Introduction
MicroRNAs (miRNAs) are small, non-coding RNAs
consisting of 19–24 nucleotides and are important in the negative
regulation of gene expression (1,2).
Alteration of miRNAs has been observed in various types of cancer
and may be involved in modulating cancer cell behaviors (2–4).
Numerous studies have shown that miRNAs are important regulators in
the diverse biological processes of cancer, including cell
proliferation, apoptosis, angiogenesis, differentiation, adhesion
and metastasis (5–8). These data emphasize the importance of
miRNAs in cancer development and provide novel insight into
understanding the molecular mechanism of tumorigenesis.
Endometrial cancer is the most common gynecologic
malignancy worldwide, and the incidence and the associated
mortality rates of this disease have increased over the last decade
(9,10). The development of endometrial
cancer is a multistep process with the accumulation of genetic and
epigenetic alterations, including those of miRNAs in, for example,
miR-205, miR-141, miR-200, miR-30c, miR-101, miR-449, miR-143 and
miR-145 (11–15). However, the exact role of miR-409
in the carcinogenesis of endometrial cancer remains to be
elucidated.
Therefore, in the present study, the expression of
miR-409 was investigated in human endometrial cancer tissues and
paired adjacent normal pancreatic tissues, and the effects of
miR-409 on cell growth and cell cycle progression were examined
in vitro. Small mothers against decapentaplegic 2 (Smad2)
was identified as a direct target of miR-409, which was confirmed
using a luciferase reporter system. These data indicated that
miR-409 directly targeted Smad2 and negatively regulated cell
proliferation and cell cycle progression of Ishikawa and HEC-1B
cells. These results suggested that miR-409 may function as a tumor
suppressor in the pathogenesis of human endometrial cancer.
Materials and methods
Patient tissue specimens
From Jan, 2016 to Jun, 2016, a total of 16 pairs of
tissue samples from 16 patients with endometrial cancer were used
in the present study, each of which consisted of human endometrial
cancer tissue and matched adjacent normal tissue from the same
patient. The matched normal tissue samples were obtained from the
distal end of the surgical excisions, distant from the tumor. The
samples were obtained from the Department of Gynecology, Maternal
and Child Healthcare Hospital (Huaian, China). All the patients
were females, the average age was 58.82±8.21-years-old. No patients
had suffered from previous tumors. The average cancer tissue size
was 4.23±1.08 mm3, and the average weight was 1.37±0.24
g. In addition, the average adjacent normal tissue size was
4.44±1.02 mm3, and the average weight was 1.45±0.28 g.
The present study was approved by the ethics committee of the
Maternal and Child Healthcare Hospital.
Cell culture and transfection
The Ishikawa, HEC-1A and HEC-1B human endometrial
cancer cell lines were purchased from America Type Culture
Collection (Manassas, VA, USA). The cells were maintained in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) in a
humidified incubator at 37°C with 5% CO2. The
transfection was performed using the Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
RNA was extracted from patient tissue specimens or
cells 48 h following transfection using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Small RNA (5 µg) was reverse transcribed into cDNA (2 µg)
using M-MLV reverse transcriptase (Promega Corporation, Madison,
WI, USA) with the specific primers (0.5 µg). The cDNA was used as
template to amplify either mature miR-409 or an endogenous control
U6 snRNA using real time PCR kit (Takara Bio, Inc., Otsu, Japan).
The reaction mixture was prepared: SYBR Premix Ex Taq II 10 µl,
forward primer 0.8 µl, reverse primer 0.8 µl, cDNA template 2 µl,
and dH2O 6.4 µl. The primer sequences used were as
follows: miR-424-forward: 5′-CAGCAGCAATTCATGT-3′, miR-424-reverse:
5′-TGGTGTCGTGGAGTCG-3′; Smad2-forward:
5′-CAGGACGGTTAGATGAGCTTGAGA-3′, Smad2-reverse:
5′-CCCACTGATCTACCGTATTTGCTG-3′; β-actin-forward:
5′-TCTGGCAACGGTGAAGGTGACA-3′, β-actin-reverse:
5′-CACCTCCCCTGTGTGGACTT-3′; U6-forward: 5′-CTCGCTTCGGCAGCACA-3′,
U6-reverse: 5′-AACGCTTCACGAATTTGCGT-3′; miR-409-forward:
5′-TATATCCAGCTGGGTGCTAATTTGCCG-3′, universal primer
5′-TGGTGTCGTGGATAC-3′. The qPCR was performed as follows: 94°C for
3 min, followed by 40 cycles of 94°C for 30 sec, 50°C for 30 sec
and 72°C for 30 sec. The RT-qPCR analysis was performed using SYBR
Premix Ex Taq (Takara Bio, Inc.) on the iQ5 Real-Time PCR detection
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
relative expression levels of miR-409 and Smad2 were defined as
follows: Quantity of miR-409/quantity of U6 within the same sample;
quantity of Smad2/quantity of β-actin within the same sample. The
2−ΔΔCq method was used to analyses the relative gene
expression (16).
Assessment of cell viability and
proliferative capacity
To determine the viability and proliferative
capacity of the cells, the cells were examined using 3-(4,
5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) and
colony formation assays, as described previously (17). Following transfection, Ishikawa and
HEC-1B cells were seeded in 96-well plates at a density of 8,000
cells per well. At different time points following transient
transfection, the cells were incubated with 10 µl MTT at a final
concentration of 0.5 mg/ml at 37°C for another 4 h. The medium was
then removed, and the precipitated formazan was dissolved in 100 µl
DMSO. Following shaking for 20 min, the absorbance at 570 nm (A570)
was detected using a uQuant Universal microplate spectrophotometer
(Bio-Tek Instruments, Inc., Winooski, VT, USA). For the colony
formation assay, the numbers of viable cell colonies were
determined 15 days following the inoculation of 150 cells/well in
triplicate into 12-well plates. The cells were stained with 0.1%
crystal violet at room temperature for 20 min. The plates were
observed under a FastScan atomic force microscope (Bruker AXS,
Bruker Corporation Santa-Barbara, CA, USA). The rate of colony
formation was calculated using the following equation: Colony
formation rate=(number of colonies/number of seeded cells)
×100%.
Cell apoptosis and cell cycle
analyses
The apoptotic ratios of the cells were determined
using the Annexin V-7-ADD apoptosis detection kit (Roche
Diagnostics, Basel, Switzerland). Briefly, 48 h following
transfection, the cells were collected and washed twice with cold
PBS buffer, resuspended in 200 µl of binding buffer, and incubated
with 20 µl of Annexin-V-R-PE for in an ice bath for 20 min in the
dark. This was followed by the addition of 10 µl 7-AAD prior to
analyzing using flow cytometry. Cells treated with DMSO were used
as a negative control. Following transfection for 48 h, the cells
were collected and fixed with 70% ethanol, stained with propidium
iodide, and analyzed by flow cytometry. The data were analyzed
using CellQuest Pro software 5.1 (BD Biosciences, Franklin Lakes,
NJ, USA). The experiments were repeated at least three times.
Western blot analysis
Total cellular extracts were extracted using
radioimmunoprecipitation assay buffer (2 µl). Adjust the density of
relative cells to 1×105/ml and inoculated with a
six-well plate. Following cell adherence to the wells, cells were
collected after 48 h. Cells were lysed with RIPA buffer; proteins
were then separated. A Bicinchoninic Acid assay was used to
determine the relative protein concentrations. Aliquotes of
proteins (50 µg in 25 µl) were separated by 10% SDS-PAGE
electrophoresis for 1.5 h under 110 V at 4°C. Polyvinylidene
difluoride membranes were blocked with 5% non-fat dry milk in
Tris-buffered saline with 0.1% Tween-20 (TBST) for 10 min at room
temperature prior to the transfer of proteins. The membrane was
incubated with rabbit anti-human phospho-Smad2 antibody (ab53100,
1:1,000, Abcam, Cambridge, UK) or rabbit anti-human GAPDH antibody
(ab9485, 1:1,000, Abcam) overnight at 4°C, rinsed three times with
TBST for 5 min, and incubated with horseradish peroxidase-labeled
goat anti-rabbit IgG (ab205718, 1:5,000, Abcam) at 37°C for 2 h.
The membrane was soaked in an enhanced chemiluminescence solution
(Sigma-Aldrich; Merck KGaA) in the darkroom for color development.
The grey value was analyzed using Image J 1.48 (National Institutes
of Health, Bethesda, MD, USA).
Target prediction and luciferase
reporter assays
Based on bioinformatics predictions using the DNA
Intelligent Analysis 2.0 (DIANA) (http://diana.imis.athena-innovation.gr/DianaTools/index.php),
miR Database (miRDB) 5.0 (http://www.mirdb.org/mirdb/policy.html), TargetScan
7.1, (http://www.targetscan.org/vert_71/) and miRwalk
databases 6.0 (http://129.206.7.150/), Smad2 was
selected as a candidate target of miR-409. The 3′untranslated
region (UTR) segments of Smad2 containing putative binding sites
for miR-409 were obtained by PCR and inserted into the pmirGLO
vector (20 µg, Promega Corporation). The wild-type reporter
construct pmirGLO/Smad2-3′UTR and the mutant reporter construct
pmirGLO/Smad2-3′UTR mut, in which the site of perfect
complementarity to miR-409 was mutated (CAA CAU U to CAA GGA A)
using site-directed mutagenesis PCR as follows: PCR-SD reactant was
mixed in a sterile centrifuge tube on ice, 2.5 U Taq DNA polymerase
and 2.5 U Taq Extender PCR Additive were added. Following PCR
amplification (denaturation at 94°C for 2 min, followed by 12
cycles of 94°C, 20 sec; 51°C 30 sec; and 72°C, 3 min, followed by
72°C for 10 min.), digestion and purification of the PCR-SDM
products, the reactant was placed on the ice for 2 min, and the 25
µl of amplification products were mixed with 1 µl Dpn I restriction
endonuclease (10 U/µl) and 1 µl of Pfu DNA polymerase (2.5 U/µl)
was centrifuged at 15,000 × g, 4°C for 1 min. Then the reactant was
incubated at 37°C for 30 min; 100 µl of H2O, 10 µl of
SDM buffer and 5 µl of ATP (10 mmol/l) were then added into the
reactant. The reactants were centrifuged at 15,000 × g, 4°C for 1
min. T4 DNA ligase (4 U/µl) was added and then the reactants was
placed at 37°C for 1 h. The heat-shocked cells were thawed lightly
on the ice and then 40 µl of cells were removed into a pre-cooled
FALCOL 2059 polypropylene tube. Ligase-treated DNA (1 µl) was then
added to the cells then incubated on ice for 30 min. Subsequently,
the reactants were placed at 42°C for 30 sec and at ice for 2 min.
The competent cells were immediately placed on the LB agar plate,
which was then incubated overnight at 37°C. The plasmid DNA was
extracted via alkaline lysis, and DNA was quantified with a UV
spectrophotometer and identified via digestion by the restriction
enzymes DpnI and HindIII. Sequence identification was
performed by Genscript Biotech (Nanjing, China). For the luciferase
reporter experiments, HEC-1B cells (2×105/ml) were
co-transfected with miR-409 mimics or miR-409 control in a 48-well
plate followed by the pmirGLO/Smad2-3′UTR reporter vector or the
pmirGLO/Smad2-3′UTR mut. Firefly luciferase and Renilla luciferase
levels were measured 48 h following transfection. Each experiment
was repeated at least three times.
Statistical analysis
All the data in the present study was analyzed by
SPSS software 22.0 (IBM Corp., Armonk, NY, USA). Data are expressed
as the mean ± standard deviation. P≤0.05 was considered to indicate
a statistically significant difference using the
Students-Newman-Keuls test.
Results
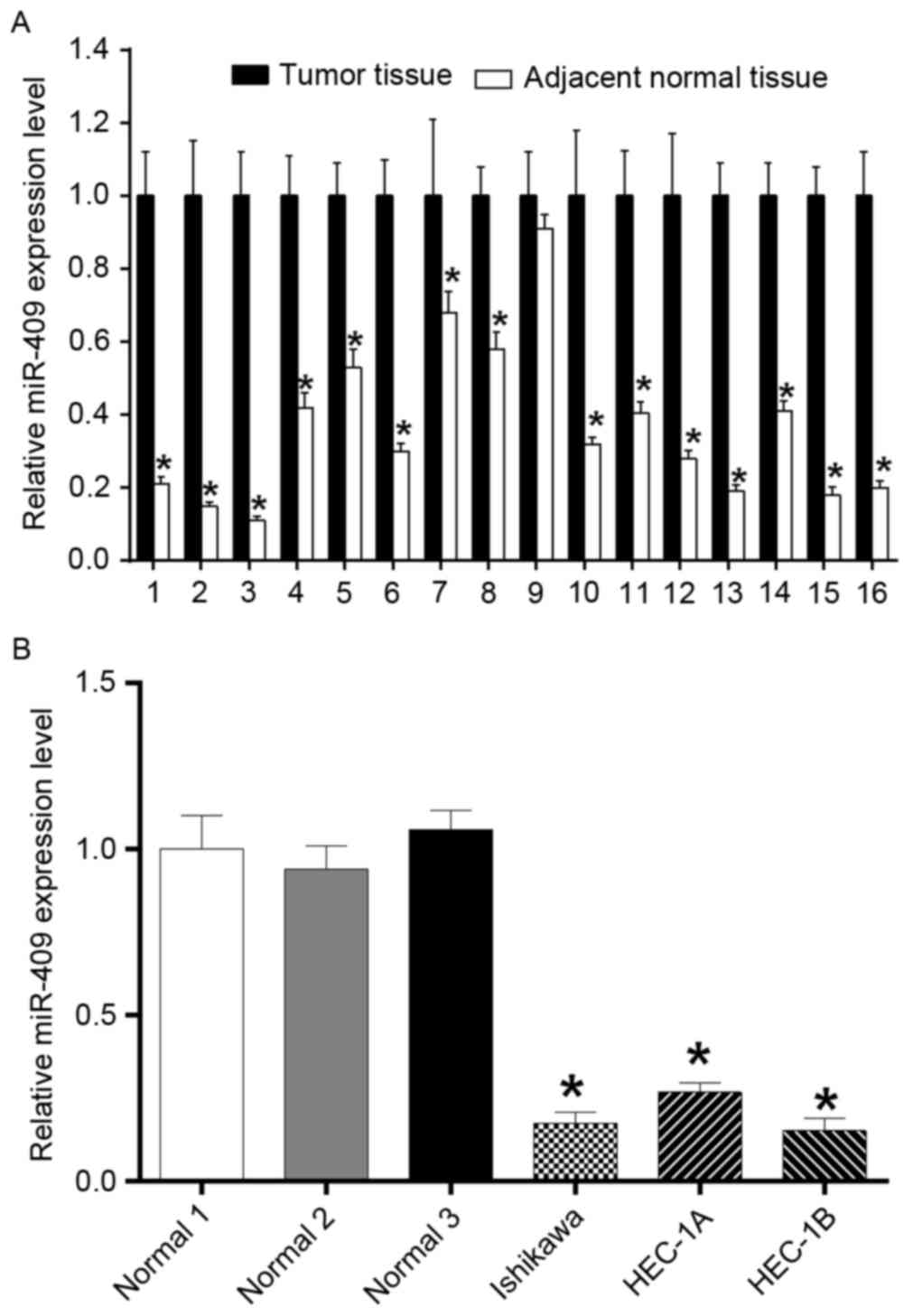
Expression of miR-409 is decreased in
endometrial cancer tissues and cell lines
To examine the role of miR-409 in the development of
endometrial cancer, the present study measured the expression of
miR-409 in 16 paired endometrial cancer samples using RT-qPCR
analysis (Fig. 1A), which showed
significantly reduced expression levels in the tumor tissues,
compared with the adjacent normal tissues. The expression of
miR-409 was significantly downregulated in the endometrial cancer
cell lines (Ishikawa, HEC-1A and HEC-1B), compared with that in the
adjacent normal tissues, also determined using RT-qPCR analysis
(Fig. 1B).
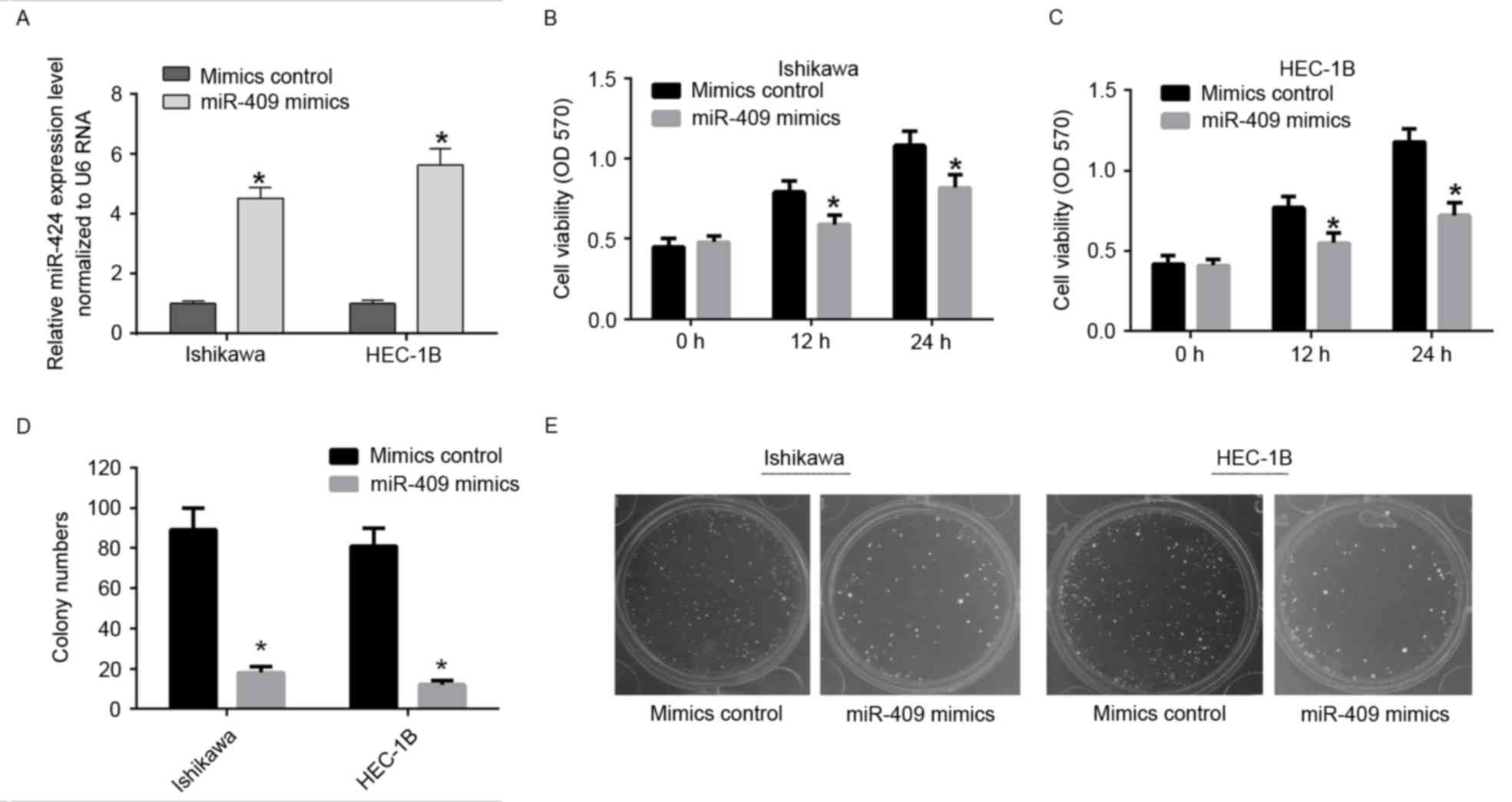
miR-409 suppresses the proliferation
of endometrial cancer cell lines
To determine the role of miR-409 in tumor cell
proliferation, the miR-409 mimics were used to induce the ectopic
expression of miR-409 in Ishikawa and HEC-1B cells (Fig. 2A). Using an MTT assay, the
overexpression of miR-409 was shown to suppress cell viability in
the Ishikawa and HEC-1B cells (Fig. 2B
and C). The colony formation rates of the Ishikawa and HEC-1B
cells transfected with miR-409 mimics, were significantly lower,
compared with those in the control group (Fig. 2D and E). These results indicated
that miR-409 suppressed the ability of Ishikawa and HEC-1B cells to
proliferate.
miR-409 initiates endometrial cancer
cell line S phase arrest and promotes apoptosis
Flow cytometry revealed that the percentages of
Ishikawa and HEC-1B cells in the of S phase of the cell cycle were
markedly higher in the miR-409 mimic-transfected groups, compared
with those in the control, suggesting that miR-409 initiated S
phase arrest (Fig. 3A). The
fluorescence-activated cell sorting analysis revealed that the
forced expression of miR-409 led to endometrial cancer cell
apoptosis. In the Ishikawa and HEC-1B cells, the percentage of
apoptotic cells was significantly increased in response to the
overexpression of miR-409, compared with that in the mimic control
(Fig. 3B). These data demonstrated
that miR-409 may inhibit proliferation by inducing S phase arrest
and promoting apoptosis in endometrial cancer.
miR-409 directly targets transcription
factor Smad2
Based on the miR-409-induced suppression of
proliferation in the endometrial cancer cells, the present study
hypothesized that miR-409 inhibited the malignancy of endometrial
cancer cells by regulating oncogenes and/or genes involved in cell
proliferation or apoptosis. Therefore, five bioinformatics
algorithms (DIANA, miRDB, TargetScan, RNA22 and miRwalk) were used
to identify potential target genes of miR-409 (Fig. 4A). As a result, Smad2, a signal
transducer and transcriptional modulator mediating multiple
signaling pathways, was predicated to have a putative miR-409
binding site within its 3′UTR (Fig.
4A) and was selected for further evaluation. The binding sites
between miR-409 and Smad2 were also conserved among species
(Fig. 4B). To confirm that miR-409
directly targeted Smad2, luciferase reporter assays were performed
to examine whether miR-409 interacts directly with its target
Smad2. A series of 3′UTR fragments were constructed, including the
wild-type Smad2 3′UTR and a binding site mutant. These fragments
were then inserted into the pmirGLO luciferase reporter plasmid. In
the Ishikawa cells, cotransfection with miR-409 and the wild-type
Smad2 3′UTR caused a significant decrease in luciferase activity,
compared with that in the control. However, the cotransfection with
mutant Smad2 3′UTR and miR-409 mimics did not alter the luciferase
intensity (Fig. 4C). The
overexpression of miR-409 reduced the mRNA and protein expression
levels of Smad2 in the Ishikawa and HEC-1B cells (Fig. 4D and E). Taken together, these
results suggested that miR-409 binds directly to the 3′UTR of
Smad2, repressing gene expression.
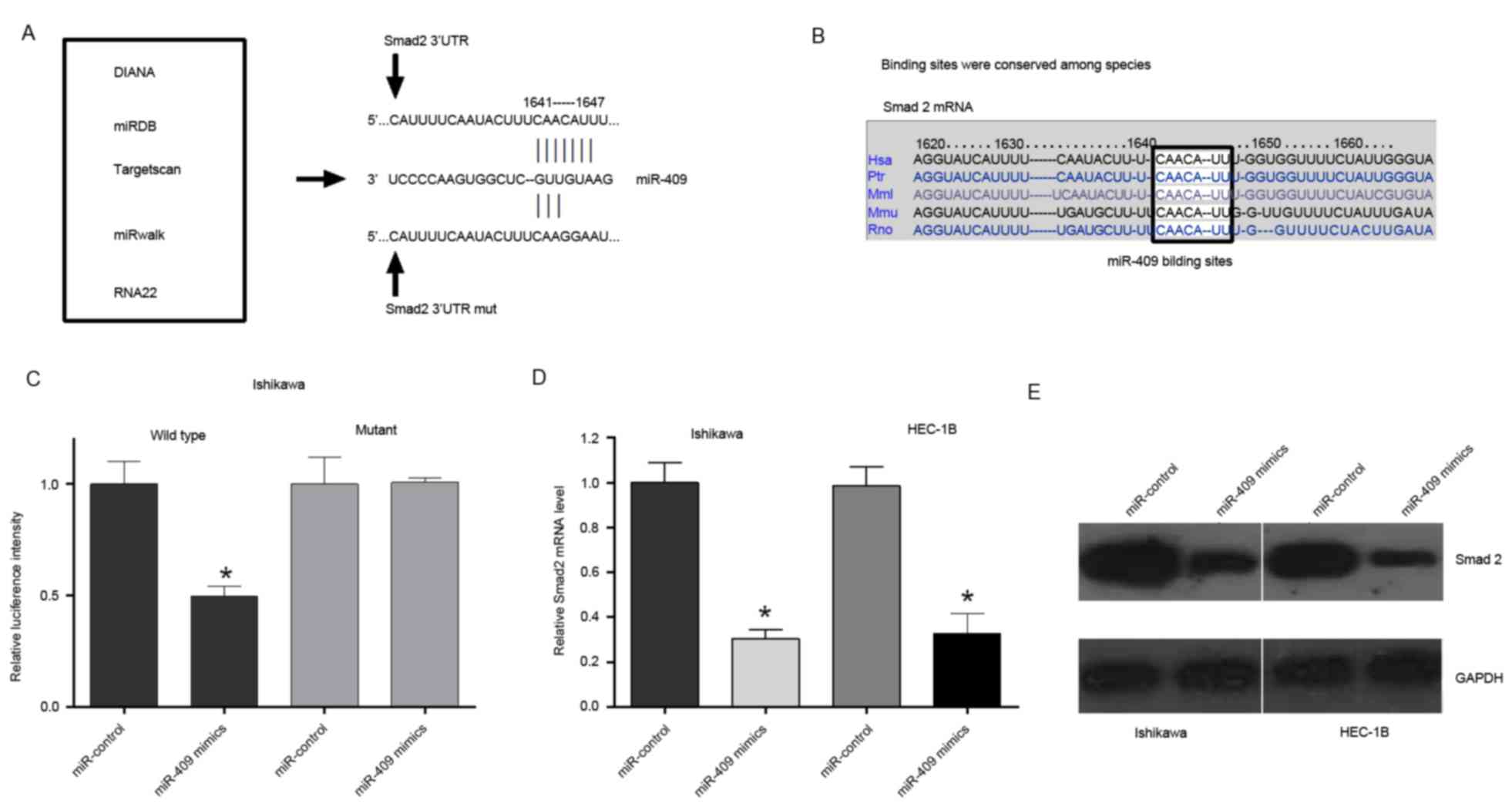 | Figure 4.miR-409 directly targets Smad2. (A)
Potential target genes of miR-409 were identified using
bioinformatics analyses (DIANA, miRDB, TargetScan, RNA22 and
miRwalk), revealing Smad2 as a potential target of miR-409.
Wild-type and mutant Smad2 3′UTRs were constructed. (B) Binding
sites between miR-409 and the Smad2 3′UTR were conserved among
species. (C) Ishikawa cells were transfected with wild-type
Smad2-3′UTR or Smad2-3′UTR-mut of the luciferase-Smad2 3′-UTR
reporter vector, miR-409 mimics and mimic control. The miR-409
mimics reduced the luciferase intensity from the luciferase-Smad2
3′-UTR reporter vector, whereas Smad2-3′UTR-mut did not alter
luciferase intensity. (D) Reverse transcription-quantitative
polymerase chain reaction analysis indicated that the expression of
Smad2 was significantly decreased in endometrial cells transfected
with miR-409 mimics. (E) Measurement of protein expression levels
of Smad2 using western blot analysis. Protein was extracted from
Ishikawa and HEC-1B cells transfected with the miR-409 mimics or
mimic control. Endogenous protein expression levels of GAPDH were
used for normalization, and the relative Smad2 protein expression
levels are shown. Smad2, small mothers against decapentaplegic 2;
miR, microRNA; UTR, untranslated region; mut, mutant; DIANA, DNA
Intelligent Analysis; miRDB, miRNA Database. *P<0.05 vs.
miR-control. |
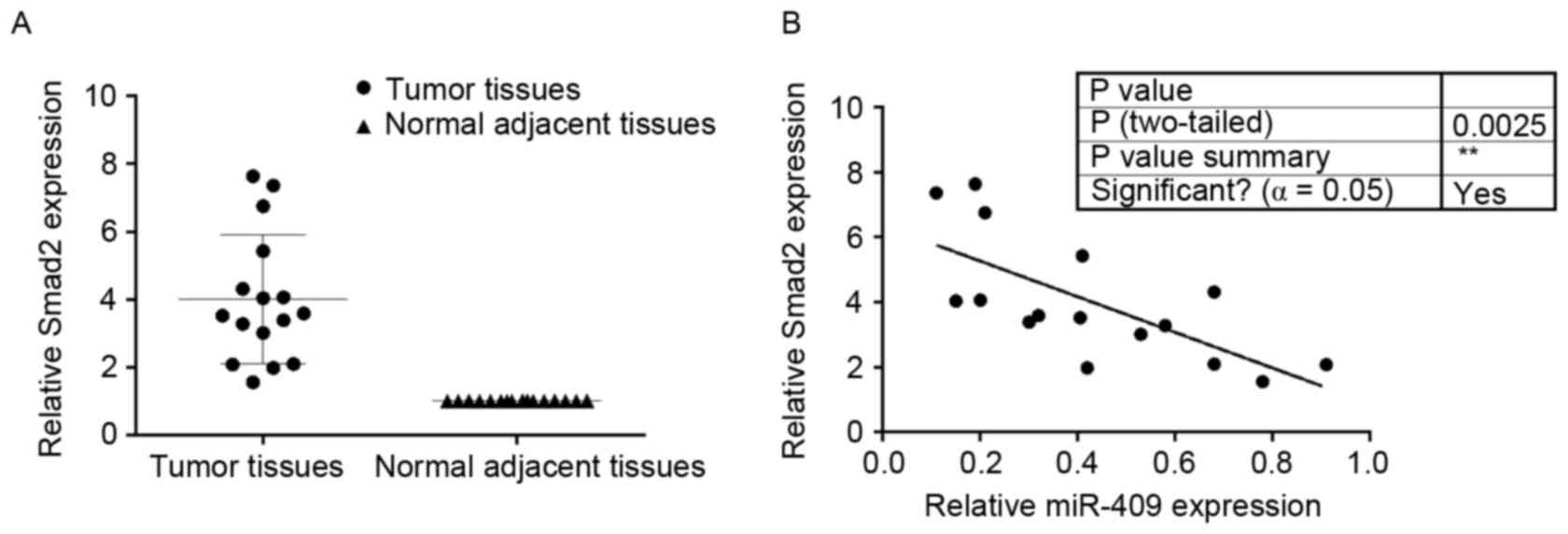
Smad2 is upregulated in endometrial
cancer tissues and is inversely correlated with miR-409
To determine the expression of Smad2 in endometrial
cancer and adjacent normal tissues, RT-qPCR analysis of Smad2 was
performed on the 16 paired endometrial cancer tissues, each
consisting of an endometrial cancer and adjacent normal tissue
specimen. In general, the expression levels of Smad2 were
significantly higher in the endometrial cancer tissues, compared
with those in the matched normal tissues (Fig. 5A). miR-409 was negatively
correlated with Smad2 in the endometrial cancer tissues (Fig. 5B).
Discussion
The elucidation of molecular and cellular mechanisms
responsible for the tumorigenesis and progression of endometrial
cancer is critical to the development of novel diagnostic and
therapeutic strategies for patients with endometrial cancer
(18,19). miRNAs are considered to be novel
candidate therapeutic agents for endometrial cancer due to their
involvement in cancer initiation and progression. For example,
miRNA-505 functions as a tumor suppressor in endometrial cancer by
targeting transforming growth factor (TGF)-α, whereas, miR-126
inhibits the migration and invasion of endometrial cancer cells by
targeting insulin receptor substrate 1. In addition, miR-490-3P may
act as a suppressor in the tumorigenesis and progression of
endometrial cancer by targeting TGF-α (16,20,21).
However, current understanding of the aberrant expression and
potential roles of miRNAs remains limited. Cohn et al
(17), reported that endometrial
cancer has a distinct miRNA profile, in which, miRNAs, including
miR-409, are significantly downregulated, compared with the miRNAs
in women without endometrial cancer.
For miR-409, it has been demonstrated that
miR-409-3p is a metastatic suppressor, and post-transcriptional
inhibition of the oncoprotein GAB1 is one of its mechanisms of
action (22). There are also
reports suggesting that miR-409-3p functions as a tumor suppressor
by inhibiting the development and metastasis of colorectal cancer,
and may become a novel diagnostic marker and target for its
treatment (23). Wan et al
(24), reported that miRNA-409-3p
functions as a tumor suppressor in human lung adenocarcinoma by
targeting c-Met. In addition, miRNA-409 suppresses tumor cell
invasion and metastasis by directly targeting radixin in gastric
cancer (25). Other studies have
indicated that stromal fibroblast-derived miR-409 promotes
epithelial-to-mesenchymal transition (EMT) and prostate
tumorigenesis, whereas miR-409-3p/-5p promotes tumorigenesis, EMT
and bone metastasis in human prostate cancer (26,27).
However, the exact mechanism underlying the effect of miR-409 in
endometrial cancer was not described. The present study found that,
compared with normal samples, miR-409 was significantly
downregulated in endometrial cancer samples, which is consistent
with the previous study. To further investigate the role of miR-409
in endometrial cancer, miR-409 mimics were used to enhance the
expression of miR-409 in Ishikawa and HEC-1B cells, and it was
found that overexpressed miR-409 suppressed the growth of Ishikawa
and HEC-1B cells and induced endometrial cancer cell apoptosis.
Smad2, which belongs to the Smad family, is similar
to the gene products of the Drosophila gene mothers against
decapentaplegic and the Caenorhabditis elegans gene Sma. It
is an important signal transducer and transcriptional modulator,
which mediates multiple signaling pathways. This protein mediates
the signal of TGF-β and regulates multiple cellular processes,
including cell proliferation, apoptosis and differentiation. A
previous study indicated that miR-212/132 functions as a tumor
suppressor by targeting SMAD2 in cervical cancer (28). Another study also revealed miR-27a
as a tumor suppressor, and identified phingosine-1-phosphate
phosphatase 1 and Smad2 as novel targets of miR-27a, linking to
Stat3 for regulating cancer cell proliferation, apoptosis and
migration in colorectal cancer (29). Yang et al (30), indicated that miR-136 may have a
tumor-suppressive effect by repressing EMT and prometastatic traits
via targeting Smad2 and Smad3. The results of the present study
demonstrated that Smad2 is a target of miR-409 in endometrial
cancer. However, as there were multiple targets of one signal
miRNA, further investigations are required to fully elucidate the
role of miR-409 in endometrial cancer.
In conclusion, the results of the present study can
be summarized as follows: i) miR-409 was downregulated in
endometrial tissues and cell lines, compared with relative normal
tissues; ii) miR-409 acted as a tumor suppressor and inhibited
endometrial cancer cell proliferation; iii) overexpression of
miR-409 led to S phase arrest of the endometrial cancer cell cycle
and induced apoptosis; iv) miR-409 directly targeted Smad2 and
showed inverse expression in endometrial cancer tissues. These
results demonstrated that miR-409 acts as a tumor suppressor in
endometrial cancer, and may serve as a potential biomarker and
novel therapeutic target for the treatment of endometrial
cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
Conception and design of the study and drafting of
the article was performed by CZ. BW performed the experiments and
analyzed data. LW made substantial contributions to the conception
and design of the present study, and conducted data analysis, and
revised the article. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of the Maternal and Child Healthcare Hospital. Written
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pillai RS, Bhattacharyya SN and Filipowicz
W: Repression of protein synthesis by miRNAs: How many mechanisms?
Trends Cell Biol. 17:118–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peters L and Meister G: Argonaute
proteins: Mediators of RNA silencing. Mol Cell. 26:611–623. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu CL, Ho JY, Chou SC and Yu DS: MiR-429
reverses epithelial-mesenchymal transition by restoring E-cadherin
expression in bladder cancer. Oncotarget. 7:26593–26603.
2016.PubMed/NCBI
|
|
4
|
Liang HQ, Wang RJ, Diao CF, Li JW, Su JL
and Zhang S: The PTTG1-targeting miRNAs miR-329, miR-300, miR-381,
and miR-655 inhibit pituitary tumor cell tumorigenesis and are
involved in a p53/PTTG1 regulation feedback loop. Oncotarget.
6:29413–29427. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Manikandan M, Deva Magendhra Rao AK,
Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R and
Munirajan AK: Oral squamous cell carcinoma: MicroRNA expression
profiling and integrative analyses for elucidation of
tumourigenesis mechanism. Mol Cancer. 15:282016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiang W, Tian Y, Jiang S, Liu S, Zhao X
and Tian D: MicroRNA-376c suppresses non-small-cell lung cancer
cell growth and invasion by targeting LRH-1-mediated Wnt signaling
pathway. Biochem Biophys Res Commun. 473:980–986. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang RJ, Li JW, Bao BH, Wu HC, Du ZH, Su
JL, Zhang MH and Liang HQ: MicroRNA-873 (miRNA-873) inhibits
glioblastoma tumorigenesis and metastasis by suppressing the
expression of IGF2BP1. J Biol Chem. 290:8938–8948. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Renjie W and Haiqian L: MiR-132, miR-15a
and miR-16 synergistically inhibit pituitary tumor cell
proliferation, invasion and migration by targeting Sox5. Cancer
Lett. 356:568–578. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jemal A, Tiwari RC, Murray T, Ghafoor A,
Samuels A, Ward E, Feuer EJ and Thun MJ; American Cancer Society, :
Cancer statistics, 2004. CA Cancer J Clin. 54:8–29. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang G, Hou X, Li Y and Zhao M: MiR-205
inhibits cell apoptosis by targeting phosphatase and tensin homolog
deleted on chromosome ten in endometrial cancer Ishikawa cells. BMC
Cancer. 14:4402014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Adila S, Zhang X, Dong Y, Li W,
Zhou M and Li T: MicroRNA expression signature profile and its
clinical significance in endometrioid carcinoma. Zhonghua Bing Li
Xue Za Zhi. 43:88–94. 2014.(In Chinese). PubMed/NCBI
|
|
13
|
Kong X, Xu X, Yan Y, Guo F, Li J, Hu Y,
Zhou H and Xun Q: Estrogen regulates the tumour suppressor
MiRNA-30c and its target gene, MTA-1, in endometrial cancer. PLoS
One. 9:e908102014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Konno Y, Dong P, Xiong Y, Suzuki F, Lu J,
Cai M, Watari H, Mitamura T, Hosaka M and Hanley SJ: MicroRNA-101
targets EZH2, MCL-1 and FOS to suppress proliferation, invasion and
stem cell-like phenotype of aggressive endometrial cancer cells.
Oncotarget. 5:6049–6062. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ye W, Xue J, Zhang Q, Li F, Zhang W, Chen
H, Huang Y and Zheng F: MiR-449a functions as a tumor suppressor in
endometrial cancer by targeting CDC25A. Oncol Rep. 32:1193–1199.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cohn DE, Fabbri M, Valeri N, Alder H,
Ivanov I, Liu CG, Croce CM and Resnick KE: Comprehensive miRNA
profiling of surgically staged endometrial cancer. Am J Obstet
Gynecol. 202(656): e651–658. 2010.
|
|
18
|
Lee II, Maniar K, Lydon JP and Kim JJ: Akt
regulates progesterone receptor B-dependent transcription and
angiogenesis in endometrial cancer cells. Oncogene. 35:5191–5201.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Choi PS, Francis JM, Imielinski
M, Watanabe H, Cherniack AD and Meyerson M: Identification of
focally amplified lineage-specific super-enhancers in human
epithelial cancers. Nat Genet. 48:176–182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao X, Zhu D, Lu C, Yan D, Li L and Chen
Z: MicroRNA-126 inhibits the migration and invasion of endometrial
cancer cells by targeting insulin receptor substrate 1. Oncol Lett.
11:1207–1212. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Devor EJ, Schickling BM, Reyes HD, Warrier
A, Lindsay B, Goodheart MJ, Santillan DA and Leslie KK: Cullin-5, a
ubiquitin ligase scaffold protein, is significantly underexpressed
in endometrial adenocarcinomas and is a target of miR-182. Oncol
Rep. 35:2461–2465. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bai R, Weng C, Dong H, Li S, Chen G and Xu
Z: MicroRNA-409-3p suppresses colorectal cancer invasion and
metastasis partly by targeting GAB1 expression. Int J Cancer.
137:2310–2322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu M, Xu A, Yuan X, Zhang Q, Fang T, Wang
W and Li C: Downregulation of microRNA-409-3p promotes
aggressiveness and metastasis in colorectal cancer: An indication
for personalized medicine. J Transl Med. 13:1952015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wan L, Zhu L, Xu J, Lu B, Yang Y, Liu F
and Wang Z: MicroRNA-409-3p functions as a tumor suppressor in
human lung adenocarcinoma by targeting c-Met. Cell Physiol Biochem.
34:1273–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng B, Liang L, Huang S, Zha R, Liu L,
Jia D, Tian Q, Wang Q, Wang C, Long Z, et al: MicroRNA-409
suppresses tumour cell invasion and metastasis by directly
targeting radixin in gastric cancers. Oncogene. 31:4509–4516. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Josson S, Gururajan M, Hu P, Shao C, Chu
GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al: miR-409-3p/-5p
promotes tumorigenesis, epithelial-to-mesenchymal transition, and
bone metastasis of human prostate cancer. Clin Cancer Res.
20:4636–4646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Josson S, Gururajan M, Sung SY, Hu P, Shao
C, Zhau HE, Liu C, Lichterman J, Duan P, Li Q, et al: Stromal
fibroblast-derived miR-409 promotes epithelial-to-mesenchymal
transition and prostate tumorigenesis. Oncogene. 34:2690–2699.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao JL, Zhang L, Guo X, Wang JH, Zhou W,
Liu M, Li X and Tang H: miR-212/132 downregulates SMAD2 expression
to suppress the G1/S phase transition of the cell cycle and the
epithelial to mesenchymal transition in cervical cancer cells.
IUBMB Life. 67:380–394. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bao Y, Chen Z, Guo Y, Feng Y, Li Z, Han W,
Wang J, Zhao W, Jiao Y, Li K, et al: Tumor suppressor microRNA-27a
in colorectal carcinogenesis and progression by targeting SGPP1 and
Smad2. PLoS One. 9:e1059912014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang Y, Liu L, Cai J, Wu J, Guan H, Zhu X,
Yuan J, Chen S and Li M: Targeting Smad2 and Smad3 by miR-136
suppresses metastasis-associated traits of lung adenocarcinoma
cells. Oncol Res. 21:345–352. 2013. View Article : Google Scholar : PubMed/NCBI
|