Introduction
Hepatocellular carcinoma (HCC) is a common type of
cancer, and its incidence is increasing rapidly, causing ~745,000
mortalities in 2012, worldwide (1–4). As
a result of the low detection rate, a number of HCC patients are at
advanced stages of the disease at the time of diagnosis with
limited sensitivity biomarkers (5). Long non-coding RNAs (lncRNAs) are a
novel class of non-coding RNAs (ncRNAs) that contribute to the
development and progression of cancer and that may serve as novel
biomarkers (6). However, the
molecular and functional mechanisms of lncRNAs in HCC remain
unknown. Therefore, studies on the role of lncRNA in HCC are
urgent. LncRNAs are >200 nucleotides in length, and previous
studies have demonstrated that lncRNAs exert important effects on
diverse cellular functions through gene expression regulation,
including chromatin modification, transcription regulation and
genomic imprinting (7,8). A number of previous studies have
demonstrated that lncRNAs participate in tumor development,
including cell cycle (9),
differentiation (10), apoptosis
(11,12), migration and invasion (13). Therefore, lncRNAs may be important
therapeutic targets for diseases.
Fer-1-like family member 4 (FER1L4) is a
tumor-associated lncRNA involved in the development of tumors, and
it has been demonstrated that FER1L4 expression is
downregulated in patients with gastric cancer (14,15).
It was reported that FER1L4 inhibits proliferation,
migration and invasion of gastrointestinal cancer (16,17).
A recent study also revealed that FER1L4 inhibits
proliferation and the cell cycle through phosphatase and tensin
homolog (PTEN) in endometrial carcinoma (18). Therefore, the interactions between
lncRNAs and protein-coding genes are popular topics in cancer
biology that provide an important theoretical basis for the
diagnosis and treatment of cancers. In the present study, the
biological mechanism of FER1L4 for proliferation in
regulating PTEN were demonstrated in HCC.
The tumor suppressor gene PTEN is located at
chromosome 10q23.31 and is frequently inactivated in cancer
(19–22). A recent study also demonstrated
that PTEN is closely linked to HCC tumorigenesis (23). As previously noted, downregulated
FER1L4 in endometrial carcinoma (EC) tissues was positively
correlated with decreased PTEN expression by using RT-qPCR assay;
FER1L4 inhibited cell proliferation, promoted cell cycle arrest at
G0/G1 phase and cell apoptosis by
upregulating PTEN expression with MTT, colony-formation and flow
cytometry detection (18). Based
on the previous study that FER1L4 serves a potential role in
regulating PTEN expression (18), it was hypothesized that
FER1L4 may also function as a PTEN regulator in
HCC.
Materials and methods
Clinical specimens
HCC cancer tissues and adjacent normal tissues (a
distance of at least 5 cm from the tumor) were collected from 35
patients with HCC at the Center Hospital of Cangzhou (Cangzhou,
China) between March 2014 and October 2015. These HCC cases without
other clinicopathological characteristics included 19 male patients
and 16 female patients with a mean age of 67.2 years (age, 36–84
years). All patients had not received preoperative treatment, such
as radiotherapy or chemotherapy. The inclusion/exclusion criteria
of HCC patients were as follows: i) All enrolled patients were
diagnosed with HCC, ii) VEGF evaluation, iii) correlation of VEGF
with OS or DFS, iv) all patients were carefully evaluated to
identify duplicate patient populations, and v) criteria used to
judge duplicate populations, such as treatment information, study
period and admission hospital. The present study was approved by
the ethics committee of the Center Hospital of Cangzhou, and
written informed consent was obtained from all patients prior to
enrolment in the study. Histological diagnosis of HCC was evaluated
according to the World Health Organization criteria. All collected
tissues were immediately stored at −80°C until use.
Cell culture
The normal hepatocyte LO2 cell line was purchased
from American Type Culture Collection (Manassas, VA, USA). The HCC
cell lines Hep3B and Huh7, and 293T cells were obtained from the
Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). LO2, Hep3B, Huh7 and 293T cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; High glucose; Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS; Sigma Aldrich; Merck KGaA, Darmstadt,
Germany) and 100 U/ml penicillin/streptomycin (Invitrogen, Thermo
Fisher Scientific, Inc.). All cells were cultured in a 37°C, 5%
CO2 cell culture incubator.
Lentiviral vector construction,
production and transfection
To obtain an FER1L4 expression vector, a
full-length FER1L4 DNA fragment was amplified by polymerase
chain reaction (PCR) using the Takara Ex Taq Hot Start
Version kit (Takara Bio, Inc., Otsu, Japan). The PCR reaction is
presented in Table I and the
reaction steps were as follows: 94°C for 5 min; 94°C for 45 sec,
60°C for 45 sec, 72°C for 1 min, for a total of 30 cycles and 72°C
for 10 min.
 | Table I.Reaction components used in the
RT-qPCR. |
Table I.
Reaction components used in the
RT-qPCR.
| Reagent | Consumption |
|---|
| Premix Ex Taq Hot
Start Version | 25.0 µl |
| PCR Forward
Primer | 1.0 µM |
| PCR Reverse
Primer | 1.0 µM |
| Template | 0.2 µl |
|
ddH2O | up to 50 µl |
The primer sequences of FER1L4 were: Forward,
5′-GATTCAGGTGGGCGGGCTGGTG-3′ and reverse,
5′-TCAGTGGCTGTGATAGGTTTA-3′. The PCR products were inserted into
the mammalian expression vector pCDNA3.1 (Invitrogen; Thermo Fisher
Scientific, Inc.). A lentiviral vector expressing Enhanced Green
Fluorescent Protein (enhanced green fluorescent protein; EGFP, Gene
Bank Accession, no. U57607) was used as an empty vector
(pCDNA3.1-expressing EGFP) was selected as a control. 293T cells
(1×106 cells/well) were seeded in 10 cm plates and
co-transfected with lentiviral packaging vectors [pMD2.G (Addgene;
cat. no. 12259), pMDL-G/P-RRE (Addgene; cat. no. 12251) and
pRSV-REV (Addgene; cat. no. 12253)] and either the constructed
pCDNA3.1-FER1L4 overexpression vector or the Control vector for 48
h. Lentiviral particles were harvested, purified and transfected
into cells (1×104 cells/well) with 8 µg/ml polybrene
(Sigma-Aldrich; Merck KGaA) in 24-well plates. Finally, the cells
with stable expression were screened using 800 µg/ml G418
(Sigma-Aldrich; Merck KGaA).
Small interfering (si)RNA
transfection
siRNA sequences were designed to target the human
FER1L4 gene and siRNAs were purchased from GenePharma
(Shanghai GenePharma Co., Ltd., Shanghai, China). The sequences of
siRNA-FER1L4 were 5′-CAGGACAGCUUCGAGUUAATT-3′ (sense) and
5′-UUAACUCGAAGCUGUCCUGTT-3′ (antisense). The sequences of the
negative control (NC) siRNA were 5′-UUCUCCGAACGUGUCACGU-3′ (sense),
5′-ACGUGACACGUUCGGAGAA-3′ (antisense). Cells (2×105
cells/well) were seeded in 6-well plates and transfected with
FER1L4 siRNAs (50 nM) or NC siRNAs (50 nM) for 48 h by using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
scientific, Inc.) according to the manufacturer's protocol.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from 35 HCC tissues or
treated cells (1×106 cells) by using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Total RNA was reverse transcribed to cDNA
using a cDNA synthesis kit (Takara Bio, Inc., Otsu, Japan)
according to the manufacturer's protocol. RT-qPCR was performed
using the standard SYBR Green PCR Master Mix kit (Takara Bio, Inc.)
on an ABI 7500 Real-Time PCR System (Applied Biosystems, Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocols. The reaction steps were as follows: Initial denaturation
at 95°C for 30 sec; followed by 40 cycles of 95°C for 5 sec and
60°C for 34 sec. Relative expression was determined based on the
2−ΔΔCq method (24).
The following primer sequences were used: FER1L4, forward
5′-CCGTGTTGGGTGCTGTTC-3′, reverse 5′-GGCAAGTCCACTGTCAGATG-3′;
PTEN, forward 5′-GTTTACCGGCAGCATCAAAT-3′, reverse
5′-CCCCCACTTTAGTGCACAGT-3′; GAPDH, forward
5′-CCTCGTCTCATAGACAAGATGGT-3′, reverse
5′-GGGTAGAGTCATACTGGAACATG-3′; GAPDH was used as an internal
control.
Western blot assay
Protein expression levels of PTEN were measured by
western blot assay. Briefly, the treated cells (1×106
cells) were lysed in Radioimmunoprecipitation Assay buffer
(Beyotime Institute of Biotechnology, Shanghai, China) containing a
protease inhibitor cocktail (Roche Diagnostics, Basel,
Switzerland). Protein concentrations were measured using a
Bicinchoninic Acid Protein Assay kit (Thermo Fisher Scientific,
Inc.). Equal concentrations of protein (30 µg) were separated by
10% SDS-PAGE and transferred onto a polyvinylidene difluoride
membrane (EMD Millipore, Billerica, MA, USA). All the membranes
were blocked with 5% skim milk (Shanghai solarbio Bioscience &
Technology Co., LTD, Shanghai, China.) for 1.5 h at room
temperature and incubated overnight at 4°C with primary antibodies
against PTEN (1:500; Abcam, Cambridge, UK, cat. no. ab31392) and
beta-actin (1:3,000; Sigma-Aldrich; Merck KGaA; cat. no. A2066)).
Subsequently, membranes were incubated with horseradish
peroxidase-conjugated goat anti-mouse (1:5,000; cat. no. CW102) and
goat anti-rabbit (1:5,000; cat. no. CW103) secondary antibody for 1
h at room temperature. The results were analyzed using an Enhanced
Chemiluminescence Detection kit (Amersham Biosciences Inc.,
Piscataway, NJ, USA) an enhanced chemiluminescence system (Amersham
Biosciences Inc.). The results were analyzed by using Image-Pro
Plus 6.0 software (National Institutes of Health, Bethesda, MD,
USA).
MTT assay
The MTT assay was used to measure the cell viability
by using MTT reagent (Cat. number M2128) according to the
manufacturer's protocol. Briefly, the treated cells (4,000
cells/well) were seeded in 96-well plates and incubated at 37°C for
0, 12, 24 and 72 h. Subsequently, 20 µl MTT (5 mg/ml) solution was
added to each well at the prescribed time and the cells were
incubated for an additional 4 h at 37°C. Dimethyl sulfoxide was
used to dissolve the formazan crystals and the absorbance was
detected at 570 nm under the micro-plate reader (BioTek
Instruments, Inc., Winooski, VT, USA).
Colony-formation assay
A colony formation assay was also performed to
measure the cell viability. Transfected cells (5×103
cells/plate) were seeded onto 6-cm culture dishes and were
maintained in the DMEM (High glucose; Invitrogen; Thermo Fisher
Scientific, Inc.) including 10% FBS for 2 weeks; the medium was
replaced every 3 days. Colonies were stained with Coomassie blue
(cat: B-2025, Sigma-Aldrich; Merck KGaA) at 37°C for 1 h, and
colony formation was calculated under the stereomicroscope by
counting the number of stained colonies.
Tumor formation in nude mice
Prior to the beginning of the experiments, specific
criteria for humane endpoints were established as previously
described, including labored breathing, reduced food or drinking
water intake, inability to stay upright, unresponsive or
unconscious to external stimuli and shaggy coat (25,26);
animal reactions were confirmed by an animal specialist. If mice
presented one or more of the above reactions, the animals were
assumed to have reached the humane endpoint. When mice met humane
endpoints, they were anesthetized with sodium pentobarbital (50
mg/kg) by intraperitoneal injection and sacrificed by cervical
dislocation immediately.
A total of 14 Female athymic BALB/c mice (age, 4–6
weeks; weight, 20–22 g) were obtained from Yangzhou University
Medical Centre (Yangzhou, China). Specific pathogen free (SPF) were
provided by the Cangzhou Central Hospital Animal Center, and they
were housed in a controlled environment (food and water available
ad libitum, 21±1°C, humidity 60%, lights on from 7:00 a.m.-7:00
p.m.). The study was approved by the ethics committee of the Cancer
Institute. All the animals were treated humanely in accordance with
the guidelines laid down by the Institutional Animal Ethics
Committee. siRNA-FER1L4- or NC-siRNA-transfected cells
(1×107 cells in 100 µl) were injected into nude mice.
Tumor volume and weights were measured and calculated at 0, 5, 10,
15, 20, 25, and 30 days; tumor volume was calculated as the volume
of a sphere: (4/3) × (π) × (r3), where radius (r) =½
diameter.
Statistical analysis
Significant differences between two groups were
analyzed by Student's t-test. FER1L4 expression was analyzed by
using one-way analysis of variance followed by a Dunnett post-hoc
test in HCC cell lines. The correlation between FER1L4 and
PTEN mRNA expression was analyzed by Pearson's correlation
coefficient. All data are presented as the mean ± standard
deviation, and analyzed by using IBM SPSS Statistics version 21
(IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate
a statistically significant difference. All experiments were
performed in triplicate.
Results
FER1L4 is expressed at low levels in
human HCC
FER1L4 mRNA expression levels were examined
in 35 pairs of HCC and paired adjacent normal tissues by RT-qPCR.
The results indicated that FER1L4 expression was decreased
in HCC tissues (n=34/35) compared with the adjacent non-cancerous
tissues (Fig. 1A). Similarly,
FER1L4 mRNA expression levels were demonstrated to be
significantly lower in Hep3B and Huh7 HCC cell lines compared with
expression in LO2 normal hepatocyte cells (P<0.001; Fig. 1B). These results suggested that
FER1L4 may function as a tumor suppressor in HCC.
FER1L4 inhibits the proliferative
ability of HCC cells
To further identify the effects of FER1L4 on
the proliferative ability of HCC cells, Hep3B cells were
transfected with lentiviral FER1L4 overexpression vectors or
siRNA-FER1L4. RT-qPCR was used to validate the efficiency of
lentiviral vector and siRNA transfections. The results demonstrated
that Hep3B cells transfected with FER1L4 overexpression
vectors expressed significantly higher levels of FER1L4
mRNA, and cells transfected with siRNA-FER1L4 expressed a
significantly lower level of FER1L4, compared with the
respective empty vector-transfected or siRNA-NC-transfected control
cells (P<0.001; Fig. 2A).
Results from MTT assays indicated that FER1L4 overexpression
significantly inhibited the proliferative ability of Hep3B cells
compared with the empty vector-transfected control cells
(P<0.001; Fig. 2B). However,
the silencing of FER1L4 expression by siRNAs significantly
promoted the proliferative ability of Hep3B cells compared with the
siRNA-NC-transfected cells (P<0.05; Fig. 2C). In addition, a colony-forming
assay was performed to examine the effects of FER1L4 on the
long-term proliferative ability of Hep3B cells. It was demonstrated
that FER1L4 over expression significantly suppressed the
long-term proliferative abilities of Hep3B cells compared with
control cells (P<0.001; Fig.
2D), whereas reduced FER1L4 expression significantly
increased the long-term proliferative abilities of Hep3B cells
compared with control cells (P<0.001; Fig. 2E).
 | Figure 2.FER1L4 inhibits the
proliferative ability of HCC cells. Hep3B HCC cells were
transfected with FER1L4-OE or with siRNA-FER1L4. (A)
Reverse transcription-quantitative polymerase chain reaction was
used to measure the mRNA expression level of FER1L4 in the
treated Hep3B cells; ***P<0.001. (B) The proliferative ability
was detected by MTT assay in Hep3B cells transfected with
FER1L4-OE or with empty vector control; ***P<0.001. (C)
The MTT assay was performed to measure the proliferation ability of
Hep3B cells transfected with siRNA-FER1L4 or siRNA-NC at 0,
12, 24, 72 h, respectively; *P<0.05. (D and E) Clonal
colony-forming experiments were performed to measure the
proliferative ability of Hep3B cells that were transfected with (D)
FER1L4-OE or the empty vector control or (E)
siRNA-FER1L4 or siRNA-NC magnification, ×100; ***P<0.001.
FER1L4, Fer-1-like family member 4; NC, negative control;
OD, optical density; OE, overexpression; siRNA, small interfering
RNA. |
Reduced FER1L4 expression promotes the
growth of HCC tumors in vivo
Based on the higher efficiency of FER1L4
siRNAs compared with the FER1L4 overexpression vector (data
not shown), FER1L4 was chosen to be silenced by siRNAs for
the in vivo study. Transfected Hep3B cells were implanted
subcutaneously into nude mice. The tumor volume was calculated at
0, 5, 10, 15, 20, 25 and 30 days; tumor volumes and longest
diameters are presented in Table
II (the values correspond to the tumors at the end of the
experiment). The results indicated that the silencing of
FER1L4 expression led to a significant increase in tumor
volume compared with siRNA-NC (P<0.001; Fig. 3A). Tumor weight was also calculated
at 30 days. The results demonstrated that the tumor weights were
2,893±345 and 1,468±321 mg in nude mice implanted with Hep3B cells
transfected with siRNA-FER1L4 and siRNA-NC at 30 days,
respectively; these data revealed that the silencing of
FER1L4 significantly decreased the tumor weight (P<0.001;
Fig. 3B).
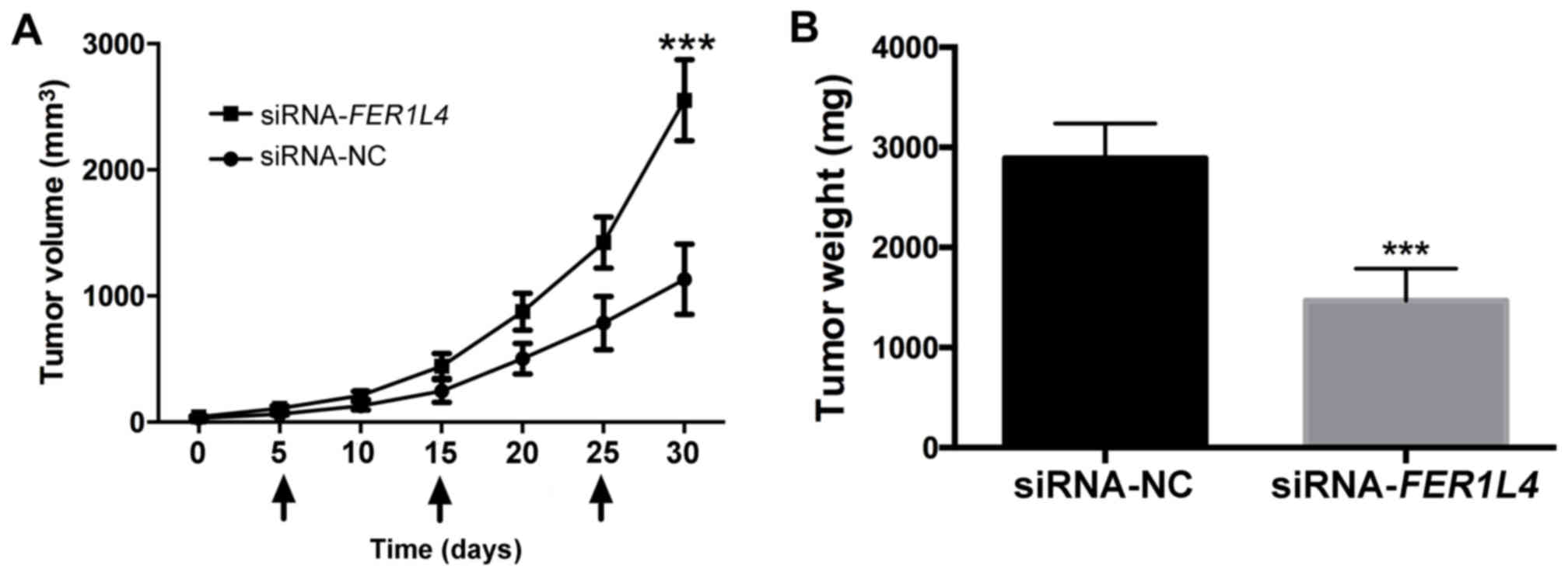 | Figure 3.Reduced FER1L4 expression
promotes the growth of HCC tumors in nude mice in vivo. (A)
Tumor volume was detected at 0, 5, 10, 15, 20, 25 and 30 days.
Arrow indicates that nude mice were injected with Hep3B cells that
were transfected with siRNA-FER1L4 or siRNA-NC at 5, 15 and
25 days. ***P<0.001. (B) When animals met humane endpoints, they
were sacrificed and tumor weights were measured; ***P<0.001.
FER1L4, Fer-1-like family member 4; HCC, hepatocellular
carcinoma; NC, negative control; siRNA, small interfering RNA. |
 | Table II.Volume and the longest diameter of
subcutaneous tumors in nude mice injected with Hep3B cells
transfected with siRNA-FER1L4 or siRNA-NC. |
Table II.
Volume and the longest diameter of
subcutaneous tumors in nude mice injected with Hep3B cells
transfected with siRNA-FER1L4 or siRNA-NC.
|
| siRNA-NC |
siRNA-FER1L4 |
|---|
|
|
|
|
|---|
| Day | Tumors (n) | Volume
(mm3) | Longest diameter
(mm) | Tumors (n) | Volume
(mm3) | Longest diameter
(mm) |
|---|
| 0 | 2 | 32.12 | 1.97 | 1 | 42.12 | 4.32 |
| 5 | 1 | 64.28 | 4.97 | 1 | 108.30 | 5.91 |
| 10 | 1 | 128.32 | 6.26 | 2 | 211.10 | 3.70 |
| 15 | 2 | 246.32 | 3.89 | 1 | 444.60 | 9.47 |
| 20 | 1 | 503.20 | 9.87 | 1 | 875.32 | 11.87 |
| 25 | 1 | 785.40 | 11.45 | 2 | 1,423.40 | 6.98 |
| 30 | 3 | 1,132.40 | 4.31 | 2 | 2,552.40 | 8.48 |
FER1L4 positively regulates PTEN
expression in HCC
To investigate the interaction between FER1L4
and PTEN, RT-qPCR and western blot assays were performed.
FER1L4 overexpression significantly upregulated the mRNA
expression level of PTEN and the silencing of FER1L4 by
siRNAs significantly downregulated the mRNA expression level of
PTEN in transfected Hep3B cells, compared with the
respective controls (both P<0.001; Fig. 4A). FER1L4 overexpression
significantly increased PTEN protein expression whereas the
silencing of FER1L4 by siRNAs significantly decreased PTEN
protein expression in transfected Hep3B cells compared with the
respective controls (both P<0.001; Fig. 4B). In addition, the mRNA expression
level of PTEN was measured in HCC tissues with either high
FER1L4 (n=21) or low FER1L4 (n=14) expression levels
(Cut off=0.435) by using SigmaPlot 10.0 (SigmaPlot Software, La
Jolla, CA, USA). The results demonstrated that high PTEN
mRNA expression may be associated with high FER1L4
expression in clinical HCC tissues (P<0.001; Fig. 4C) and the expressions were
positively correlated (R2=0.6058; P<0.01; Fig. 4D).
Discussion
Previous studies have demonstrated that a number of
ncRNAs serve important roles in human diseases by activating or
repressing genes (24,27,28).
Additional studies have demonstrated that the dysregulated
expression or the dysfunction of specific ncRNAs might drive
tumorigenesis (29). There are
three forms of ncRNAs that are especially important for the
regulation of gene expression: MicroRNAs (miRNAs), lncRNAs and
circular RNAs (30), of which
miRNAs have gained much attention and have been considered to serve
important roles in cancer through the transcriptional regulation of
specific target mRNAs (31).
lncRNAs are one of the emerging fields in ncRNA research that have
been reported to participate in various biological processes,
including transcription, mRNA splicing, RNA decay and translation
(32–34). Previous studies indicated that
lncRNAs may be associated with the development and progression of
human cancer. For example, one study demonstrated that
FER1L4 serves as a potential biomarker of gastric cancer
with lymph node invasion (35).
FER1L4 was reported to inhibit proliferation in endometrial
carcinoma (18) and FER1L4
was expressed at a low level in gastric cancer patients (15). Another study demonstrated that
FER1L4 inhibits cancer cell growth by regulating PTEN
expression (17). In the preset
study, FER1L4 was revealed to be expressed at a low level in
human HCC tissues. FER1L4 was demonstrated to inhibit the
proliferative ability of HCC cells in vitro, and silencing
of FER1L4 promoted the proliferative ability of HCC in
vivo. Therefore, the results of the present study strongly
supported the potential tumor suppressor role of FER1L4 in
cancer. Additionally, it may serve as a target for HCC therapy.
A number of previous studies have demonstrated that
PTEN serves crucial roles in the progression of tumors, including
cell proliferation, differentiation, migration and apoptosis
(36–38). Additional studies suggested that
PTEN is associated with miRNA (miR)-21 and affects the
prognosis of colorectal cancer (39), whereas PTEN may inhibit the
epithelial-mesenchymal transition and invasive ability of tongue
cancer cells (40). In addition,
low expression of PTEN is also considered to be a potential
biomarker for resistance to human epidermal growth factor receptor
2-targeted therapy in advanced gastric cancer (41). Recent work has demonstrated that
there is connection between lncRNA and PTEN by the
lncRNA-GAS5/PTEN/miR-103 axis in endometrial cancer cells (42). In addition, a previous study
reported that the FER1L4/PTEN axis has significant effect in
regulating the function of endometrial carcinoma (18). In the present study, it was
demonstrated that PTEN was expressed at high levels in HCC
tissues with high FER1L4 expression. Additionally, both
overexpression and knockdown of FER1L4 may be able to
regulate PTEN expression in HCC. Therefore, the results of
the present study indicated that FER1L4 may also act as a
PTEN regulator in HCC, inhibiting the proliferative ability
of HCC cells.
In conclusion, the present study results may be
summarized as follows: i) FER1L4 was expressed at a low
level in human HCC tissues; ii) FER1L4 inhibited the
proliferative ability of HCC cells; iii) PTEN was positively
associated with FER1L4 expression in HCC; and iv)
FER1L4 silencing by siRNAs promoted the growth of HCC tumors
in vivo. Therefore, FER1L4 may be a potential
therapeutic target for HCC.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
XS and GQZ contributed to study design, the majority
of the experiments and data collection. CYL and CDL contributed to
data analysis and drafted the manuscript. GQZ and CYL were
responsible for figure preparation, improving the manuscript and
helping to conceive the study. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of the Center Hospital of Cangzhou, (Cangzhou, China).
Written informed consent was obtained from all patients prior to
enrolment in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Li Z, Zhang C, Lou C, Yan F, Mao Y, Hong X
and Zhang Y: Comparison of percutaneous cryosurgery and surgical
resection for the treatment of small hepatocellular carcinoma.
Oncol Lett. 6:239–245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Asia-Pacific Working Party on Prevention
of Hepatocellular Carcinoma: Prevention of hepatocellular carcinoma
in the Asia-Pacific region: Consensus statements. J Gastroenterol
Hepatol. 25:657–663. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: Incidence, mortality, survival, and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Conti F, Dall'Agata M, Gramenzi A and
Biselli M: Biomarkers for the early diagnosis of bacterial
infection and the surveillance of hepatocellular carcinoma in
cirrhosis. Biomark Med. 9:1343–1351. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang
G and Zhang Y: lncRNA GAS(5) enhances G1 cell cycle arrest via
binding to YBX1 to regulate p21 expression in stomach cancer. Sci
Rep. 5:101592015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu X, Li D, Zhang W, Guo M and Zhan Q:
Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6
mRNA decay. EMBO J. 31:4415–4427. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lakhotia SC: Long non-coding RNAs
coordinate cellular responses to stress. Wiley Interdiscip Rev RNA.
3:779–796. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Paralkar VR and Weiss MJ: A new ‘Linc’
between noncoding RNAs and blood development. Genes Dev.
25:2555–2558. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dhamija S and Diederichs S: From junk to
master regulators of invasion: lncRNA functions in migration, EMT
and metastasis. Int J Cancer. 139:269–280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song H, Sun W, Ye G, Ding X, Liu Z, Zhang
S, Xia T, Xiao B, Xi Y and Guo J: Long non-coding RNA expression
profile in human gastric cancer and its clinical significances. J
Transl Med. 11:2252013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Z, Shao YF, Tan L, Shi HJ, Chen SC and
Guo JM: Clinical significance of the low expression of FER1L4 in
gastric cancer patients. Tumor Biol. 35:9613–9617. 2014. View Article : Google Scholar
|
|
16
|
Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F
and Yan D: Long non-coding RNA Fer-1-like protein 4 suppresses
oncogenesis and exhibits prognostic value by associating with
miR-106a-5p in colon cancer. Cancer Sci. 106:1323–1332. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xia T, Chen SC, Jiang Z, Shao Y, Jiang X,
Li P, Xiao B and Guo J: Long noncoding RNA FER1L4 suppresses cancer
cell growth by acting as a competing endogenous RNA and regulating
PTEN expression. Sci Rep. 5:134452015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiao Q and Li H: LncRNA FER1L4 suppresses
cancer cell proliferation and cycle by regulating PTEN expression
in endometrial carcinoma. Biochem Biophys Res Commun. 478:507–512.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Malaney P and Dave V: Loss of PTEN
cooperates with mutant KRAS initiating EMT and increased stemness
in a mouse model of lung cancer. Mol Cancer Res. 12:B092014.
View Article : Google Scholar
|
|
20
|
Takashi K, Naozumi H, Kazuyoshi I, Koji S,
Daisuke A, Tomomi O and Yoshinori H: Gene modulation of
phosphorylation sites in tumor suppressor PTEN inhibits
hypoxia-induced phenotype changes through epithelial-mesenchymal
transition (EMT) in lung cancer. Am J Respirat Crit Care Med.
183:A35032011.
|
|
21
|
Cho BC, Kim SM, Hong YK and Kim H:
Increased PTEN instability-mediated Akt activation confers acquired
resistance to cetuximab and increased migration/invasion potentials
in non-small cell lung cancer. Clin Exp Metastas. 28:230. 2011.
|
|
22
|
Sun Y, Tian H and Wang L: Effects of PTEN
on the proliferation and apoptosis of colorectal cancer cells via
the phosphoinositol-3-kinase/Akt pathway. Oncol Rep. 33:1828–1836.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang D, Zhou PH, Wang W, Wang X, Li J,
Sun X and Zhang L: MicroRNA-616 promotes the migration, invasion
and epithelial-mesenchymal transition of HCC by targeting PTEN.
Oncol Rep. 35:366–374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lindl T, Gross U, Ruhdel I, von Aulock S
and Völkel M: Guidance on determining indispensability and
balancing potential benefits of animal experiments with costs to
the animals with specific consideration of EU directive 2010/63/EU.
ALTEX. 29:219–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ray MA, Johnston NA, Verhulst S, Trammell
RA and Toth LA: Identification of markers for imminent death in
mice used in longevity and aging research. J Am Assoc Lab Anim Sci.
49:282–288. 2010.PubMed/NCBI
|
|
27
|
Broeckx BJG, Hitte C, Coopman F, Verhoeven
GE, De Keulenaer S, De Meester E, Derrien T, Alfoldi J,
Lindblad-Toh K, Bosmans T, et al: Improved canine exome designs,
featuring ncRNAs and increased coverage of protein coding genes.
Sci Rep. 5:128102015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qu ZP and Adelson DL: Identification and
comparative analysis of ncRNAs in human, mouse and zebrafish
indicate a conserved role in regulation of genes expressed in
brain. PLoS One. 7:e522752012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ferro M, Altieri V, Montanaro V and
Cimmino A: Ultraconserved region (Ucrs) encoding ncrnas involvement
in bladder cancer tumorigenesis: A new(P)layer in the ‘dark
matter’. Anticancer Res. 33:22662013.
|
|
30
|
Hayes EL and Lewis-Wambi JS: Mechanisms of
endocrine resistance in breast cancer: An overview of the proposed
roles of noncoding RNA. Breast Cancer Res. 17:402015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Louro R, Smirnova AS and Verjovski-Almeida
S: Long intronic noncoding RNA transcription: Expression noise or
expression choice? Genomics. 93:291–298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang M, Huang J, He J, Yi Q, Zhang Z, Tu
L, Ma D and Yan J: FER1L4: A potential plasma biomarker to identify
gastric cancer with lymph node invasion. Int J Clin Exp Pathol.
9:1982–1988. 2016.
|
|
36
|
Lotan TL, Carvalho FLF, Peskoe SB, Hicks
JL, Good J, Fedor H, Humphreys E, Han M, Platz EA, Squire JA, et
al: PTEN loss is associated with upgrading of prostate cancer from
biopsy to radical prostatectomy. Mod Pathol. 28:128–137. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dawson H, Koelzer V, Karamitopoulou E,
Lugli A and Zlobec I: The role of the tumor suppressor PTEN in
colorectal cancer is highly dependent on the tumor area. Lab
Invest. 95:156a2015.
|
|
38
|
Keniry M and Parsons R: The role of PTEN
signaling perturbations in cancer and in targeted therapy.
Oncogene. 27:5477–5485. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yazdani Y, Farazmandfar T, Azadeh H and
Zekavatian Z: The prognostic effect of PTEN expression status in
colorectal cancer development and evaluation of factors affecting
it: miR-21 and promoter methylation. J Biomed Sci. 23:92016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xie SM, Lu ZY, Lin YZ, Shen LJ and Yin C:
Upregulation of PTEN suppresses invasion in Tca8113 tongue cancer
cells through repression of epithelial-mesenchymal transition
(EMT). Tumor Biol. 37:6681–6689. 2016. View Article : Google Scholar
|
|
41
|
Zhang X, Park JS, Park KH, Kim KH, Jung M,
Chung HC, Rha SY and Kim HS: PTEN deficiency as a predictive
biomarker of resistance to HER2-targeted therapy in advanced
gastric cancer. Oncolo. 88:76–85. 2015. View Article : Google Scholar
|
|
42
|
Guo C, Song WQ, Sun P, Jin L and Dai HY:
LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in
endometrial cancer cells. J Biomed Sci. 22:1002015. View Article : Google Scholar : PubMed/NCBI
|


















