reverse transcription was as
follows: 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. qPCR
was carried out on an ABI 7900 thermocycler (Applied Biosystems;
Thermo Fisher Scientific, Inc.), using a TaqMan MicroRNA PCR Kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 50°C for 2 min, 95°C for
10 min; 40 cycles of denaturation at 95°C for 15 sec and
annealing/extension at 60°C for 60 sec. For determination of VEGF
mRNA expression levels, total RNA was converted into cDNA with a
PrimeScript RT Reagent Kit (Takara Bio, Dalian, China), and then
cDNA was subjected to qPCR using a SYBR Premix Ex Taq™
Kit (Takara Bio, Dalian, China). The temperature protocol for
reverse transcription was as follows: 37°C for 15 min and 85°C for
5 sec. The cycling conditions for qPCR were as follows: 5 min at
95°C, followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec.
U6 small nuclear (sn)RNA and β-actin were used as endogenous
controls for miR-944 and VEGF mRNA, respectively. These genes were
all of human origin and the primers designed were as follows:
miR-944, 5′-GCGGCGGAAATTATTGTACATC-3′ (forward) and
5′-ATCCAGTGCAGGGTCCGAGG-3′ (reverse); U6 snRNA,
5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′
(reverse); VEGF, 5′-CGTGTACGTTGGTGCCCGCT-3′ (forward) and
5′-TCCTTCCTCCTGCCCGGCTC-3′ (reverse); and β-actin,
5′-ACAGAGCCTCGCCTTTGCCGATC-3′ (forward) and
5′-ATCCTTCTGACCCATGCCCACCA-3′ (reverse). All data were analyzed by
the 2−ΔΔCq method (21).
MTT assay
Following 24 h of incubation at 37°C with 5%
CO2, transfected cells (~1×106 cells/well)
were harvested using 0.25% trypsin-0.02% EDTA solution and prepared
into single-cell suspensions. Subsequently, 3×103 cells
were plated into 96-well plates with six parallel replicates and
incubated at 37°C in a humidified atmosphere containing 5%
CO2. MTT assay was conducted at 24, 48 and 72 h
following incubation. At every time point, 20 µl of MTT solution (5
mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added
into each well. The supernatant was gently discarded following 4 h
of incubation at 37°C. Subsequently, cells were lysed in 150 µl
dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) at 37°C for 10 min.
At every time point, a microplate reader was used to detect optical
density values at a wavelength of 490 nm (Bio-Rad Laboratories,
Inc., Hercules, CA, USA).
Transwell invasion assay
At 48 h post-transfection, cells were collected and
suspended in FBS-free DMEM. A total of 1×105 transfected
cells (200 µl) in FBS-free DMEM were seeded into the upper chamber
of Transwell inserts that were precoated with Matrigel (BD
Biosciences, San Jose, CA, USA). The lower chambers of Transwell
inserts were filled with 500 µl of DMEM supplemented with 10% FBS.
Following 24 h of incubation, noninvasive cells located on the
upper surface of the chamber were gently removed with a cotton
swab. Invasive cells were fixed in 100% methanol at room
temperature for 20 min and then stained with 0.5% crystal violet
solution (Sigma-Aldrich; Merck KGaA) at room temperature for 20
min. Following washing three times with PBS (Gibco; Thermo Fisher
Scientific, Inc.), the number of invasive cells in five randomly
selected fields per insert was quantified under a Leica inverted
microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Bioinformatics analysis
The putative targets of miR-944 were predicted using
TargetScan Human 7.1 (www.targetscan.org/vert_71/) and miRDB (mirdb.org/).
Luciferase reporter assay
Bioinformatics analysis predicted that VEGF is a
potential target gene of miR-944. Luciferase reporter assays were
performed to determine whether miR-944 can interact with the
3′-untranslated region (UTR) of VEGF. The wild-type (Wt) fragment
of the VEGF 3′-UTR containing miR-944 binding sites and the mutant
(Mut) fragment of VEGF 3′UTR lacking complementarity with miR-944
were chemically synthesized by Shanghai GenePharma Co., Ltd, and
inserted into pMIR-GLOTM Luciferase vectors (Promega Corporation,
Madison, WI, USA). Constructs containing the Wt or Mut fragments of
VEGF 3′UTR were designated as pMIR-VEGF-3′-UTR Wt or
pMIR-VEGF-3′-UTR Mut, respectively. To conduct the luciferase
reporter assay, cells were plated into 24-well plates at a density
of 1.0×105 cells/well. Lipofectamine 2000 was used to
cotransfect MG-63 and U2OS cells with pMIR-VEGF-3′-UTR Wt (0.2 µg)
or pMIR-VEGF-3′-UTR Mut (0.2 µg) and miR-944 mimics (50 pmol) or
miR-NC (50 pmol). At 48 h post-transfection, cells were harvested,
and luciferase activity was analyzed with a dual-Luciferase
Reporter assay system (Promega Corporation) in accordance with the
manufacturer's protocol. The activity of firefly luciferase was
normalized to that of Renilla luciferase.
Western blot analysis
Proteins were isolated from tissues or cultured
cells using cold radioimmunoprecipitation lysis buffer (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), and total protein
concentrations were evaluated using bicinchoninic assay kits
(Beyotime Institute of Biotechnology, Haimen, China) in accordance
with the manufacturer's protocol. Equal amounts of protein (30
µg/lane) were separated through 10% SDS-PAGE and then transferred
onto polyvinylidene membranes (Sigma-Aldrich; Merck KGaA).
Following blocking at room temperature for 2 h in 5% fat-free milk
in Tris-buffered saline containing 0.1% Tween-20 (TBST), the
membranes were incubated overnight at 4°C with primary antibodies
against VEGF (cat. no. ab1316; 1:1,000 dilution) or GAPDH (cat. no.
ab125247; 1:1,000 dilution; both from Abcam, Cambridge, UK).
Following three washes with TBST, the membranes were incubated with
goat anti-mouse horseradish peroxidase-conjugated secondary
antibodies (cat. no. ab205719; 1:5,000 dilution; Abcam) at room
temperature for 2 h. Protein signals were visualized by enhanced
chemiluminescence Protein Detection kit (Pierce; Thermo Fisher
Scientific, Inc.). The densities of protein signals were analyzed
with ImageJ software (version 1.48; National Institutes of Health,
Bethesda, MD, USA).
Statistical analysis
All results were expressed as the mean ± standard
deviation from at least three independent experiments. All
statistical tests were conducted using SPSS software (version 18.0;
SPSS, Inc., Chicago, IL, USA). Differences between two groups were
analyzed with paired Student's t-test, and one-way analysis of
variance was performed for multiple groups with Student Newman
Keuls as a post hoc test. The correlation between miR-944 and VEGF
mRNA expression levels was examined using Spearman's correlation
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
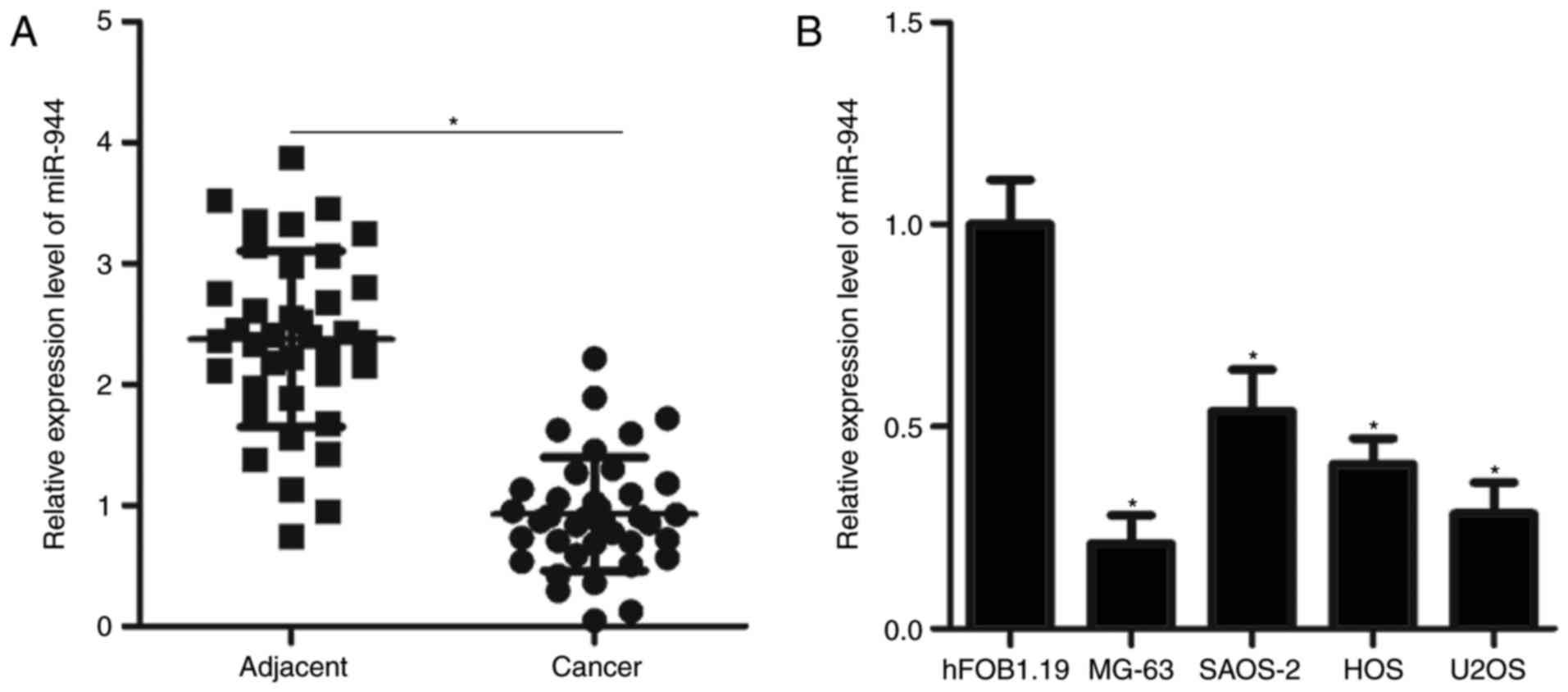
MiR-944 expression is downregulated in
cancer tissues and cell lines
MiR-944 expression in 38 pairs of OS cancer and
adjacent tissues was investigated by RT-qPCR. As illustrated in
Fig. 1A, miR-944 expression in
cancer tissues was decreased compared with that in adjacent tissues
(P<0.05). MiR-944 expression levels were also investigated in
four OS cell lines and the normal human osteoblast cell line
hFOB1.19. Results demonstrated that miR-944 expression was
downregulated in all four OS cell lines compared with in hFOB1.19
(P<0.05; Fig. 1B). These
results suggested that miR-944 is downregulated in OS, and this
downregulation may be associated with the development of OS.
MiR-944 overexpression inhibits OS
cell proliferation and invasion
Given that miR-944 was decreased in OS, it was
hypothesized that miR-944 may have a tumor-suppressive role in OS.
To confirm this hypothesis, MG-63 and U2OS cells were selected,
which exhibited lower miR-944 expression than the other cell lines,
for further experiments. MG-63 and U2OS cells were transfected with
miR-944 mimics or miR-NC. RT-qPCR analysis demonstrated that
miR-944 expression levels in MG-63 and U2OS cells transfected with
miR-944 mimics were increased compared with that in MG-63 and U2OS
cells transfected with miR-NC (P<0.05; Fig. 2A). MTT assay results revealed that
the upregulation of miR-944 expression levels significantly reduced
the proliferation of MG-63 and U2OS cells (P<0.05; Fig. 2B). Furthermore, Transwell invasion
assays indicated that miR-944 overexpression decreased the invasive
capabilities of MG-63 and U2OS cells (P<0.05; Fig. 2C and D). These results suggested
that miR-944 may act as a tumor suppressor in OS in
vitro.
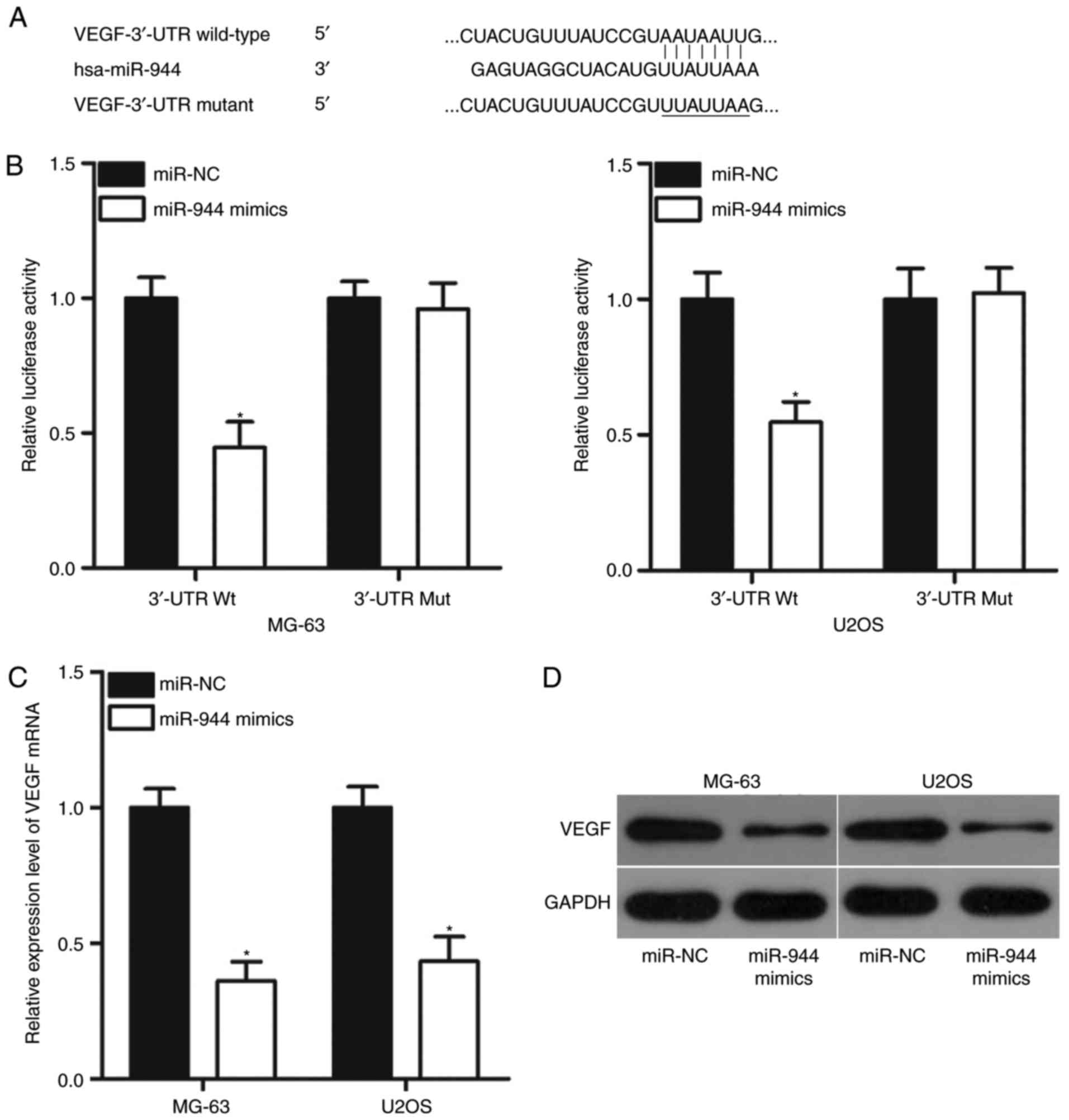
VEGF is a direct target of miR-944 in
OS
Bioinformatics analysis was performed to predict the
putative targets of miR-944 and to delineate the mechanism
responsible for the tumor-suppressing role of miR-944 in OS. As
demonstrated in Fig. 3A, the
3′-UTR of VEGF contains a highly conserved binding site for
miR-944. Thus, VEGF was selected for investigation to further
verify that it serves a crucial role in OS onset and development
(22–25). Subsequently, whether the putative
binding site has functional relevance was investigated by
luciferase reporter assays. As speculated, overexpression of
miR-944 decreased the luciferase activity of the pMIR-VEGF-3′-UTR
Wt group (P<0.05) but failed to affect that of the
pMIR-VEGF-3′-UTR Mut group (Fig.
3B). This result demonstrated that miR-944 could directly
interact with the 3′-UTR of VEGF. In addition, RT-qPCR and western
blot analysis results indicated that the overexpression of miR-944
significantly suppressed VEGF mRNA (P<0.05; Fig. 3C) and protein (P<0.05; Fig. 3D) expression levels in MG-63 and
U2OS cells.
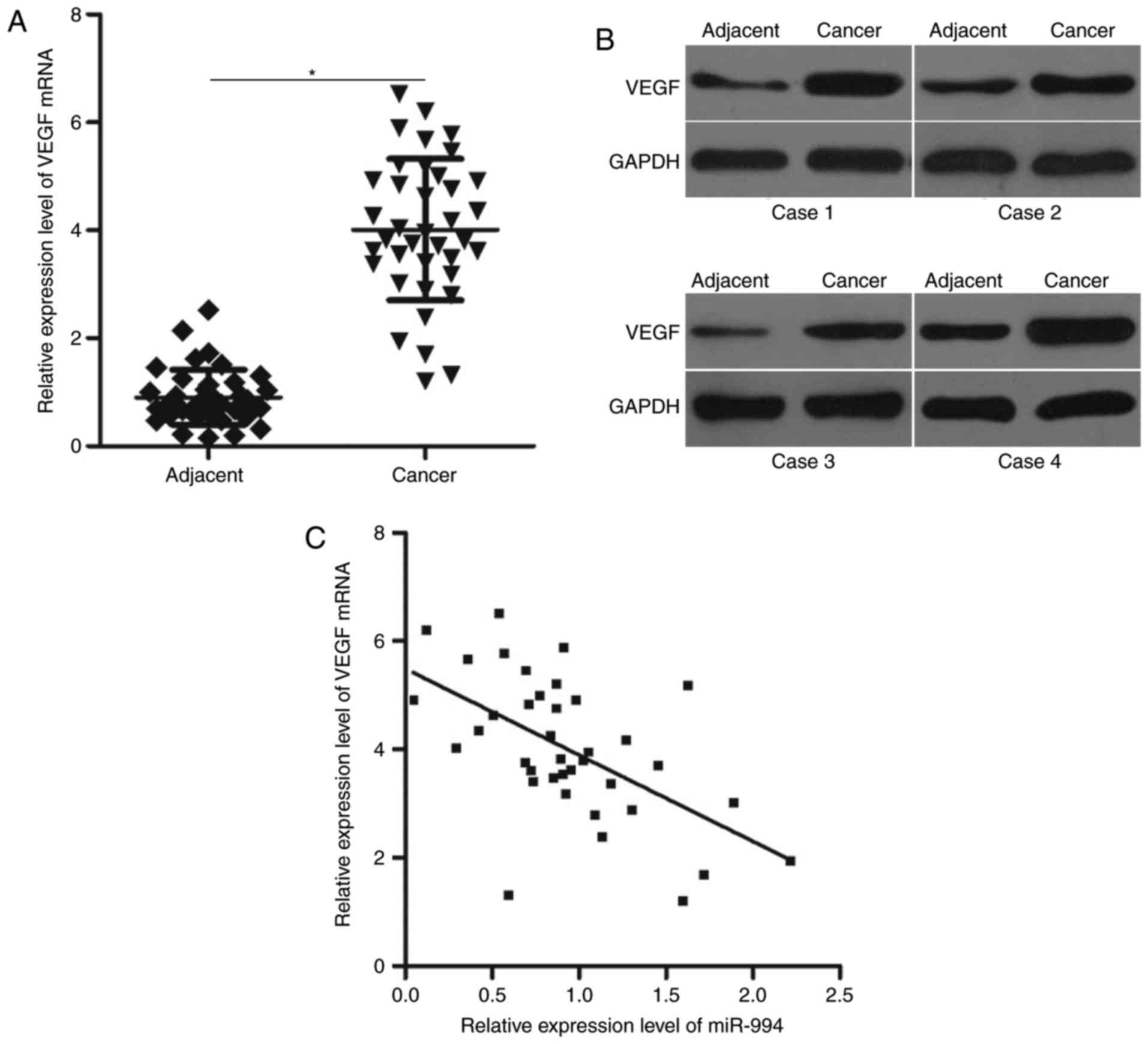
VEGF is upregulated in cancer tissues,
and its expression is negatively correlated with miR-944
expression
To further clarify the association between miR-944
and VEGF in OS, VEGF mRNA expression was investigated in 38 pairs
of cancer tissues and adjacent tissues. RT-qPCR analysis revealed
that the mRNA level of VEGF was increased in cancerous compared
with adjacent tissues (P<0.05; Fig.
4A). In addition, the results of western blot analysis
demonstrated that VEGF protein was overexpressed in cancer tissues
compared with adjacent tissues (Fig.
4B). Furthermore, Spearman's correlation analysis revealed that
miR-944 and VEGF mRNA levels were inversely correlated in cancer
tissues (r=−0.5692, P=0.0002; Fig.
4C).
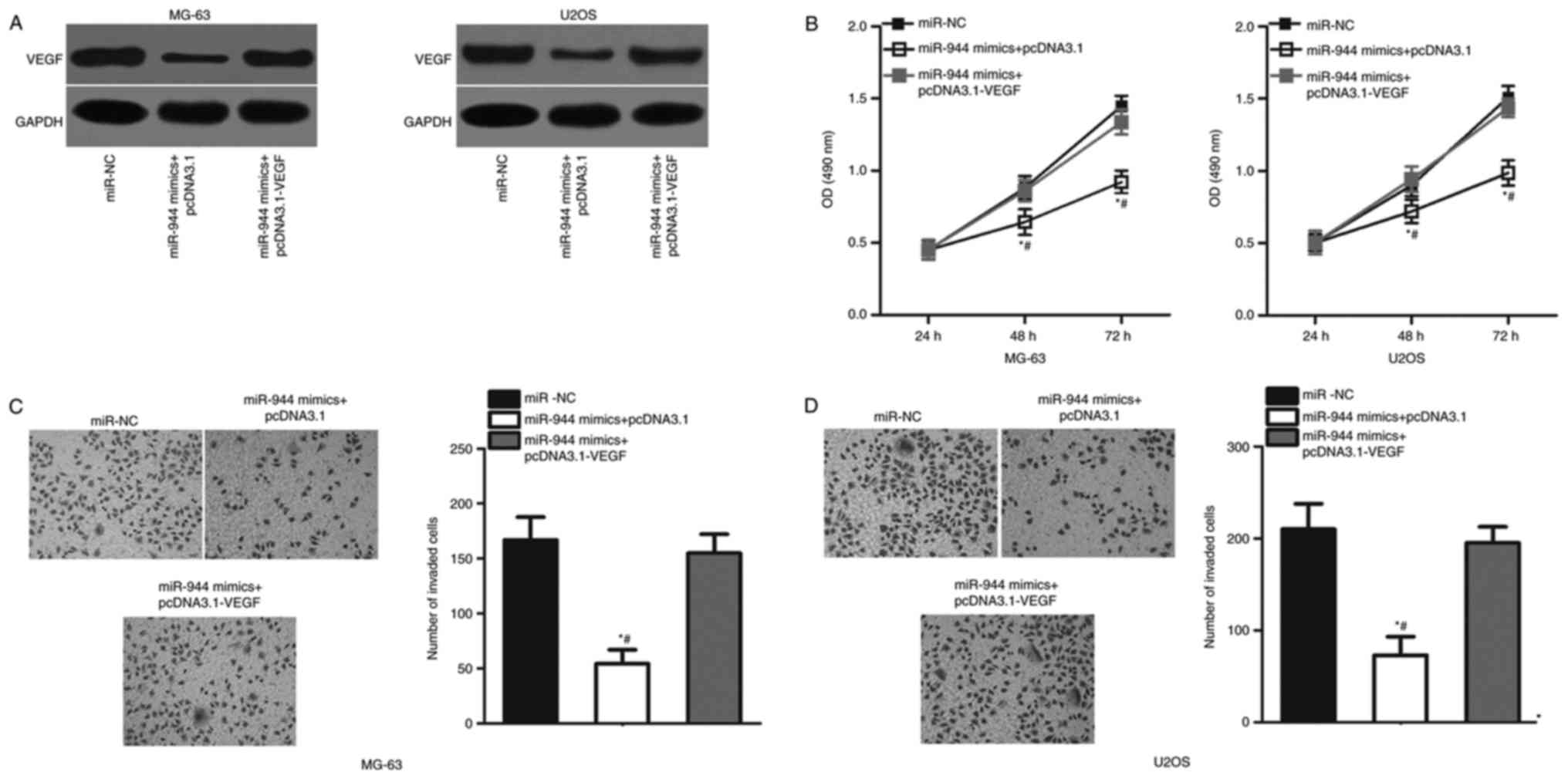
Overexpression of VEGF expression
levels abolishes the inhibitory effects of miR-944 overexpression
in OS cells
To further verify whether VEGF mediates the role of
miR-944 in OS, MG-63 and U2OS cells were cotransfected with miR-944
mimics and empty pcDNA3.1 plasmid or pcDNA3.1-VEGF. The
miR-944-mediated downregulation of VEGF protein expression levels
was almost reversed in MG-63 and U2OS cells cotransfected with
pcDNA3.1-VEGF (P<0.05; Fig.
5A). Additionally, overexpression of VEGF expression
counteracted the suppressive effects of miR-944 overexpression on
proliferation (P<0.05; Fig. 5B)
and invasion (P<0.05; Fig. 5C and
D) of MG-63 and U2OS cells. These results suggested that
miR-944 exhibits a tumor suppressive role in OS by inhibiting VEGF
expression in vitro.
Discussion
Emerging evidence has demonstrated that the
dysregulation of miRNA serves crucial roles in the tumorigenesis
and tumor development of OS, mainly by affecting various
pathological behaviors (26,27).
Therefore, better knowledge of miRNAs in OS may provide novel
insights into the pathogenesis of OS, and may help the development
of promising therapeutic targets for treating patients with this
disease. The present study demonstrated that miR-944 was
significantly downregulated in cancer tissues and cell lines.
Further investigation indicated that overexpression of miR-944
expression inhibited cell proliferation and invasion in OS cell
lines. Additionally, VEGF was validated as a direct target of
miR-944 in OS. VEGF was upregulated in cancer tissues and this
upregulation was inversely correlated with miR-944 expression.
Furthermore, overexpression of VEGF reversed the tumour-suppressive
effects of miR-944 on cell proliferation and invasion in OS in
vitro. These results demonstrated that miR-944 can exert tumor
suppressive-like behavior in OS by directly targeting VEGF. Hence,
miR-944 may be identified as an effective target for the treatment
of patients with OS, but further investigation is required.
Previous studies have demonstrated that miR-944 is
aberrantly expressed in several malignant phenotypes of human
cancer (18–20). For example, miR-944 is upregulated
in endometrial cancer tissues and cell lines. Its increased level
is significantly correlated with pathological classification by the
International Federation of Gynaecology and Obstetrics Stages of
Endometrial Cancer (18). He et
al (18) demonstrated that the
overexpression of miR-944 increases endometrial cancer cell
proliferation, cell cycle progression and reduces apoptosis in
vitro. Conversely, knockdown of miR-944 suppresses the tumour
growth of endometrial cancer in vivo, while Kaplan-Meier
analysis indicates that high miR-944 expression levels are
associated with poor overall survival of patients with endometrial
cancer (18). Furthermore, miR-944
has been demonstrated to be overexpressed in cervical cancer, and
Xie et al (19)
demonstrated that downregulating miR-944 attenuates cell
proliferation and motility in cervical cancer, and breast cancer.
In addition, He et al (20)
revealed that ectopic expression of miR-944 promotes cell growth
and metastasis in breast cancer. Additionally, the same study
demonstrated that downregulation of miR-944 increased the
cytotoxicity of cisplatin in cisplatin-resistant breast cancer
cells. However, miR-944 is notably downregulated in colorectal
cancer tissues and cell lines. Low expression level of miR-944 is
associated with malignant clinical parameters and poor prognosis of
patients with colorectal cancer (28). In the same study, miR-944 was
identified as a tumour suppressor in colorectal cancer as it
inhibits cell migration and invasion. Downregulation of miR-944 has
also been observed in gastric cancer (29) where it has been reported that
resumed expression of miR-944 decreases metastasis and
epithelial-mesenchymal transition, and in non-small cell lung
cancer, where miR-944 overexpression inhibits cell proliferation
(30). These findings suggest that
the functional roles of miR-944 in human malignancies have tissue
specificity and miR-944 should be investigated as a potential
therapeutic target for the aforementioned cancer types, but this
needs to be investigated further.
Identification of the direct targets of miR-944 may
provide a molecular basis for investigation of the regulatory
mechanisms of miR-944. Several targets of miR-944 have been
validated and include cell adhesion molecule 2 (18) in endometrial cancer, C2 and WW
domain containing E3 ubiquitin protein ligase 2 (19) and S100P binding protein (19) in cervical cancer, B cell lymphoma 2
interacting protein 3 (20) in
breast cancer, metastasis associated in colon cancer 1 (28) in colorectal cancer, and Eph
receptor A7 (30) in non-small
cell lung cancer. VEGF, also known as VEGFA, was demonstrated to be
a direct target gene of miR-944 in OS, in the present study. It is
aberrantly overexpressed in a variety of human malignancies, such
as gastric cancer (31), thyroid
cancer (32), bladder cancer
(33), and colorectal cancer
(34). Overexpression of VEGF has
also been reported in OS, and this upregulation is strongly
correlated with Enneking stage and distant metastasis (22). OS patients with high VEGF
expression levels exhibit an unfavorable prognosis compared with
patients with low VEGF levels (23). VEGF serves an important role in OS
initiation and progression via regulating cell proliferation,
apoptosis, migration, invasion, metastasis, and angiogenesis
(24,25). Hence, knocking down VEGF expression
levels may be an attractive therapeutic approach in the therapy of
patients with OS.
In conclusion, the present study demonstrated that
miR-944 was significantly downregulated in cancer tissues and cell
lines. Upregulating miR-944 inhibited cellular proliferation and
invasion in OS cells by directly targeting VEGF. These results
suggest a theoretical basis for the application of miR-944 in
treating patients with OS. However, several limitations of the
present study should be recognised. The number of cases in the
present study was small. In addition, normal bone tissues were not
used as a control. Furthermore, the effects of miR-944 on an OS
animal model were not studied. Therefore, future in vitro
and in vivo experiments need to be performed in order to
validate the present findings.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ML and TY designed this research, wrote and revised
the manuscript. TY, SZ and JZ performed RT-qPCR, western blot
analysis and MTT assay. GL, CL and YW conducted the Transwell
invasion and luciferase reporter assay. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Jining No. 1 People's Hospital, and was performed
according to the principles of the Declaration of Helsinki. Written
informed consent was also provided from all patients enrolled in
the present study.
Patient consent for publication
Written informed consent was provided.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gianferante DM, Mirabello L and Savage SA:
Germline and somatic genetics of osteosarcoma-connecting aetiology,
biology and therapy. Nat Rev Endocrinol. 13:480–491. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fagioli F, Aglietta M, Tienghi A, Ferrari
S, del Prever Brach A, Vassallo E, Palmero A, Biasin E, Bacci G,
Picci P and Madon E: High-dose chemotherapy in the treatment of
relapsed osteosarcoma: An Italian sarcoma group study. J Clin
Oncol. 20:2150–2156. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wada T, Isu K, Takeda N, Usui M, Ishii S
and Yamawaki S: A preliminary report of neoadjuvant chemotherapy
NSH-7 study in osteosarcoma: Preoperative salvage chemotherapy
based on clinical tumor response and the use of granulocyte
colony-stimulating factor. Oncology. 53:221–227. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tan ML, Choong PF and Dass CR:
Osteosarcoma: Conventional treatment vs. gene therapy. Cancer Biol
Ther. 8:106–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ghosh T, Varshney A, Kumar P, Kaur M,
Kumar V, Shekhar R, Devi R, Priyanka P, Khan MM and Saxena S:
MicroRNA-874-mediated inhibition of the major G1/S phase
cyclin, CCNE1, is lost in osteosarcomas. J Biol Chem.
292:21264–21281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li D, Wang H, Song H, Xu H, Zhao B, Wu C,
Hu J, Wu T, Xie D, Zhao J, et al: The microRNAs miR-200b-3p and
miR-429-5p target the LIMK1/CFL1 pathway to inhibit growth and
motility of breast cancer cells. Oncotarget. 8:85276–85289.
2017.PubMed/NCBI
|
|
13
|
Huang RS, Zheng YL, Zhao J and Chun X:
microRNA-381 suppresses the growth and increases cisplatin
sensitivity in non-small cell lung cancer cells through inhibition
of nuclear factor-kB signaling. Biomed Pharmacother. 98:538–544.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao F, Pu Y, Qian L, Zang C, Tao Z and
Gao J: MiR-20a-5p promotes radio-resistance by targeting NPAS2 in
nasopharyngeal cancer cells. Oncotarget. 8:105873–105881. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao Y, Liu X and Lu YX: MicroRNA-143
regulates the proliferation and apoptosis of cervical cancer cells
by targeting HIF-1α. Eur Rev Med Pharmacol Sci. 21:5580–5586.
2017.PubMed/NCBI
|
|
16
|
He Y and Yu B: MicroRNA-93 promotes cell
proliferation by directly targeting P21 in osteosarcoma cells. Exp
Ther Med. 13:2003–2011. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin W, Zhu X, Yang S, Chen X, Wang L,
Huang Z, Ding Y, Huang L and Lv C: MicroRNA-203 inhibits
proliferation and invasion, and promotes apoptosis of osteosarcoma
cells by targeting Runt-related transcription factor 2. Biomed
Pharmacother. 91:1075–1084. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He Z, Xu H, Meng Y and Kuang Y: miR-944
acts as a prognostic marker and promotes the tumor progression in
endometrial cancer. Biomed Pharmacother. 88:902–910. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xie H, Lee L, Scicluna P, Kavak E, Larsson
C, Sandberg R and Lui WO: Novel functions and targets of miR-944 in
human cervical cancer cells. Int J Cancer. 136:E230–E241. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He H, Tian W, Chen H and Jiang K: MiR-944
functions as a novel oncogene and regulates the chemoresistance in
breast cancer. Tumour Biol. 37:1599–1607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Y, Zhang F, Zhang Z, Wang D, Cui B,
Zeng F, Huang L, Zhang Q and Sun Q: High expression levels of Cyr61
and VEGF are associated with poor prognosis in osteosarcoma. Pathol
Res Pract. 213:895–899. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu XW, Wu TY, Yi X, Ren WP, Zhou ZB, Sun
YQ and Zhang CQ: Prognostic significance of VEGF expression in
osteosarcoma: A meta-analysis. Tumour Biol. 35:155–160. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mei J, Gao Y, Zhang L, Cai X, Qian Z,
Huang H and Huang W: VEGF-siRNA silencing induces apoptosis,
inhibits proliferation and suppresses vasculogenic mimicry in
osteosarcoma in vitro. Exp Oncol. 30:29–34. 2008.PubMed/NCBI
|
|
25
|
Peng N, Gao S, Guo X, Wang G, Cheng C, Li
M and Liu K: Silencing of VEGF inhibits human osteosarcoma
angiogenesis and promotes cell apoptosis via VEGF/PI3K/AKT
signaling pathway. Am J Transl Res. 8:1005–1015. 2016.PubMed/NCBI
|
|
26
|
Kushlinskii NE, Fridman MV and Braga EA:
Molecular mechanisms and microRNAs in osteosarcoma pathogenesis.
Biochemistry (Mosc). 81:315–328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sampson VB, Yoo S, Kumar A, Vetter NS and
Kolb EA: MicroRNAs and potential targets in osteosarcoma: Review.
Front Pediatr. 3:692015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wen L, Li Y, Jiang Z, Zhang Y, Yang B and
Han F: miR-944 inhibits cell migration and invasion by targeting
MACC1 in colorectal cancer. Oncol Rep. 37:3415–3422. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pan T, Chen W, Yuan X, Shen J, Qin C and
Wang L: miR-944 inhibits metastasis of gastric cancer by preventing
the epithelial-mesenchymal transition via MACC1/Met/AKT signaling.
FEBS Open Bio. 7:905–914. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu M, Zhou K and Cao Y: MicroRNA-944
affects cell growth by targeting EPHA7 in non-small cell lung
cancer. Int J Mol Sci. 17:pii: E1493. 2016. View Article : Google Scholar
|
|
31
|
Ji YN, Wang Q, Li Y and Wang Z: Prognostic
value of vascular endothelial growth factor A expression in gastric
cancer: A meta-analysis. Tumour Biol. 35:2787–2793. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tian XF, Zhang XW, Chen RX, Wang JW and
Zhang D: Clinical significance of expression of VEGF and bFGF in
thyroid carcinoma. Zhonghua Wai Ke Za Zhi. 42:864–866. 2004.(In
Chinese). PubMed/NCBI
|
|
33
|
Huang YJ, Qi WX, He AN, Sun YJ, Shen Z and
Yao Y: Prognostic value of tissue vascular endothelial growth
factor expression in bladder cancer: A meta-analysis. Asian Pac J
Cancer Prev. 14:645–649. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wen L, Wang R, Lu X and You C: Expression
and clinical significance of vascular endothelial growth factor and
fms-related tyrosine kinase 1 in colorectal cancer. Oncol Lett.
9:2414–2418. 2015. View Article : Google Scholar : PubMed/NCBI
|