Introduction
Coronary artery disease (CAD), which is the most
common type of cardiovascular disease, is the most prevalent cause
of mortality worldwide (1).
Recently, the incidence of CAD was demonstrated to have increased
in China (2). Typically, the
underlying mechanism of CAD involves a section of the coronary
artery inter wall developing atherosclerosis. There are a variety
of risk factors associated with CAD, including hypertension,
obesity, smoking, family history of the disease (3), diabetes, lack of exercise, depression
(4) and high blood lipids
(5,6). Although a number of advances in the
understading of associated pathogeny, diagnosis and treatment of
CAD have been described previously, current knowledge of the
molecular mechanisms underlying CAD is insufficient for the
development of improved treatment and diagnostic strategies for
patients with CAD, and thus requires further investigation.
Identification of the potential therapeutic targets for CAD remains
clinically important.
DEGs have important roles in complex human diseases.
MicroRNA (miRNAs) are a class of small noncoding RNA molecules that
serve a crucial role in RNA silencing and the post-transcriptional
regulation of gene expression by binding to the 3′-untranslated
region of their target genes (7,8). The
identification of DEGs and DEMs associated with CAD may help to
elucidate the underlying molecular mechanisms as well as discover
novel biomarkers and therapies.
Recently, the present study performed a
comprehensive analysis of global mRNA and miRNA expression profile
datasets, including data from normal individuals and patients with
CAD. A total of 350 mRNAs and 66 miRNAs were dysregulated in
patients with CAD. Using these data, 2 networks of miRNA-gene
associations were established. Through the identification of 26
miRNA-target DEGs, potential underlying molecular mechanisms and
therapeutic targets for CAD were investigated.
Materials and methods
mRNA and miRNA expression
datasets
In the present study, the GSE28829 mRNA (9) and GSE59421 miRNA (10) expression datasets were downloaded
from the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo). In the GSE28829
dataset, 16 atherosclerotic plaque samples from patients and 13
control samples were included and analyzed using the Affymetrix
Human Genome U133 Plus 2.0 Array. In the GSE59421 dataset, the
expression levels in platelets were identified in 33 male patients
with premature CAD and 37 age- and sex-matched healthy controls,
and were quantified using the agilent-021827 Human miRNA Microarray
(V3) (miRBase release 12.0 miRNA ID version). These 2 datasets were
indexed and loaded into Entrez GEO Profiles and Entrez GEO
DataSets, which allows users to query and analyze the data using
simple Boolean queries, and provides weblinks to free public raw
data, accurate information, reliable data platforms and other
National Center for Biotechnology Information resources wherever
possible.
Microarray data mining
Analysis of differentially expressed genes (DEGs)
was conducted for the mRNA and miRNA microarray data using GEO2R
Online Analytical tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/). DEGs and
differentially expressed miRNAs (DEMs) were identified using an
adjusted P-value <0.05 and a |log fold change (FC)| >1 as
criteria.
Establishment of the miRNA-gene
regulatory network
miRWalk2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
is a web-based tool developed by Dweep et al (11,12)
that supplies the largest available collection of predicted and
experimentally verified miRNA-target interactions with various
novel and unique features. In the present study, target genes of
the DEMs were identified using miRWalk2.0. Furthermore, Cytoscape
version 3.5.1 software (13) was
used to establish the miRNA-gene regulatory network.
miRNA-target DEG protein-protein
interactions
Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING; http://string-db.org/) is a database of known and
predicted protein-protein interactions (14). In the present study, the
protein-protein interactions were screened using STRING.
Functional enrichment analysis and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analysis
Based on the Database for Visualization, Annotation
and Integrated Analysis (DAVID; david.abcc.ncifcrf.gov/.) (15), the enriched Gene Ontology (GO)
terms for miRNA-target DEGs were identified (only the GO terms that
were enriched in ≥5 genes were included). GO analysis is a commonly
used approach for functional studies of large-scale transcriptomic
data (16). The KEGG pathway
database (17) contains
information on networks of molecules or genes.
miRNA-target DEGs chromosomal
location
The Map Viewer (https://www.ncbi.nlm.nih.gov/projects/mapview/)
provides a wide variety of genome mapping and sequencing data
(18). The locations of the
miRNA-target DEGs on the chromosomes were identified using this
resource.
Results
A total of 350 DEGs and 66 DEMs are
identified
The available numerical mRNA and miRNA expression
values were used to identify DEGs and DEMs. A total of 59
upregulated and 291 downregulated DEGs, and 30 upregulated and 36
downregulated DEMs were identified in patients with CAD compared
with the normal controls. The top 10 DEGs and DEMs are presented in
Table I (P<0.05; |logFC|
>1).
 | Table I.Top 10 most differentially expressed
genes and differentially expressed miRNAs. |
Table I.
Top 10 most differentially expressed
genes and differentially expressed miRNAs.
| Gene | P-value | logFC | miRNA | P-value | logFC |
|---|
| C2 |
1.6×10−10 | −1.25626406 | hsa-miR-221 |
1.13×10−4 | −1.2728544 |
| DENND2D |
5.16×10−10 | −1.15465237 | hsa-miR-1246 |
2.52×10−4 | −1.6266646 |
| ADAP2 |
2.45×10−9 | −1.22371715 | hsa-miR-425 |
8.73×10−4 | −1.1111774 |
| JAK3 |
3.09×10−9 | −1.09039561 | hsa-miR-505 |
1.54×10−3 | −1.1639686 |
| SLAMF8 |
4.26×10−9 | −2.07254568 | hsa-miR-598 |
1.93×10−3 | −1.2433251 |
| CD68 |
4.51×10−9 | −1.13556898 | hsa-miR-431* |
3.26×10−3 | 1.0564151 |
| LINC01094 |
6.25×10−9 | −1.10497076 | hsa-miR-25 |
3.85×10−3 | −1.0901694 |
| SERPINA1 |
8.24×10−9 | −1.73250228 | hsa-miR-30e |
4.66×10−3 | −1.1414787 |
| VAMP8 |
8.69×10−9 | −1.82538528 | hsa-miR-370 |
5.2×10−3 | 1.2500556 |
| C3AR1 |
1.14×10−8 | −2.22628842 |
hsa-miR-1274a |
5.36×10−3 | −1.27226 |
miRNA-gene regulation network and
protein interactions of 57 overlapping DEGs
Using miRWalk2.0, 3,588 target genes of the DEMs
were identified, and 57 of these target genes were demonstrated to
overlap with the identified DEGs. In the group of 57 overlapping
DEGs, a protein-protein interaction network of 26 DEGs was
established using STRING. Furthermore, 26 miRNA-gene pairs were
obtained among the 26 aforementioned DEGs and 19 DEMs. The 19 DEMs
were demonstrated to regulate multiple genes, thereby forming a
considerable network (Fig. 1).
GO and KEGG pathway analysis
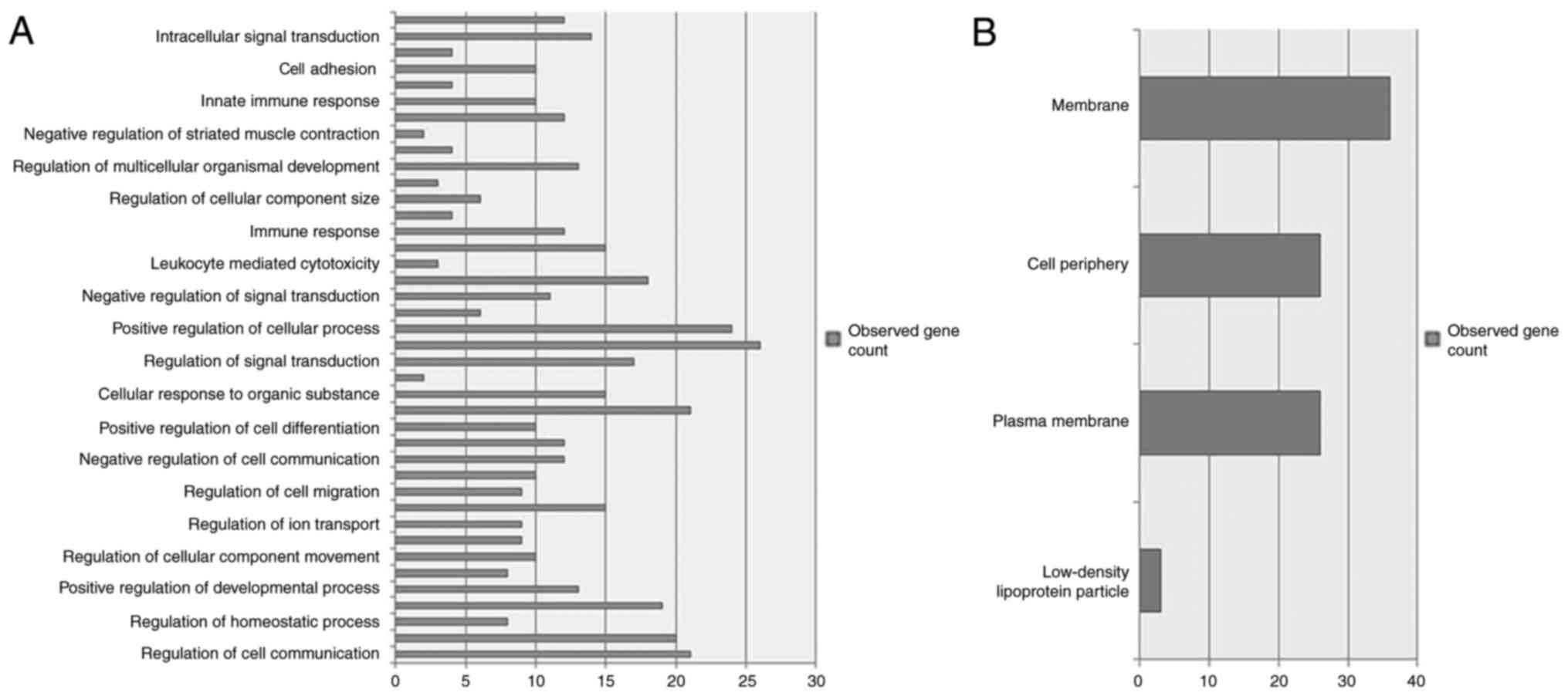
Using DAVID, 40 enriched GO process terms were
obtained for miRNA-targeting DEGs. The five most significantly
enriched GO terms, including ‘regulation of cell communication’,
‘regulation of signaling’, ‘regulation of response to stimulus’,
‘positive regulation of biological process’ and ‘positive
regulation of cellular process’, are presented in Fig. 2A. A total of 4 enriched GO
component terms were obtained for miRNA-target DEGs, including
‘low-density lipoprotein particle’, ‘plasma membrane’, ‘cell
periphery’ and ‘membrane’ (Fig.
2B). KEGG pathway analysis identified 3 genes [hexokinase 2
(HK2), phosphatidylinositol-4,5-biphosphate 3-kinase catalytic
subunit γ (PIK3CG), and suppressor of cytokine signaling 3 (SOCS3)]
that were present in Type II diabetes mellitus network and 3 other
genes, ATPase Na+/K+ transporting subunit α 2, HK2 and PIK3CG that
were present in the carbohydrate digestion and absorption
network.
Furthermore, the miRNA-gene regulatory network of
the 26 miRNA-gene pairs was established using Cytoscape version
3.5.1 software (Fig. 3). The 26
regulatory pairs between DEMs and their target genes were
identified, including hsa-miR-199a-5p-apolioprotein E (APOE),
hsa-miR-106a-branched chain amino acid transaminase 1 (BCAT1),
hsa-miR-1234-cytohesin 1 interacting protein (CYTIP),
hsa-miR-1249-macrophage scavenger receptor 1 (MSR1),
hsa-miR-142-3p-ecotropic viral integration site 2B (EVI2B),
hsa-miR-142-3p-myristoylated alanine rich protein kinase C
substrate (MARCKS),
hsa-miR-142-3p-Phosphatidylinositol-4,5-bisphosphate 3-kinase
catalytic subunit gamma isoform (PIK3CG), hsa-miR-152-MAF basic
leucine zipper (BZIP) transcription factor B (MAFB),
hsa-miR-215-DEP domain containing MTOR interacting protein
(DEPTOR), hsa-miR-215-HOP homeobox (HOPX), hsa-miR-22-colony
stimulating factor 1 receptor (CSF1R), hsa-miR-221-coronin 1A
(CORO1A), hsa-miR-221-Suppressor of cytokine signaling 3 (SOCS3),
hsa-miR-221-T cell lymphoma invasion and metastasis 1 (TIAM1),
hsa-miR-30b-immunoglobulin superfamily member 6 (IGSF6),
hsa-miR-30c-C-X-C motif chemokine ligand 16 (CXCL16),
hsa-miR-30e-Serpin family E member 2 (SERPINE2),
hsa-miR-323a-3p-phospholipase A2 group VII (PLA2G7),
hsa-miR-329-ecotropic viral integration site 2A (EVI2A),
hsa-miR-33a-chromosome 15 open reading frame 48 (C15orf48),
hsa-miR-33a-FYN binding protein 1 (FYB), hsa-miR-376-death
associated protein kinase 1 (DAPK1), hsa-miR-484-hematopoietic
cell-specific Lyn substrate 1 (HCLS1), hsa-miR-484-HK2,
hsa-miR-486-3p-netrin 1 (NTN1) and hsa-miR-616-lysozyme (LYZ).
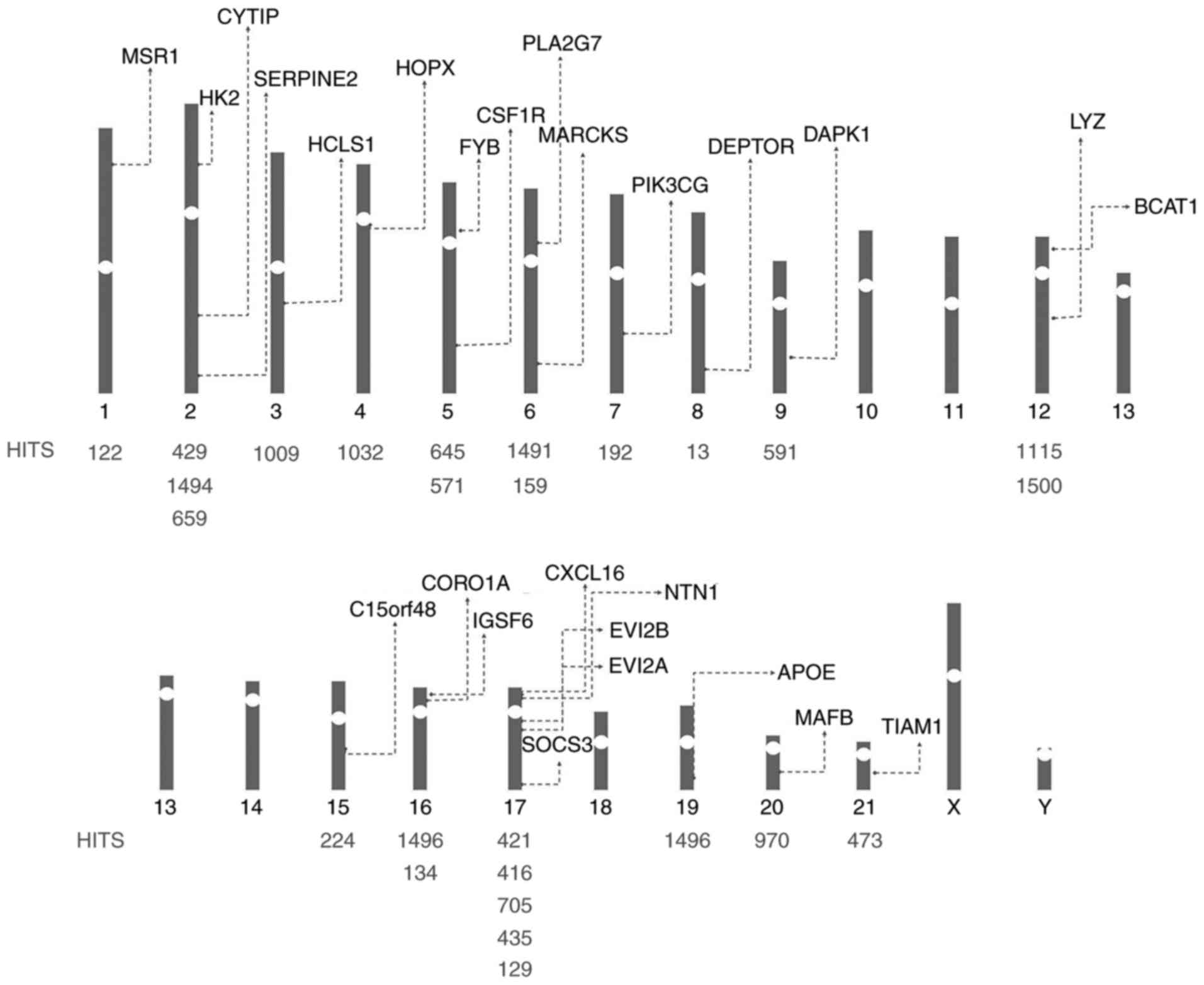
miRNA-target DEG chromosomal
location
The 26 DEGs in the miRNA-gene pairs were located on
the chromosomes by Map Viewer (Fig.
4). The chromosomal locations of these genes were
inhomogeneous, and they were primarily distributed on chromosomes
17, 2, 5, 6, 12, 16, 1, 3, 4, 7, 8, 9, 15, 19, 20, and 21. None of
the associated genes were distributed on sex chromosomes.
Discussion
miRNAs are crucial and evolutionarily conserved
components of gene regulation (19). Up- or downregulated expression
levels of specific miRNAs were observed in diseased human hearts,
implying their involvement in cardiomyopathies (20). CAD is an ischemic heart disease
that occurs when part of the arterial internal wall develops
atherosclerosis, which is associated with lipid retention and
inflammation. Murine miRNA-712 is a potential biomarker for
atherosclerosis (21). However,
the detailed mechanism underlying the mechanism by which miRNAs
regulate the expression of CAD-associated genes and gene products
requires thorough investigation.
In the present study, a total of 59 upregulated and
291 downregulated DEGs, and 30 upregulated and 36 downregulated
DEMs were identified in patients with CAD compared with normal
controls. The GO analysis indicated that several significantly
enriched terms from the categories of regulation of cell
communication, regulation of signaling, regulation of response to
stimulus, positive regulation of biological process, and positive
regulation of cellular process were markedly over-represented in
patients with CAD, suggesting that these biological processes are
important topics for future study.
The present study also identified the interactions
and associations among proteins, which were encoded by 26 DEGs in
the miRNA-gene pairs. In addition, the 19 miRNAs (which target the
26 DEGs) also regulate a large quantity of genes.
Apolipoprotein E is encoded by the APOE gene, and
serves as a shuttle lipid in the bloodstream (22). Maintaining constant levels of
cholesterol is very important for avoiding cardiovascular disease.
Carriers of the APOEe4 allele exhibit increased risks of developing
atherosclerosis; an abnormal expression of APOE is associated with
fatty deposition, which increases the risk of heart attack and
stroke (23). In addition,
previous studies have identified that mutated APOE causes an
increased risk of Alzheimer's disease in individuals; however, the
mechanism is unclear (24). Thus,
it may be suggested that the APOE gene is associated with
Alzheimer's disease and CAD. The BCAT1 gene encodes a form of
transaminase to catalyze the branched-chain amino acid
transamination, which when defective is prone to 2 types of
disease; hypervalinemia and hyperleucine-isoleucinemia (25,26).
The coding products of the MSR1 gene are membrane glycoproteins,
which are associated with atherosclerosis, Alzheimer's disease and
host defense (27). DEPTOR has
been considered as an endogenous regulator overexpressed in
multiple myeloma cells (28).
Increased levels of CSF1R were identified in microglia in
Alzheimer's disease (29). A
number of types of cytokines [interleukin (IL)6, IL10, interferon
(IFN)-γ] may induce the expression of SOCS3 gene. Knocking out the
SOCS3 gene prevents insulin resistance in obesity (30). The IGSF6 gene is associated with
inflammatory bowel disease (31).
HK2 is the first and rate-limiting enzyme in the
Embden-Meyerhof-Parnas pathway (32). DAPK1 is a positive mediator of
γ-interferon-induced programmed cell death. Diseases identified to
be associated with HCLS1 include spherocytosis, type 1 and
congenital hemolytic anemia (33).
The product of the HCLS1 gene serve a role in antigen receptor
signaling for clonal expansion and deletion in lymphoid cells
(34). Platelet-activating factor
acetylhydrolase deficiency may be caused by the defective PLA2G7
gene, in which the associated pathways are the phospholipase-C
pathway and sweet taste receptor signaling (35). Oxidation of low-density
lipoproteins is an initial step of atherogenesis that generates
pro-inflammatory phospholipids, including platelet-activating
factor and its analogs (36). GO
annotations associated with EVI2A include transmembrane signaling
receptor activity (37). Mutations
in SERPINE2 may cause a range of diseases; the α-antitrypsin
deficiency is one of the most common hereditary diseases associated
with this gene (38). CXCL16
expression is induced by the inflammatory cytokines IFN-γ and tumor
necrosis factor-α (39). In
patients with systemic lupus erythematosus, the differential
expression of the FYB gene encodes a protein that participates in
the T-cell signaling cascade and in IL-2A expression modulation
(40). The C15orf48 gene was first
identified in a study of human esophageal squamous cell carcinoma
tissues (41). The HOPX gene is
involved in the malignant conversion of placental trophoblasts
(42). The protein encoded by
CYTIP is weakly expressed in resting natural killer and T cells,
which contain 2 leucine zipper domains and a putative C-terminal
nuclear targeting signal (43).
CYTIP is associated with amplification and expansion of oncogenic
pathways, and exhibits metastatic traits (43). The gene product of the EVI2B gene
has been studied extensively regarding its role in
neurofibromatosis, leukemia, and myeloid leukemia (44). The MARCKS gene product serves
important roles in cell shape, cell motility, secretion,
transmembrane transport, regulation of cell cycle and neural
development (45). The PIK3CG gene
product is an enzyme that phosphorylates phosphoinositides on the
3-hydroxyl group of the inositol ring, PIK3CG SNPs (rs1129293 and
rs17398575) increase the risk of ischemia in patients suffering
from coronary heart disease (44).
MAFB is a bZIP transcription factor that serves an important role
in the regulation of lineage-specific hematopoiesis, and rs2902940A
alleles of the MAFB gene result in decreased serum ApoAI levels in
controls and an increased risk of CAD (46). The encoded nuclear protein
represses ETS1-mediated transcription of erythroid-specific genes
in myeloid cells (47). CORO1A has
been implicated in T-cell mediated immunity and mitochondrial
apoptosis (48). TIAM1 has been
identified as a specific activator of the Rho-like GTPase Rac and
is implicated in the regulation of different cell biological
functions, including cell polarity, adhesion, migration, invasion,
metastasis, and carcinogenesis (49). NTN1 is included in a family of
laminin-associated secreted proteins (50). The LYZ protein exhibits
antibacterial activity against certain bacterial species. Missense
mutations in the LYZ gene have been identified in heritable renal
amyloidosis. miRNAs serve as stimulating factors at various stages
of atherosclerosis to regulate a number of genes, thereby
regulating endothelial cells, vascular smooth muscle cells, and the
function of macrophages, which control the occurrence of
atherosclerosis (51).
The chromosomal locations of these aforementioned 26
genes are inhomogeneous, and they are primarily distributed on
chromosomes 17, 2, 5, 6, 12, 16, 1, 3, 4, 7, 8, 9, 15, 19, 20 and
21. No miRNA-target DEGs were identified on the sex chromosomes,
which suggested that CAD may be independent of sex. in CAD.
Finally, the results of the study may be a useful resource for
understanding the functions of miRNA and potential therapeutic
targets in the pathophysiology of CAD, but additional molecular
analyses are required. The affected interactions of the 26
miRNA-gene pairs identified in the present study may be validated
in vitro by dual-luciferase reporter gene assays, and the
functions of the 26 miRNAs and their target genes will be confirmed
in vivo and in vitro, so that potential therapeutic
targets may be identified and effective targeting of the affected
biological pathways may be achieved for the treatment and improved
prognosis of CAD.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Key Programs of the Educational Commission of Hebei Province of
China (grant no. ZD2018076) and Youth Science and Technology
Project of Health and Municipal Commission of Population and Family
of Hebei Province of China (grant no. 20180811).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
repository [GSE28829 dataset, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE28829;
GSE59421 dataset, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59421].
Authors' contributions
WW performed the bioinformatics analyses. ZX mined
and downloaded the data from the Gene Expression Omnibus database.
XZ analyzed and interpreted the data. XC identified differentially
expressed genes targeted by microRNAs associated with coronary
heart disease and analyzed the results. WW was a major contributor
in the writing of the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mendis S, Puska P and Norrving B: Global:
Atlas on cardiovascular disease prevention and control. Geneva:
World Health Organization in collaboration with the World Heart
Federation and the World Stroke Organization. 3–18. 2011.
|
|
2
|
Ding D, Wang M, Su D, Hong C, Li X, Yang
Y, Zhang Y, Hu G and Ling W: Body mass index, high-sensitivity
C-reactive protein and mortality in Chinese with coronary artery
disease. PLoS One. 10:e01357132015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Centers for Disease Control and Prevention
(CDC): Prevalence of coronary heart disease-United States,
2006–2006. MMWR Morb Mortal Wkly Rep. 60:1377–1381. 2011.PubMed/NCBI
|
|
4
|
GBD 2015 Disease and Injury Incidence and
Prevalence Collaborators: Global, regional, and national incidence,
prevalence, and years lived with disability for 310 diseases and
injuries, 1990–1990: A systematic analysis for the global burden of
disease study 2015. Lancet. 388:1545–1602. 2016.PubMed/NCBI
|
|
5
|
Kontos MC, Diercks DB and Kirk JD:
Emergency department and office-based evaluation of patients with
chest pain. Mayo Clin Proc. 85:284–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dai X, Wiernek S, Evans JP and Runge MS:
Genetics of coronary artery disease and myocardial infarction.
World J Cardiol. 8:1–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zimmerman AL and Wu S: MicroRNAs, cancer
and cancer stem cells. Cancer Lett. 300:10–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Doring Y, Manthey HD, Drechsler M, Lievens
D, Megens RT, Soehnlein O, Busch M, Manca M, Koenen RR, Pelisek J,
et al: Auto-antigenic protein-DNA complexes stimulate plasmacytoid
dendritic cells to promote atherosclerosis. Circulation.
125:1673–1683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kok MG, Halliani A, Moerland PD, Meijers
JC, Creemers EE and Pinto-Sietsma SJ: Normalization panels for the
reliable quantification of circulating microRNAs by RT-qPCR. FASEB
J. 29:3853–3862. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of 3 genomes. J Biomed Inform. 44:839–837.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:697. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hulsegge I, Kommadath A and Smits MA:
Globaltest and GOEAST: Two different approaches for Gene Ontology
analysis. BMC Proc. 3 Suppl 4:S102009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wolfsberg TG: Using the NCBI Map Viewer to
browse genomic sequence data. Curr Protoc Hum Genet Chapter.
18:Unit 18.5. 2011.
|
|
19
|
Peterson KJ, Dietrich MR and McPeek MA:
MicroRNAs and metazoan macroevolution: Insights into canalization,
complexity, and the Cambrian explosion. Bioessays. 31:736–747.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tatsuguchi M, Seok HY, Callis TE, Thomson
JM, Chen JF, Newman M, Rojas M, Hammond SM and Wang DZ: Expression
of microRNAs is dynamically regulated during cardiomyocyte
hypertrophy. J Mol Cell Cardiol. 42:1137–1141. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Insull W Jr: The pathology of
atherosclerosis: Plaque development and plaque responses to medical
treatment. Am J Med. 122 1 Suppl:S3–S14. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Phillips MC: Apolipoprotein E isoforms and
lipoprotein metabolism. IUBMB Life. 66:616–623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eichner JE, Dunn ST, Perveen G, Thompson
DM, Stewart KE and Stroehla BC: Apolipoprotein E polymorphism and
cardiovascular disease: A HuGE review. Am J Epidemiol. 155:487–495.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mahley RW, Weisgraber KH and Huang Y:
Apolipoprotein E4: A causative factor and therapeutic target in
neuropathology, including Alzheimer's disease. Proc Natl Acad Sci
USA. 103:5644–5651. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chang IW, Wu WJ, Wang YH, Wu TF, Liang PI,
He HL, Yeh BW and Li CF: BCAT1 overexpression is an indicator of
poor prognosis in patients with urothelial carcinomas of the upper
urinary tract and urinary bladder. Histopathology. 68:520–532.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goto M, Miyahara I, Hirotsu K, Conway M,
Yennawar N, Islam MM and Hutson SM: Structural determinants for
branched-chain aminotransferase isozyme-specific inhibition by the
anticonvulsant drug gabapentin. J Biol Chem. 280:37246–37256. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ashkenas J, Penman M, Vasile E, Acton S,
Freeman M and Krieger M: Structures and high and low affinity
ligand binding properties of murine type I and type II macrophage
scavenger receptors. J Lipid Res. 34:983–1000. 1993.PubMed/NCBI
|
|
28
|
Peterson TR, Laplante M, Thoreen CC,
Sancak Y, Kang SA, Kuehl WM, Gray NS and Sabatini DM: DEPTOR is an
mTOR inhibitor frequently overexpressed in multiple myeloma cells
and required for their survival. Cell. 137:873–886. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mitrasinovic OM, Grattan A, Robinson CC,
Lapustea NB, Poon C, Ryan H, Phong C and Murphy GM Jr: Microglia
overexpressing the macrophage colony-stimulating factor receptor
are neuroprotective in a microglial-hippocampal organotypic
coculture system. J Neurosci. 25:4442–4451. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jorgensen SB, O'Neill HM, Sylow L,
Honeyman J, Hewitt KA, Palanivel R, Fullerton MD, Öberg L,
Balendran A, Galic S, et al: Deletion of skeletal muscle SOCS3
prevents insulin resistance in obesity. Diabetes. 62:56–64. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
King K, Moody A, Fisher SA, Mirza MM,
Cuthbert AP, Hampe J, Sutherland-Craggs A, Sanderson J, MacPherson
AJ, Forbes A, et al: Genetic variation in the IGSF6 gene and lack
of association with inflammatory bowel disease. Eur J Immunogenet.
30:187–190. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ahn KJ, Kim J, Yun M, Park JH and Lee JD:
Enzymatic properties of the N- and C-terminal halves of human
hexokinase II. BMB Rep. 42:350–355. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Feinstein E, Druck T, Kastury K, Berissi
H, Goodart SA, Overhauser J, Kimchi A and Huebner K: Assignment of
DAP1 and DAPK-genes that positively mediate programmed cell death
triggered by IFN-gamma-to chromosome regions 5p12.2 and 9q34.1,
respectively. Genomics. 29:305–307. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
van Rossum AG, Schuuring-Scholtes E, van
Buuren-van Seggelen V, Kluin PM and Schuuring E: Comparative genome
analysis of cortactin and HS1: The significance of the F-actin
binding repeat domain. BMC Genomics. 6:152005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mohler ER III, Ballantyne CM, Davidson MH,
Hanefeld M, Ruilope LM, Johnson JL and Zalewski A: Darapladib
Investigators: The effect of darapladib on plasma
lipoprotein-associated phospholipase A2 activity and cardiovascular
biomarkers in patients with stable coronary heart disease or
coronary heart disease risk equivalent: The results of a
multicenter, randomized, double-blind, placebo-controlled study. J
Am Coll Cardiol. 51:1632–1641. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ninio E, Tregouet D, Carrier JL, Stengel
D, Bickel C, Perret C, Rupprecht HJ, Cambien F, Blankenberg S and
Tiret L: Platelet-activating factor-acetylhydrolase and
PAF-receptor gene haplotypes in relation to future cardiovascular
event in patients with coronary artery disease. Hum Mol Genet.
13:1341–1351. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cawthon RM, O'Connell P, Buchberg AM,
Viskochil D, Weiss RB, Culver M, Stevens J, Jenkins NA, Copeland NG
and White R: Identification and characterization of transcripts
from the neurofibromatosis 1 region: The sequence and genomic
structure of EVI2 and mapping of other transcripts. Genomics.
7:555–565. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Janciauskiene SM, Bals R, Koczulla R,
Vogelmeier C, Köhnlein T and Welte T: The discovery of
α1-antitrypsin and its role in health and disease. Respir Med.
105:1129–1139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Abel S, Hundhausen C, Mentlein R, Schulte
A, Berkhout TA, Broadway N, Hartmann D, Sedlacek R, Dietrich S,
Muetze B, et al: The transmembrane CXC-chemokine ligand 16 is
induced by IFN-gamma and TNF-alpha and shed by the activity of the
disintegrin-like metalloproteinase ADAM10. J Immunol.
172:6362–6372. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Addobbati C, Brandão LA, Guimarães RL,
Pancotto JA, Donadi EA, Crovella S, Segat L and Sandrin-Garcia P:
FYB gene polymorphisms are associated with susceptibility for
systemic lupus erythemathosus (SLE). Hum Immunol. 74:1009–1014.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zimmer A, Bouley J, Le Mignon M, Pliquet
E, Horiot S, Turfkruyer M, Baron-Bodo V, Horak F, Nony E, Louise A,
et al: A regulatory dendritic cell signature correlates with the
clinical efficacy of allergen-specific sublingual immunotherapy. J
Allergy Clin Immunol. 129:1020–1030. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Asanoma K, Kato H, Inoue T, Matsuda T and
Wake N: Analysis of a candidate gene associated with growth
suppression of choriocarcinoma and differentiation of trophoblasts.
J Reprod Med. 49:617–626. 2004.PubMed/NCBI
|
|
43
|
Heufler C, Ortner D and Hofer S: Cybr,
CYTIP or CASP: An attempt to pinpoint a molecule's functions and
names. Immunobiology. 213:729–732. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zjablovskaja P, Kardosova M, Danek P,
Angelisova P, Benoukraf T, Wurm AA, Kalina T, Sian S, Balastik M,
Delwel R, et al: EVI2B is a C/EBPα target gene required for
granulocytic differentiation and functionality of hematopoietic
progenitors. Cell Death Differ. 24:705–716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Prieto D and Zolessi FR: Functional
diversification of the four MARCKS family members in zebrafish
neural development. J Exp Zool B Mol Dev Evol. 328:119–138. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang Q, Yin RX, Zhou YJ, Cao XL, Guo T and
Chen WX: Association of polymorphisms in the MAFB gene and the risk
of coronary artery disease and ischemic stroke: A case-control
study. Lipids Health Dis. 14:792015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dwyer J, Li H, Xu D and Liu JP:
Transcriptional regulation of telomerase activity: Roles of the Ets
transcription factor family. Ann N Y Acad Sci. 1114:36–47. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kerber RA, OBrien E and Cawthon RM: Gene
expression profiles associated with aging and mortality in humans.
Aging Cell. 8:239–250. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu Z, Gakhar L, Bain FE, Spies M and
Fuentes EJ: The Tiam1 guanine nucleotide exchange factor is
auto-inhibited by its pleckstrin homology coiled-coil extension
domain. J Biol Chem. 292:17777–17793. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ren XR, Hong Y, Feng Z, Yang HM, Mei L and
Xiong WC: Tyrosine phosphorylation of netrin receptors in netrin-1
signaling. Neurosignals. 16:235–345. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jean E, Ebbo M, Valleix S, Benarous L,
Heyries L, Grados A, Bernit E, Grateau G, Papo T, Granel B, et al:
A new family with hereditary lysozyme amyloidosis with gastritis
and inflammatory bowel disease as prevailing symptoms. BMC
Gastroenterol. 14:1592014. View Article : Google Scholar : PubMed/NCBI
|