Introduction
Bladder cancer is a common genitourinary disease
worldwide, particularly in the male population, with ~74,000 newly
diagnosed cases in the United States in 2014 (1,2).
Despite advances in surgical techniques, including the widespread
application of minimally invasive surgery as well as an improved
understanding of multimodal treatments involving chemotherapy,
radiotherapy, and immunotherapy, the 5-year cancer-specific
survival for patients with advanced bladder cancer remains ~50%
without any improvement in the past two decades (3–6).
Furthermore, the decision making in clinical practice depends on
the results of cystoscopy examination, imaging and
histopathological examination, which cannot provide sufficient
information regarding patients' prognosis (7). Therefore, it is necessary to
investigate novel molecular biomarkers that may aid increased
accuracy of prognostic evaluation.
Long non-coding RNAs (lncRNAs), which can be located
in the nucleus or cytoplasm, are RNA molecules of >200
nucleotides with limited protein-coding potential (8,9).
With the help of microarray and next-generation sequencing,
accumulating evidence has confirmed that a number of dysregulated
lncRNAs in bladder cancer tissues and cell lines may serve critical
roles in tumor formation, progression, and/or metastasis, e.g.,
urothelial cancer associated 1 (UCA1), maternally expressed gene 3
(MEG3), H19 (10–13), and these lncRNAs when detected in
cancerous tissues and body fluids may serve as biomarkers for early
diagnosis, as therapeutic targets, and/or prognostic markers of
bladder cancer. For example, associated studies have identified
that the upregulated lncRNA-UCA1 in bladder cancer may contribute
to tumor proliferation, patient mortality and tumor invasiveness,
and its detection in the urine sediment may improve the sensitivity
and specificity of bladder cancer diagnoses (14,15).
In addition, certain upregulated lncRNAs in bladder
cancer, including lncRNA-n336928, cervical carcinoma expressed
proliferating cell nuclear antigen-regulatory lncRNA and promoter
of cyclin-dependent kinase inhibitor 1 antisense DNA
damage-activated RNA, have not only been reported to have the
ability to promote tumorigenesis, but have also been implicated in
poor prognosis and may serve as independent prognostic factors
(7,16,17).
Therefore, lncRNAs as novel biomarkers could be useful in clinical
procedures involving the diagnosis and prognosis of bladder
cancer.
In the present study, based on previous
high-throughput sequencing applied to 10 pairs of tissue samples,
the aim was to screen RNAs for significantly differentially
expressed lncRNAs that could be novel molecular biomarkers for the
prognosis of bladder cancer. It is noteworthy that differential
expression patterns of lncRNA-n346372 were identified in bladder
cancer tissues and corresponding normal tissues; furthermore, the
association of this lncRNA with clinical variables and prognosis of
patients with bladder cancer were investigated.
Patients and methods
Patients, tissue specimens and cell
lines
A total of 60 patients with bladder cancer and
paired normal tissues adjacent to the tumor were included in the
present study; all patients provided written informed consent.
Following a radical cystectomy, all the resected specimens were
snap-frozen in liquid nitrogen immediately and then stored at −80°C
(in a freezer) until analysis. Initially, 10 pairs out of the total
number of samples, including five pairs of non-muscle-invasive
bladder cancer (NMIBC; Ta three cases; T1 two cases) and five pairs
of muscle-invasive bladder cancer (MIBC; T2a three cases; T2b two
cases) were analyzed by high-throughput transcriptome sequencing.
The present study's protocol was approved by the Ethics Committee
of Changhai Hospital of the Second Military Medical University
(Shanghai, China).
T24 bladder cancer cells were purchased from the
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China), and were cultured in RPMI-1640 medium
with 10% fetal bovine serum (FBS; both Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and 1% ampicillin (100
units/ml) and streptomycin (100 units/ml; Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C in a humidifed atmosphere of 95% air and
5% CO2. T24 cells were plated at a density of
5×104 per well in 6-well plates.
Clinical data collection
Clinical parameters in the present study, including
age, sex, smoking history, tumor size, tumor number, tumor stage
and histological grade, are summarized in Table I. The 2002 Tumor Node Metastasis
criteria were adopted to evaluate the tumor tissue stage and the
2004 World Health Organization classification was employed to
evaluate the histological grade (18); the pathological diagnosis of each
specimen was made independently by two pathologists. In particular,
there were 15 cases diagnosed with NMIBC and 45 cases of MIBC, and
out of all the samples 17 cases were classified as low-grade
bladder cancer and 43 cases as high-grade bladder cancer. Follow-up
information came from outpatient visits and regular telephone
interviews.
 | Table I.Correlation analysis between ln346372
expression level and clinical-pathological data of all enrolled
patients in the present study. |
Table I.
Correlation analysis between ln346372
expression level and clinical-pathological data of all enrolled
patients in the present study.
|
|
| Ln346372
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | Group | Low, n (%) | High, n (%) | Total | P-value |
|---|
| Age, (years) | ≤60 | 4 (26.7) | 11 (73.3) | 15 | 0.133 |
|
| >60 | 22 (48.9) | 23 (51.1) | 45 |
|
| Sex | Male | 21 (39.6) | 32 (60.4) | 53 | 0.377 |
|
| Female | 4 (57.1) | 3 (42.9) | 7 |
|
| Smoking history | No | 20 (50.0) | 20 (50.0) | 40 | 0.064 |
|
| Yes | 5 (25.0) | 15 (75.0) | 20 |
|
| Tumor size
(cm) | ≤3 | 6 (50.0) | 6 (50.0) | 12 | 0.608 |
|
| >3 | 19 (39.6) | 29 (60.4) | 48 |
|
| Tumor number | Unifocal | 4 (50.0) | 4 (50.0) | 8 | 0.513 |
|
| Multifocal | 21 (40.4) | 31 (59.6) | 52 |
|
| Tumor stage | ≤T1 | 11 (73.3) | 4 (26.7) | 15 | 0.004 |
|
| T2-T4 | 14 (31.1) | 31 (68.9) | 45 |
|
| Tumor grade | Low | 14 (82.4) | 3 (17.6) | 17 | <0.001 |
|
| High | 11 (25.6) | 32 (74.4) | 43 |
|
High-throughput transcriptome
sequencing
Total RNA was extracted from 10 pairs of samples
using the TRIzol reagent (Thermo Fisher Scientific, Inc.), followed
by quantification and a purity check on a NanoDrop-2000 (Thermo
Fisher Scientific, Inc., Wilmington, DE, USA) and an integrity
check based on formaldehyde denaturing 2% agarose gel
electrophoresis (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Following DNA digestion with DNase I (Bio-Rad Laboratories, Inc.),
enrichment of total RNA was conducted to isolate mRNA or removal of
ribosomal RNAs using magnetic beads with oligo (dT) (New England
BioLabs, Inc., Ipswich, MA, USA). Mixed with Fragmentation Buffer
(Ambion; Thermo Fisher Scientific, Inc.), the enriched mRNAs were
broken down into short fragments, which could serve as
reverse-transcription templates for the subsequent cDNA synthesis.
These short fragments were purified and resolved in Elution buffer
(Ambion; Thermo Fisher Scientific, Inc.) for end repair and
single-nucleotide A (adenine) addition. After being attached to
adapters and following agarose gel electrophoresis analysis, they
were selected as suitable templates for polymerase chain reaction
(PCR) amplification using OneTaq® DNA polymerase (New
England BioLabs, Inc.). The thermocycling conditions were as
follows: 95°C for 30 sec, followed by 30 cycles of denaturation at
95°C for 5 sec, annealing at 60°C for 30 sec, and extension at 72°C
for 30 sec. Subsequently, the Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc., Santa Clara, CA, USA) and the ABI StepOnePlus
Real-Time PCR System (Agilent Technologies, Inc.) were used for
quantification and quality evaluation of the established sample
library and were designated as the quality control steps. Finally,
the library was sequenced on a HiSeq™ 2000 instrument
(Illumina, Inc., San Diego, CA, USA). The National Center for
Biotechnology Information (NCBI) database (www.ncbi.nlm.nih.gov/genome/gdv/browser/?context=genome&acc=GCF_000001405.38)
was used to search sequence assignment of lncRNA-n346372.
Reverse transcription-quantitative PCR
(RT-qPCR)
Following extraction from bladder cancer tissues and
corresponding normal tissues with the TRIzol reagent (Thermo Fisher
Scientific, Inc.), total RNA was reverse-transcribed to cDNA using
the PrimeScript RT Reagent kit (Takara Bio, Inc., Otsu, Japan) at
37°C for 15 min and 95°C for 5 sec, followed by storage at 4°C.
RT-qPCR was conducted using SYBR® Premix Ex
Taq™ (Takara Bio, Inc.) with an Applied Biosystems Step
One Plus (Agilent Technologies, Inc.). The thermocycling parameters
were as follows: 95°C for 30 sec, followed by 30 cycles of
denaturation at 95°C for 5 sec, annealing at 60°C for 30 sec, and
extension at 72°C for 30 sec. A total of 20 µl PCR reaction mixture
was prepared for incubation in a 96-well optical plate and melting
curve analysis was carried out to assess the specificity of the PCR
products. The sequences of primers were as follows: n346372 Forward
primer, 5′-ATGTGATGGTGAGTAGGAG-3′, and reverse primer,
5′-GAAGCAGTGTCATTTAGAGCA-3′; β-actin forward primer,
5′-GAGCTACGAGCTGCCTGACG-3′, and reverse primer,
5′-CCTAGAAGCATTTGCGGTGG-3′. The relative expression level of
lncRNA-n346372 was calculated by the 2−ΔΔCq method
(19) with β-actin as an
endogenous gene for normalization.
Fluorescence in situ hybridization
(FISH)
A Ribo™ lncRNA FISH kit (cat. no. C10910) purchased
from Guangzhou RiboBio Co., Ltd. (Guangzhou, China) was employed
for RNA FISH to identify the location and expression of
lncRNA-n346372 in T24 bladder cancer cells and patients tissue
samples, according to the manufacturer's protocol. In brief, cells
were cultured (and 4-mm-thick tissue slices were prepared), and
fixed in 4% paraformaldehyde for 10 min at room temperature. Cells
and tissues were permeabilized with Triton-100 for 5 min at 4°C
(Beyotime Institute of Biotechnology, Shanghai, China), and
Cy3-labelled n346372 and DAPI-labelled 18S-RNA probes (Guangzhou
RiboBio Co., Ltd.) were detected. In the dark, DNA was stained with
DAPI for 10 min at room temperature, followed by washing in PBS
three times every 5 min. Slides were mounted and examined by
confocal fluorescence microscopy (×200; Olympus Corporation, Tokyo,
Japan).
Statistical analysis
The data were presented as the mean ± standard
deviation of at least three independent experiments. All
statistical analyses were conducted using the SPSS software,
version 22.0 (IBM Corp., Armonk, NY, USA). Continuous variables
were analyzed by the Student's t test and categorical variables
were subjected to the χ2 test. The overall survival
rates were calculated by the Kaplan-Meier method with the log-rank
test to assess the level of significance between two the survival
curves. Univariate and multivariate Cox regression models were
employed to investigate independent predictive factors of overall
survival. Heat maps were generated with MeV 4.8 software
(mev.tm4.org). P<0.05 was considered to indicate a
statistically significant difference.
Results
Numerous lncRNAs are significantly
upregulated in bladder cancer tissues compared with matched
adjacent normal tissues
According to the results of the high-throughput
sequencing applied to 10 pairs of bladder cancer tissues and
matched adjacent non-cancerous tissues, with P<0.05, a fold
change value >2, and false discovery rate <0.05 as the
screening strategy, a total of 169 lncRNAs were identified in
accordance with the above criteria. These 169 lncRNAs were
demonstrated to exhibit significantly differential expression
between bladder cancer tissues and matched adjacent normal tissues,
including 32 upregulated lncRNAs exhibited in a heat map
(P<0.05; Fig. 1) and 137
downregulated lncRNAs. Notably, in the upregulated group, eight
lncRNAs with the fold change >3.0 were identified; in descending
order they were n346372, n337945, n341831, n344768, n377154,
n387149, n385823 and n373941.
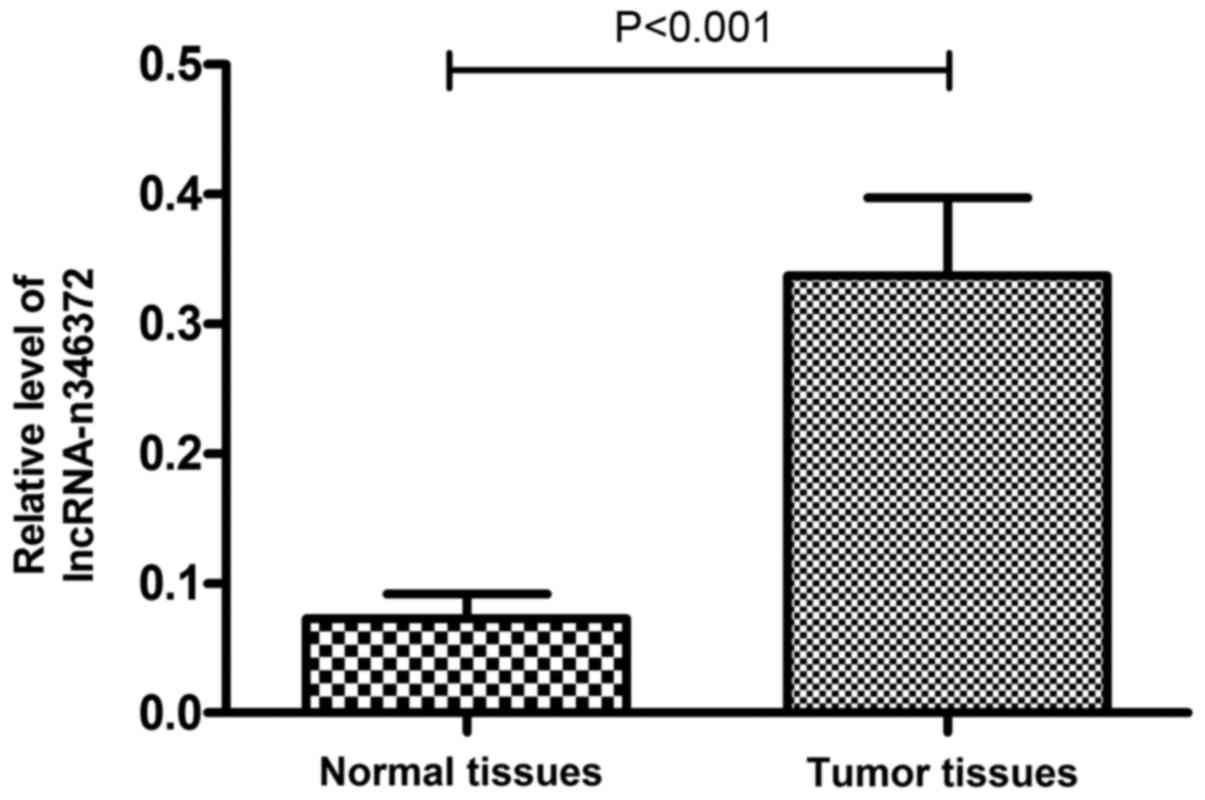
Relative expression level of n346372
increases in bladder cancer tissues compared with the matched
adjacent normal tissues
The following verification was performed on 60 pairs
of tissue samples and the results of the RT-qPCR experiment
demonstrated that the relative expression level of n346372 was
significantly increased in bladder cancer tissues compared with
matched adjacent normal tissues; this result was consistent with
the sequencing data (P<0.001; Fig.
2). Furthermore, following the RNA FISH assay of T24 bladder
cancer cells and 10 pairs of tissue samples, it was demonstrated
that upregulated n346372 was located in the cytoplasm (Fig. 3), and the fluorescent signal of
n346372 in bladder cancer tissues exceeded that in matched adjacent
normal tissues (Fig. 4), providing
further evidence of the differential expression trend between
bladder cancer tissues and corresponding normal tissues.
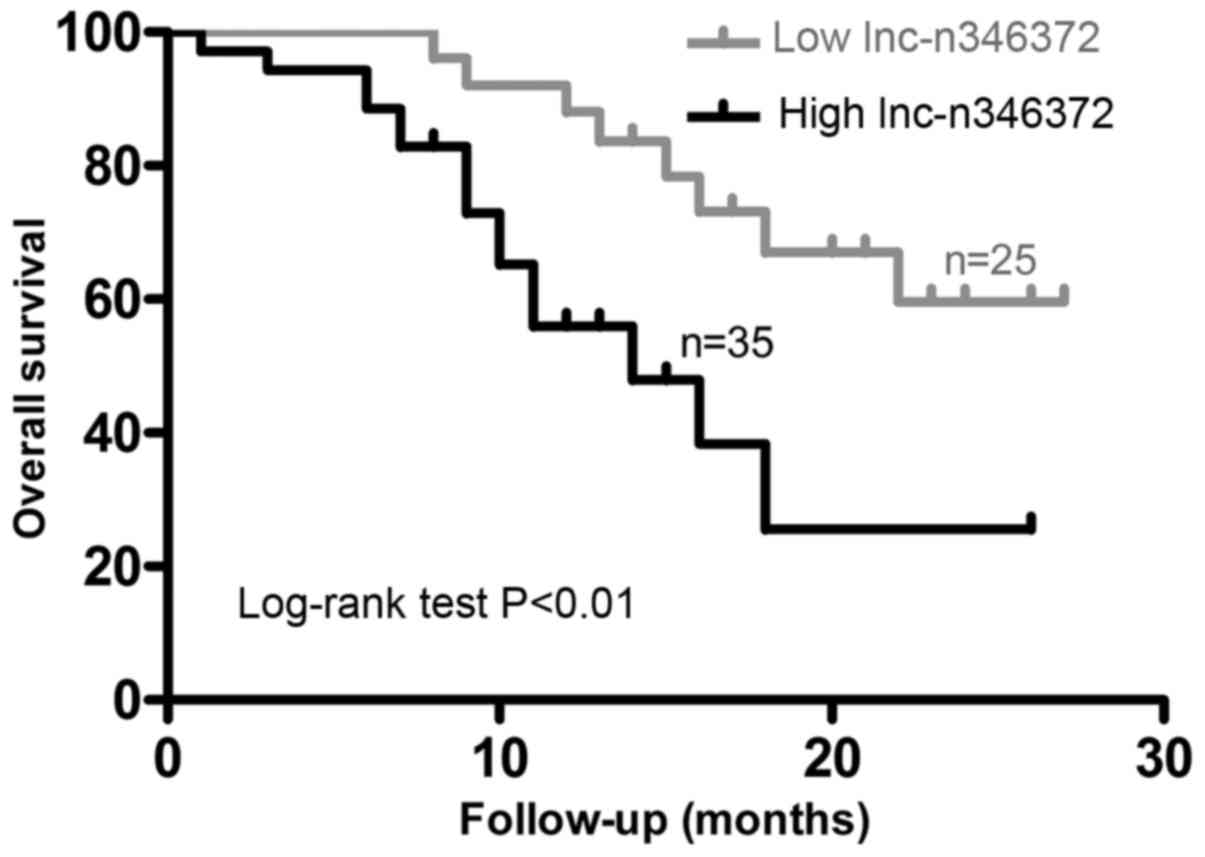
Overexpression of n346372 in bladder
cancer tissues is associated with a poor prognosis and can serve as
an independent prognostic factor of overall survival
Initially, the median level of tumor n346372 in all
enrolled patients was selected as a cut-off level and on this
basis, the study population was divided into two groups:
Low-expression (n=25) and high-expression (n=35). First, it was
demonstrated that ~68.9% of patients with the diagnosis of MIBC
exhibited high n346372 expression in the tumor, whereas only 26.7%
of patients with NMIBC exhibited such a high level in the tumor.
Similarly, 74.4% of patients with poorly differentiated tumor
tissue overexpressed n346372, while only 17.6% of patients with a
low histological grade of the tumor overexpressed n346372.
Following analysis of the relative level of n346372 and certain
common clinical variables, the overexpression of n346372 in bladder
cancer was identified to be positively associated with advanced
tumor stage and higher histological grade (P<0.05; Table I). Furthermore, following
examination of the overall survival rate with the Kaplan-Meier
method used in terms of the relative level of n346372, the results
demonstrated that the overall survival in the low-expression group
was significantly improved compared with the high-expression group
(P<0.05; Fig. 5). Univariate
and multivariate Cox regression analyses were performed to
determine the prognostic significance of n346372 expression levels,
and the univariate analysis indicated that the n346372 expression,
tumor stage, and tumor grade were significantly associated with the
overall survival of bladder cancer patients (P<0.01; Table II), whereas the multivariate
analysis demonstrated that apart from tumor stage and histological
grade, the n346372 expression level was also an independent
prognostic factor of bladder cancer (P<0.05; Table II).
 | Table II.Univariate and multivariate Cox
regression analysis for prognostic factors predicting overall
survival of patients with bladder cancer. |
Table II.
Univariate and multivariate Cox
regression analysis for prognostic factors predicting overall
survival of patients with bladder cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Covariant | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| n346372
expression | 5.563 | 1.992–15.536 | 0.001 | 3.729 | 1.127–12.346 | <0.001 |
| Age (year) | 0.998 | 0.369–2.696 | 0.997 |
|
|
|
| Sex | 1.682 | 0.570–4.961 | 0.346 |
|
|
|
| Smoking
history | 0.938 | 0.369–2.384 | 0.893 |
|
|
|
| Tumor size
(cm) | 1.658 | 0.388–7.091 | 0.495 |
|
|
|
| Tumor number | 1.507 | 0.511–4.443 | 0.457 |
|
|
|
| Tumor stage | 16.420 | 2.148–125.526 | 0.007 | 8.338 | 1.066–65.201 | 0.043 |
| Tumor grade | 13.816 | 1.854–102.945 | 0.010 | 8.187 | 1.043–64.245 | 0.045 |
Discussion
Accumulating evidence has confirmed that despite
initially being regarded as spurious transcriptional noise, lncRNAs
exert strong regulatory action on diverse biological processes,
particularly cellular development and metabolism. LncRNAs can also
be expressed abnormally in a variety of solid tumors as well as
serving as ideal molecular biomarkers for the diagnosis and
prediction of the prognosis of a number of different types of
cancer (20–25). Additionally, a handful of
dysregulated lncRNAs identified in previous studies serve important
roles in bladder cancer pathogenesis and may increase diagnostic
efficacy and prognostic accuracy (26–31).
For example, protein sprouty homolog 4-intronic transcript 1, HOX
antisense intergenic RNA and metastasis-associated lung
adenocarcinoma transcript 1 have been demonstrated to be
substantially upregulated in bladder cancer, and further
investigation indicates that these upregulated lncRNAs in bladder
cancer are closely associated with an advanced pathological stage,
recurrence of the Ta/T1 stage and poor survival, respectively.
Furthermore, a small interfering RNA-mediated knockdown of these
genes significantly inhibits the biological functions of bladder
cancer cells, including their proliferation, invasiveness and
migration capability (32–33). In contrast, several lncRNAs
downregulated in bladder cancer have also been reported, for
example, MEG3, a gene encoding a lncRNA, which is expressed in
normal tissues while its expression is lacking in a number of cell
lines as well as in bladder cancer; additionally, the
downregulation of lncRNA-MEG3 can activate autophagy and increase
cell proliferation and is negatively associated with bladder cancer
formation (10,12,34).
In the present study, significant overexpression of
lncRNA-n346372 was first identified in bladder cancer tissues
compared with paired adjacent normal tissues; the gene locus of
this lncRNA is located on chromosome 22 (chr22: 16189344-16192955)
with a mature transcript of 228 bp in length. The relative level of
n346372 was validated by RT-qPCR in 60 pairs of bladder cancer
tissues in addition to the results of FISH in tissue samples,
suggesting that the overexpression of n346372 in bladder cancer may
be involved in certain pathways contributing to tumorigenesis
and/or cancer progression. Nonetheless, sequence assignment based
on the NCBI database revealed that this region contains no
protein-coding genes and the specific regulatory function and a
possible target gene(s) remain unclear; therefore, future studies
at the cellular and molecular levels need to be conducted to
investigate the detailed molecular mechanism of action of
n346372.
Notably, following the analysis of the expression
level of n346372 and clinical variables, the results demonstrated
that 68.9% of the patients with an advanced tumor stage (T2-T4) are
more likely to exhibit upregulation of n346372 in the tumor, and
the two parameters exhibit a positive association, indicating that
n346372 may promote the process of muscle invasion in bladder
cancer. Additionally, 74.4% of patients with a high histological
grade tended to highly express this lncRNA in the tumor and a
similar association between upregulated n346372 and poor
differentiation was identified, suggesting that the overexpression
of n346372 may contribute to tumor progression and poor prognosis
of patients with bladder cancer. To prove this hypothesis, a
comparison of overall survival between the two groups was conducted
and the results revealed that patients in the high-expression group
are more likely to have a poor prognosis. Finally, multivariate
analysis indicated that n346372 expression correlates with the
prognosis of bladder cancer as well as the tumor stage and
histological grade described in a series of guidelines. These data
demonstrated that n346372 may serve as an independent prognostic
factor of bladder cancer. There are certain limitations to the
present study. Independent verification of the results of the
present study is not guaranteed because of the small sample size
used. A larger number of samples need to be subsequently analyzed
for validation. In addition, it is necessary to perform independent
studies to confirm the prognostic value of n346372 in bladder
cancer.
In conclusion, to the best of the authors' knowledge
this is the first study describing differential expression of
n346372 in bladder cancer tissues compared with paired adjacent
normal tissues. A positive association of n346372 overexpression
with advanced tumor stage and poor differentiation was also noted.
Furthermore, bladder cancer patients with upregulated n346372 are
more likely to have an unfavorable prognosis in contrast with
patients who exhibit downregulation and consequently, n346372 was
demonstrated in the present study to hold promise as an independent
prognostic factor of bladder cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Nature Science Foundation of China (grant no. 8150101083).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AL, ZZ and CX designed the study. AL performed the
experiments. ZZ drafted this manuscript. CX critically revised the
manuscript. WX coordinated the experiments, interpreted the data
and prepared the manuscript. SQ performed the statistical analysis.
MH collected and analyzed the clinical samples. SZ designed the
RNA-FISH and high-throughput transcriptome sequencing
experiments.
Ethics approval and consent to
participate
The present study's protocol was approved by the
Ethics Committee of Changhai Hospital of the Second Military
Medical University.
Patient consent for publication
All of the patients provided written informed
consent.
Competing interests
The authors declare they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
FISH
|
fluorescence in situ
hybridization
|
|
lncRNA
|
long non-coding RNA
|
|
MIBC
|
muscle-invasive bladder cancer
|
|
NMIBC
|
non-muscle-invasive bladder cancer
|
|
RT-qPCR
|
reverse-transcription quantitative
polymerase chain reaction
|
References
|
1
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Witjes JA, Compérat E, Cowan NC, De Santis
M, Gakis G, Lebret T, Ribal MJ, Van der Heijden AG and Sherif A:
European Association of Urology: EAU guidelines on muscle-invasive
and metastatic bladder cancer: Summary of the 2013 guidelines. Eur
Urol. 65:778–792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen J, Wang L, Tang Y, Gong G, Liu L,
Chen M, Chen Z, Cui Y, Li C, Cheng X, et al: Maspin enhances
cisplatin chemosensitivity in bladder cancer T24 and 5637 cells and
correlates with prognosis of muscle-invasive bladder cancer
patients receiving cisplatin based neoadjuvant chemotherapy. J Exp
Clin Cancer Res. 35:22016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Powles T, Eder JP, Fine GD, Braiteh FS,
Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, et
al: MPDL3280A (anti-PD-L1) treatment leads to clinical activity in
metastatic bladder cancer. Nature. 515:558–562. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alfred WJ, Lebret T, Compérat EM, Cowan
NC, De Santis M, Bruins HM, Hernández V, Espinós EL, Dunn J,
Rouanne M, et al: Updated 2016 EAU guidelines on muscle-invasive
and metastatic bladder cancer. Eur Urol. 71:462–475. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mohammed AA, El-Tanni H, El-Khatib HM,
Mirza AA, Mirza AA and Alturaifi TH: Urinary bladder cancer:
biomarkers and target therapy, new era for more attention. Oncol
Rev. 10:3202016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen T, Xie W, Xie L, Sun Y, Zhang Y, Shen
Z, Sha N, Xu H, Wu Z, Hu H and Wu C: Expression of long noncoding
RNA lncRNA-n336928 is correlated with tumor stage and grade and
overall survival in bladder cancer. Biochem Biophys Res Commun.
468:666–670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Necsulea A, Soumillon M, Warnefors M,
Liechti A, Daish T, Zeller U, Baker JC, Grützner F and Kaessmann H:
The evolution of lncRNA repertoires and expression patterns in
tetrapods. Nature. 505:635–640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang M, Lu W, Huang Y, Shi J, Wu X, Zhang
X, Jiang R, Cai Z and Wu S: Downregulation of the long noncoding
RNA TUG1 inhibits the proliferation, migration, invasion and
promotes apoptosis of renal cell carcinoma. J Mol Histol.
47:421–428. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Q, Su M, Lu G and Wang J: The
complexity of bladder cancer: Long noncoding RNAs are on the stage.
Mol Cancer. 12:1012013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan J, Li X, Wu W, Xue M, Hou H, Zhai W
and Chen W: Long non-coding RNA UCA1 promotes cisplatin/gemcitabine
resistance through CREB modulating miR-196a-5p in bladder cancer
cells. Cancer Lett. 382:64–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ying L, Huang Y, Chen H, Wang Y, Xia L,
Chen Y, Liu Y and Qiu F: Downregulated MEG3 activates autophagy and
increases cell proliferation in bladder cancer. Mol Biosyst.
9:407–411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hua Q, Lv X, Gu X, Chen Y, Chu H, Du M,
Gong W, Wang M and Zhang Z: Genetic variants in lncRNA H19 are
associated with the risk of bladder cancer in a Chinese population.
Mutagenesis. 31:531–538. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang C, Li X, Wang Y, Zhao L and Chen W:
Long non-coding RNA UCA1 regulated cell cycle distribution via CREB
through PI3-K dependent pathway in bladder carcinoma cells. Gene.
496:8–16. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Chen W, Yang C, Wu W, Wu S, Qin X
and Li X: Long non-coding RNA UCA1a(CUDR) promotes proliferation
and tumorigenesis of bladder cancer. Int J Oncol. 41:276–284.
2012.PubMed/NCBI
|
|
16
|
Zhan Y, Lin J, Liu Y, Chen M, Chen X,
Zhuang C, Liu L, Xu W, Chen Z, He A, et al: Up-regulation of long
non-coding RNA PANDAR is associated with poor prognosis and
promotes tumorigenesis in bladder cancer. J Exp Clin Cancer Res.
35:832016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhan Y, Li Y, Guan B, Chen X, Chen Z, He
A, He S, Gong Y, Peng D, Liu Y, et al: Increased expression of long
non-coding RNA CCEPR is associated with poor prognosis and promotes
tumorigenesis in urothelial bladder carcinoma. Oncotarget.
8:44326–44334. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hayn MH, Hussain A, Mansour AM, Andrews
PE, Carpentier P, Castle E, Dasgupta P, Rimington P, Thomas R, Khan
S, et al: The learning curve of robot-assisted radical cystectomy:
Results from the International Robotic Cystectomy Consortium. Eur
Urol. 58:197–202. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Struhl K: Transcriptional noise and the
fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol.
14:103–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gupta Chandra S and Tripathi Nandan Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Harries LW: Long non-coding RNAs and human
disease. Biochem Soc Trans. 40:902–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou M, Zhao H, Wang Z, Cheng L, Yang L,
Shi H, Yang H and Sun J: Identification and validation of potential
prognostic lncRNA biomarkers for predicting survival in patients
with multiple myeloma. J Exp Clin Cancer Res. 34:1022015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Q, Su M, Lu G and Wang J: The
complexity of bladder cancer: Long noncoding RNAs are on the stage.
Mol Cancer. 12:1012013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen J, Miao Z, Xue B, Shan Y, Weng G and
Shen B: Long non-coding RNAs in urologic malignancies: Functional
roles and clinical translation. J Cancer. 28:1842–1855. 2016.
View Article : Google Scholar
|
|
28
|
Li LJ, Zhu JL, Bao WS, Chen DK, Huang WW
and Weng ZL: Long noncoding RNA GHET1 promotes the development of
bladder cancer. Int J Clin Exp Pathol. 7:7196–7205. 2014.PubMed/NCBI
|
|
29
|
Wang HM, Lu JH, Chen WY and Gu AQ:
Upregulated lncRNA-UCA1 contributes to progression of lung cancer
and is closely related to clinical diagnosis as a predictive
biomarker in plasma. Int J Clin Exp Med. 8:11824–11830.
2015.PubMed/NCBI
|
|
30
|
Zhu YP, Bian XJ, Ye DW, Yao XD, Zhang SL,
Dai B, Zhang HL and Shen YJ: Long noncoding RNA expression
signatures of bladder cancer revealed by microarray. Oncol Lett.
7:1197–1202. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao XL, Zhao ZH, Xu WC, Hou JQ and Du XY:
Increased expression of SPRY4-IT1 predicts poor prognosis and
promotes tumor growth and metastasis in bladder cancer. Int J Clin
Exp Pathol. 8:1954–1960. 2015.PubMed/NCBI
|
|
32
|
Yan TH, Lu SW, Huang YQ, Que GB, Chen JH,
Chen YP, Zhang HB, Liang XL and Jiang JH: Upregulation of the long
noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder
cancer. Tumour Biol. 35:10249–10257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|