Introduction
The increase of cancer occurrence is a health burden
on society worldwide (1–3). More and better prediction models,
including biomarkers or more complex bioinformatics, are required
to provide early diagnosis and effective therapy for cancer.
Over the past decade, accumulating evidence has
identified that >90% of the human genome is transcribed,
whereas, <2% may be subsequently translated, which indicates
that the majority of the genome generates many thousands of
non-coding RNA (ncRNA) transcriptions (4). Benefiting from the immense technical
advances in high-throughput sequencing of transcripts (5), numerous functional ncRNAs have been
verified to direct post-transcriptional gene expression or guide
RNA modifications rather than encode proteins in human cancer
(5,6).
Long non-coding RNAs (lncRNAs), ncRNA members, have
been classically characterized as regulatory RNA molecules >200
nucleotides, without detectable open-reading frames to encode
proteins (7,8). Based on the transcript length,
lncRNAs may be further classified as long-intergenic non-coding
RNA, very long intergenic non-coding RNA, macroRNA or
promoter-associated long RNA (9).
Aberrant expressions of carcinogenic or tumour-suppressive lncRNAs
have been identified in a broad spectrum of cancer types. Homeobox
(HOX) transcript antisense RNA (HOTAIR) (10) serves as a pro-oncogenic capability
marker, whereas, X inactive specific transcript (11) serves as a tumour suppressor. In
addition, lncRNAs are involved in various biological processes,
including cell proliferation (12), migration (10), differentiation (13), immune response (14) and apoptosis (15). Furthermore, as modulators in
epigenetic processes, lncRNAs may adjust gene expression in
chromatin modification, transcription, and post-transcriptional
processing (16). In the nucleus,
lncRNAs may serve as an organisational framework involved in
interactions between proteins and between protein or DNA (10,17),
enhancing gene transcription from the enhancer regions (enhancer
RNA) (18) or their neighbouring
loci (ncRNA-a) (19). In the
cytoplasm, lncRNAs serve as a sponge to titrate proteins (12,20)
or microRNAs (miRNAs/miRs) (21).
These characteristics suggest the important roles of lncRNA
applications in the diagnostic, prognostic and therapeutic
evaluation of cancer.
The lncRNA termed colon cancer-associated
transcript-1 (CCAT1), additionally termed LOC100507056 or
cancer-associated region long non-coding RNA-5, has received
increased attention among cancer-associated lncRNAs (22,23).
Since its identification, a number of previous studies demonstrated
that CCAT1 is significantly upregulated in a number of malignancies
and serves a pivotal role in tumourigenesis; it is thus of great
value for diagnostic screening and therapy in cancer (24–26).
In the present review, the currently available
studies of the clinical importance and functional regulatory
mechanisms of lncRNA CCAT1 in various human cancer types are
discussed.
Structure characterisation of CCAT1
CCAT1 was originally identified by Nissan et
al (22) as a highly specific
biomarker upregulated in colon malignancy. The CCAT1 gene is mapped
to chromosome 8q24.21, which is described as a ‘hot spot’
containing single-nucleotide polymorphisms strongly involved in
numerous cancer types (Figs. 1 and
2) (22,27,28).
Furthermore, CCAT1 spans a region of 2,628 base pairs in length and
has two isoforms: CCAT1-S and CCAT1-L. CCAT1-L overlaps with CCAT1;
however, CCAT1-L is exclusively positioned in the nucleus, whereas,
the short isoform-CCAT1-S is cytoplasmic (23,29).
Additionally, downregulated CCAT1-L results in the simultaneous
disruption of CCAT1-S, suggesting that CCAT1-S may be developed
from CCAT1-L and that there may be a positive association between
them (23).
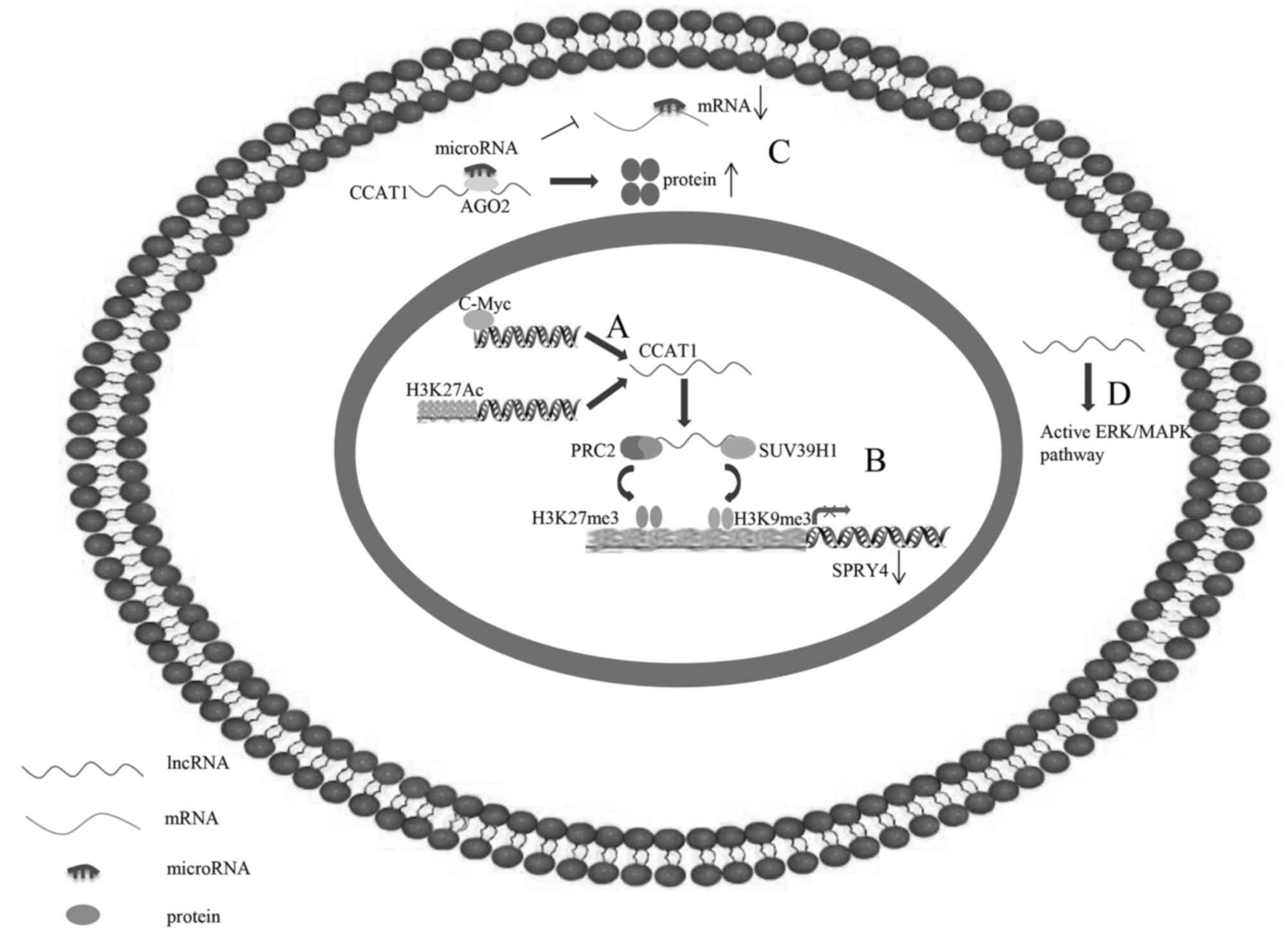 | Figure 2.Underlying regulatory mechanisms of
CCAT1 in human cancer. (A) C-Myc is able to directly bind to E-box
element in CCAT1 promoter regions to activate CCAT1 transcription.
(B) CCAT1, which may be activated by H3K27-acetylation, is able to
serve as a scaffold for PRC2 and SUV39H1, and modulate the histone
methylation of promoter of SPRY4, thereby epigenetically silencing
tumour suppressor gene SPRY4. (C) CCAT1 additionally functions as
competing endogenous RNA by sponging microRNA to free its target
mRNA for protein production. (D) CCAT1 may activate the ERK/MAPK
signalling pathway. CCAT1, colon cancer-associated transcript 1;
ERK/MAPK, extracellular signal-regulated kinase/mitogen-activated
protein kinase; lncRNA, long non-coding RNA; PRC2, polycomb
repressive complex 2; SPRY4, sprouty RTK signalling antagonist 4;
SUV39H1, suppressor of variegation 3–9 homolog 1. |
Functions and mechanisms of CCAT1
CCAT1 has been reported to be significantly
upregulated in various cancer tissues, including colorectal cancer
(CRC), lung cancer (LC), gastric cancer (GC) and hepatocellular
carcinoma (HCC), and is closely involved in proliferation, cell
cycle, apoptosis, migration, invasion, chemoresistance and
epithelial-to-mesenchymal transition (EMT) in various tumour cells
(Table I). In addition, CCAT1 is
positively associated with tumourigenesis, tumour invasion depth,
lymph node metastasis, higher tumour node metastasis (TNM) stage
and poor survival (Table II).
Mechanistically, CCAT1, activated by c-Myc, may regulate target
gene expression by binding protein to epigenetically modulate the
promoter histone methylation of target gene expression in the
nucleus (Fig. 2A and B), or
serving as competing endogenous RNA (ceRNA) to sponge microRNA
(Fig. 2C) and through involvement
in the extracellular signal-regulated kinase/mitogen-activated
protein kinase (ERK/MAPK) signaling pathway (Fig. 2D) in the cytoplasm. Notably c-Myc
activates CCAT1, which enhances c-Myc expression via let-7,
indicating there may be a feedback loop between them (30,31).
 | Table I.Functional characterization of CCAT1
in various tumours. |
Table I.
Functional characterization of CCAT1
in various tumours.
| First author,
year | Tumour type | Expression | Functional
role | Associated
gene | Protein
binding | Role | (Refs.) |
|---|
| Nissan, 2012; He,
2014; Jing, 2017; Alaiyan, 2013; Ye, 2015; Kim, 2014; Ozawa, 2017;
Zhao, 2015; Kam, 2014; McCleland, 2016 | Colorectal
cancer | Upregulated | Proliferation,
migration, invasion, EMT, cell cycle | c-Myc | / | Oncogenic | (22,30,40–46,48) |
| Zhuang, 2016;
Zhang, 2017 | LSCC | Upregulated | Proliferation, cell
cycle, migration and invasion, EMT | let-7, HMGA2,
c-Myc, mir218, ZFX | / | Oncogenic | (31,52) |
| Zhang, 2016 | Esophageal squamous
cell carcinoma | Upregulated | Proliferation,
migration | miR-7, HOXB13,
H3K27Ac, SPRY4 | EZH2, SUV39H1,
H3K27me3, H3K9me3 | Oncogenic | (55) |
| Zhang, 2014;
Mizrahi, 2015; Yang, 2013; Zhou, 2016; Liu, 2017 | Gastric cancer | Upregulated | Proliferation and
migration, invasion | c-Myc, ERK/MAPK,
miR-490, hnRNPA1 | / | Oncogenic | (56–60) |
| Deng, 2015; Wang,
2017; Zhu, 2015; Zhu, 2015; Dou, 2017; | Hepatocellular
carcinoma | Upregulated | Proliferation,
migration, invasion, cell cycle | let-7, HMGA2,
c-Myc, miR-200b, mir490-3p, CDK1 | / | Oncogenic | (25,68–72) |
| Ma, 2015 | Gallbladder
cancer | Upregulated | Proliferation,
migration, invasion | miR-218-5p,
Bmi1 | / | Oncogenic | (75) |
| Jiang, 2017; Zhang,
2017 |
Cholangiocarcinoma | Upregulated | Migration,
invasion, EMT | miR-152 | / | Oncogenic | (77,78) |
| Yu, 2016 | Pancreatic
cancer | Upregulated | proliferation, cell
cycle, and migration, EMT | c-Myc | / | Oncogenic | (81) |
| White, 2014; Luo,
2014; Cabanski, 2015; Hu, 2017; Lu, 2016; Lu, 2017; Chen, 2016 | Lung cancer | Upregulated | Proliferation,
migration, invasion, apoptosis, chemoresistance, EMT, cell
cycle | let-7, Bcl-xl;
miR-218-5p, Bmi1, c-Myc, miR-130a-3p, SOX4 | / | Oncogenic | (86–92) |
| Cao, 2017; Liu,
2013 | Epithelial ovarian
cancer | Upregulated | EMT, migration and
invasion | miR-152,
miR-130b | / | Oncogenic | (93,94) |
| Zhang, 2015; Lai,
2017 | Breast cancer | Upregulated | / | miR-148b | / | Oncogenic | (96,97) |
| Jia, 2017 | Cervical
cancer | Upregulated | Proliferation,
migration and invasion | / | / | Oncogenic | (98) |
| Zhao, 2016 | Endometrial
carcinoma | Upregulated | / | / | / | Oncogenic | (99) |
| Gao, 2017 |
Medulloblastoma | Upregulated | Proliferation, cell
cycle, metastasis | MAPK | / | Oncogenic | (101) |
| Chen, 2016 | Acute myeloid
leukemia | Upregulated | Proliferation,
monocytic differentiation | miR-155, c-Myc | / | Oncogenic | (102) |
| Zhao, 2017 | Osteosarcoma | Upregulated | Proliferation,
migration, invasion | miR-148a,
PIK3IP1 | / | Oncogenic | (103) |
| Wang, 2017 | Nasopharynx
cancer | Upregulated | / | miR-181a,
CPEB2 | / | Oncogenic | (104) |
| Zhang, 2017 | Retinoblastoma | Upregulated | Proliferation,
migration, Invasion, apoptosis, cell cycle | miR-218-5p | / | Oncogenic | (26) |
| Lv, 2017 | Melanoma | Upregulated | Proliferation and
invasion | miR-33a | / | Oncogenic | (105) |
| Wang, 2016; Cui,
2017 | Glioma | Upregulated | Proliferation,
migration, EMT and apoptosis | microRNA-410,
miR-181b | / | Oncogenic | (106,107) |
| Ma, 2017 | HNSCC | Upregulated | / | / | / | Oncogenic | (100) |
 | Table II.Clinical significance of CCAT1 in
various tumours. |
Table II.
Clinical significance of CCAT1 in
various tumours.
| First author,
year | Tumour type | Overexpression of
lncCCAT1 | Refs. |
|---|
| Nissan, 2012; He,
2014; Jing, 2017; Alaiyan, 2013; Kim, 2014; Ye, 2015; Ozawa, 2017;
Zhao, 2015; Kam, 2014; McCleland, 2016 | Colorectal
cancer | Clinical stage,
lymph nodes metastasis, local infiltration depth, vascular
invasion, CA19-9 level, RFS and OS | (22,30,40–46,48) |
| Zhuang, 2016;
Zhang, 2017 | Laryngeal squamous
cell carcinoma | T3-4 grade,
advanced clinical stage | (31,52) |
| Wu, 2016 | Esophageal squamous
cell carcinoma | OS, TNM stage,
histological grade | (24) |
| Zhang, 2017; Zhang,
2014; Mizrahi I, 2015; Yang, 2013; Zhou B, 2016 | Gastric cancer | Growth of primary
tumour, lymph node metastasis, metastatic disease, TNM grade, OS
and RFS | (55–59) |
| Deng, 2015; Wang,
2017; Zhu, 2015; Zhu, 2015; Dou, 2017; Dou, 2017 | Hepatocellular
carcinoma | Tumour size,
Edmondson-Steiner grade, liver cirrhosis, tumour number, prognosis
(OS, RFS, disease-free survival), vascular invasion, microvascular
invasion, capsular formation, AFP | (25,68–72) |
| Ma, 2015 | Gallbladder
cancer | tumour status,
lymph node invasion and advanced TNM stage | (75) |
| Jiang, 2017; Zhang,
2017 |
Cholangiocarcinoma | Histological
differentiation, lymph node invasion, TNM stage, OS | (77,78) |
| Yu, 2016 | Pancreatic
cancer | Tumourigenesis,
metastasis | (81) |
| White, 2014; Luo,
2014; Cabanski, 2015; Hu, 2017; Lu, 2016; Lu, 2017; Chen,
2016; | Lung cancer | Prognosis
biomarker, OS times, TNM stage | (86–92) |
| Cao, 2017; Liu,
2013 | Epithelial ovarian
cancer | FIGO stage,
histological grade, lymph node metastasis and poor survival | (93,94) |
| Zhang, 2015; Lai,
2017 | Breast cancer | Differentiation
grade, TNM stage, and lymph node metastases | (96,97) |
| Jia, 2017 | Cervical
cancer | / | (98) |
| Zhao, 2016 | Endometrial
carcinoma | FIGO stage, lymph
node metastasis, OS | (99) |
| Gao, 2017 |
Medulloblastoma | / | (101) |
| Chen, 2016 | Acute myeloid
leukemia | / | (102) |
| Zhao, 2017 | Osteosarcoma | / | (103) |
| Wang, 2017 | Nasopharynx
cancer | Paclitaxel
resistant | (104) |
| Zhang, 2017 | Retinoblastoma | / | (26) |
| Lv, 2017 | Melanoma | / | (105) |
| Wang, 2016; Cui,
2017 | Glioma | Pathological
grade | (106,107) |
| Ma, 2017 | HPV-associated head
and neck squamous cell carcinoma | Modified tumour
inflammation and immunity microenviron ment, myeloid-derived
suppressor cell recruitment and cancer development | (100) |
lncRNA CCAT1 in human cancer
CRC
CRC, the second-leading cause of mortality in the
United States, is a principal global health issue (1,32).
With the revelation of novel molecular and epigenetic mechanisms,
lncRNAs, including HOX transcript antisense RNA (33), colon cancer-associated transcript 2
(34), metastasis-associated lung
adenocarcinoma transcript 1 (35),
carcinoembryonic antigen (36) and
LINC00152 (37), have become
biological targets for diagnostic, therapeutic and prognostic
applications in patients with CRC. However, they all have
limitations in the early diagnosis of CRC (38). Therefore, it is vital to identify
novel bio-targets associated with CRC tumourigenesis (39).
Through reverse transcription-quantitative
polymerase chain reaction analysis, Nissan et al (22) first demonstrated that CCAT1
expression levels in the mucosa of colon adenocarcinoma were
significantly higher compared with normal colon tissues. A recent
genome-wide association analysis in CRC demonstrated the same
result (40). In addition, in the
later stages of the disease, CCAT1 is strongly expressed in early
stages of tumourigenesis, including tumour-proximal colonic
epithelium and adenomatous polyps, which was demonstrated by
Alaiyan et al (41) across
the colon adenocarcinoma sequence. Elevated CCAT1 expression levels
are positively associated with advanced clinical stages, lymphatic
metastasis, local invasive depth, vascular invasion, CA19-9,
recurrence-free survival (RFS) and overall survival (OS) (30,42–44).
Furthermore, CCAT1 expression levels are significantly increased in
the peripheral blood of patients with CRC. In particular, at a mild
phase, increased CCAT1 combined with increased plasma HOTAIR was
able to more powerfully diagnose patients with CRC from a group of
healthy controls (22,45). In addition, CCAT1-specific peptide
nucleic acid-based molecular beacons have been identified as a
diagnostic marker to detect CRC in vitro, ex vivo and in
situ (46). However, because
of the relatively small number of specimens in this previous study,
extensive and multi-centre randomized controlled trials are
required.
In vitro, CCAT1 is overexpressed in
CRC-derived cells compared with normal colon-derived fibroblasts.
Decreased CCAT1 was able to repress proliferation, migration,
invasion and EMT, and led to G0/G1 cell-cycle arrest in CRC cell
lines (30,42,43).
Previous studies suggested that c-Myc, which is a
pivotal transcriptional regulator significantly amplified in
various types of cancer, may directly combine with E-box elements
in the CCAT1 promoter regions to activate CCAT1 transcription
(Fig. 2A) (30,42).
Additionally, CCAT1 may serve as an enhancer-templated RNA to
predict bromodomain and extraterminal (BET)-mediated c-Myc
regulation, and BET inhibition JQ1 sensitivity in CRC, which has
been observed in specific previous studies with certain
haematological malignancies (47,48).
This data suggests that the oncogene CCAT1 may serve
as a novel biomarker for the early diagnosis and prognosis of CRC.
In particular, it may be ideal for those patients who are sensitive
to BET inhibitor-JQ1 in the treatment of CRC.
Laryngeal squamous cell carcinoma
(LSCC)
LSCC is the second most common head and neck
malignancy (resulting in high mortality rates) worldwide (49,50).
Despite the progress achieved in the diagnosis and therapy of LSCC
in the past few decades, the survival rate has not noticeably
increased (51). Therefore, novel
molecular targets for LSCC are urgently required.
CCAT1 expression was higher in LSCC compared with
matched normal tissues, and it was associated with advanced
clinical stage (31,52). Mechanistically, CCAT1
overexpression promotes LSCC cell proliferation and invasion by
suppressing let-7 expression and enhancing its target genes Myc and
HMGA2, or by enhancing the zinc finger protein, X-linked, by
sponging microRNA-218 (Fig. 3)
(31,52).
 | Figure 3.CCAT1 interacts with various target
genes by sponging different miRs. Bcl-xl, B cell lymphoma
extra-large; CCAT1, colon cancer-associated transcript 1; CDK,
cyclin-dependent kinase; CPEB, cytoplasmic polyadenylation element
binding protein; HOXB, homeobox protein-B; HMGA, high mobility
group AT-Hook; hnRNPA, heterogeneous nuclear ribonucleoprotein A;
ITPKB, inositol-trisphosphate 3-kinase B; miR, microRNA; SOX,
(sex-determining region Y)-box; ZFX, zinc finger protein
X-linked. |
Esophageal squamous cell carcinoma
(ESCC)
ESCC is developing at the fastest rate among all
cancer types in East Asia (53,54).
Therefore, a deeper understanding of the molecular basis underlying
ESCC is required to improve diagnosis and treatment.
CCAT1 is frequently increased in ESCC, which partly
results from H3K27-acetylation activation of promoter (Fig. 2B), and it is an independent
prognostic factor for advanced histological grade of patients with
ESCC (55). Furthermore,
proliferation and migration, in vitro and in vivo,
are significantly supressed following knockdown of CCAT1 (55). Zhang et al (55) identified that in the nucleus, CCAT1
has the role of a modular scaffold for polycomb repressive complex
2 and Suppressor Of Variegation 3–9 Homolog 1. This combination
modulates the histone methylation of sprouty RTK signalling
antagonist 4 (SPRY4) promoter, thereby epigenetically silencing
tumour suppressor genes SPRY4 (Fig.
2B). In the cytoplasm, CCAT1 upregulates HOXB13 as a molecular
decoy for miR-7, thereby facilitating cell viability and migration
(Fig. 3).
GC
GC is one of the most lethal malignancies worldwide
(1). The expression of CCAT1 is
notably upregulated in GC compared with normal tissue (56,57).
Notably, the CCAT1 expression levels in adjacent normal tissues
from GC cases were higher compared with a negative control group,
and recurrent GC tissues demonstrated the highest expression levels
among these groups (57).
Furthermore, CCAT1 overexpression is positively associated with
metastasis, TNM grade, OS and RFS in patients with GC (58–60).
In vitro, abnormal CCAT1 expression levels
promote GC cell proliferation, migration and invasion (58,59).
Similar to CRC, c-Myc activates the promoter and increases CCAT1
expression levels by directly binding to E-box elements (58). Zhang et al (56) demonstrated that CCAT1 was involved
in the ERK/MAPK signalling pathway to promote the growth of GC
(Fig. 2D). Another previous study
suggested that CCAT1 may additionally function as ceRNA by sponging
miR-490 and free miR-490 target heterogeneous nuclear RNP A1
(hnRNPA1) for tumourigenesis in GC (Fig. 3) (59); the relevance has been verified in
breast, colorectal, lung and glioma cancer (61–65).
HCC
HCC is the third leading cause of tumour-induced
mortality worldwide and accounts for a large proportion of
mortalities in China (3,66). Despite recent progress in
experimental oncology, patients with HCC continue to have poor
long-term prognosis (67).
Therefore, it is crucial to identify reliable biomarkers of HCC to
develop novel clinical strategies and increase the survival rates
of patients with HCC.
CCAT1 expression levels are higher in HCC compared
with pair-matched healthy hepatic tissues, particularly in highly
metastatic HCC (25,68). Upregulation of CCAT1 has been
identified to be positively associated with tumour size, liver
cirrhosis, tumour number, vascular invasion, microvascular
invasion, capsular formation, Edmondson-Steiner grade and α foetal
protein, and it is an independent risk factor for disease-free
survival and OS (25,68–70).
Additionally, CCAT1 overexpression significantly accelerates HCC
cell proliferation, migration and invasion, in vitro
(25,68,69).
Similar to CRC and LSCC, CCAT1 activated by c-Myc
(70), promotes HCC proliferation
and metastasis by functioning as a let-7 sponge to supress its
endogenous targets, HMGA2 and c-Myc (25). Furthermore, the CCAT1/miR-200b and
CCAT1/miR-490-3p/cyclin-dependent kinase 1 regulatory pathway may
additionally promote HCC progression (Fig. 3) (71,72).
These findings implicate the potential role of CCAT1 in HCC
therapies.
Gallbladder cancer (GBC)
Gallbladder cancer (GBC) is the most common cancer
of the biliary tract, and has a particularly high incidence in
Chile, Japan and northern India (73). Despite the great efforts made to
identify novel molecular abnormalities that contribute to GBC, many
remain unknown (74).
CCAT1 is upregulated in GBC and is positively
correlated with tumour status, lymph node invasion and advanced TNM
stage (75). CCAT1 was able to
improve the proliferation, migration and invasion of GBC cells
in vitro (75). These
effects are dependent on its competitive binding to miR-218-5p,
thereby regulating Bmi1 (Fig. 3)
(75).
Cholangiocarcinoma (CCA)
Without typical symptoms and sensitive indicators,
the diagnosis of the majority of CCA cases typically occurs at a
late stage, with poor prognosis (76). Therefore, identifying more
efficient markers and examining the molecular mechanism underlying
the carcinogenesis and progression of CCA is urgently required.
It was observed that CCAT1 expression levels are
elevated in CCA compared with the adjacent normal controls, and it
is positively associated with histological differentiation, lymph
node invasion, TNM stage and OS in patients with CCA (77). A previous mechanistic study
identified that CCAT1 led to migration, invasion and EMT activation
by binding to miR-152 in CCA cells (78).
Pancreatic cancer (PC)
PC is a lethal digestive system malignancy (1) due to the late detection of the
disease and the lack of effective therapies for terminally staged
tumours (79); only 25% of
patients with metastatic PC have a five-year survival rate
(80). Therefore, a comprehensive
understanding of the molecular mechanisms underlying PC
tumourigenesis is urgently required to identify novel therapeutic
targets.
CCAT1 expression levels are notably higher in PC
specimens and PC cell lines compared with matched noncancerous
controls (81). In addition, the
silencing of CCAT1 inhibited proliferation and migration, extending
the cell cycle progression and decreasing cyclin D1 expression in
PC cells (82). In CRC, GC and
HCC, c-Myc was able to activate CCAT1 expression by targeting its
promoter at the E-box, thereby contributing to tumourigenesis and
metastasis in PC, suggesting that CCAT1 may serve as a potential
therapeutic target for PC.
LC
LC is one of the frequent causes of cancer mortality
worldwide, resulting in more than one million mortalities annually
(83,84). Despite advancements in clinical and
experimental oncology, effective diagnostic and prognostic
biomarkers and alternative treatment options are still required due
to the late diagnosis and quick onset of chemoresistance (85).
Overexpression of CCAT1 is evident in non-small cell
lung cancer (NSCLC) and is associated with reduced OS times,
advanced disease stage and lymph node involvement (86–88).
Knockdown of CCAT1 suppressed the proliferation, migration and
invasion, and reversed the EMT of NSCLC cells (88). Furthermore, CCAT1 was able to
enhance cisplatin resistance of NSCLC cells through the
CCAT1/miR-130a-3p/ sex-determining region Y-box (SOX)4 axis
(Fig. 3) (89).
As cigarette smoking is a key risk factor for LC, Lu
et al (90) determined that
CCAT1 was able to regulate neoplastic activity by epigenetically
silencing miR-218 and acting through Bmi1, thereby promoting cell
cycle progression in the cigarette smoke extract (CSE)-induced
carcinogenesis of human bronchial epithelial (HBE) cells (Fig. 3). In a subsequent study, the
authors additionally demonstrated that CCAT1 bound let-7c and
subsequently upregulated c-Myc, which was able to promote
CSE-transformed HBE cell proliferation and invasion (Fig. 3) (91).
CCAT1 is upregulated in the docetaxel-resistant lung
adenocarcinoma (LAD) cell line, and suppression of CCAT1 inhibits
cell proliferation, enhances apoptosis, decreases chemoresistance,
and reverses the docetaxel-resistant LAD cells-induced EMT
phenotype (92). Furthermore,
CCAT1 exerted the oncogenic function in LAD cells, partially by
competitive sponging (let-7c) to prevent the inhibition of B cell
lymphoma-extra-large, thereby resulting in chemoresistance and EMT
in docetaxel-resistant LAD cells (Fig.
3).
These results suggested that CCAT1 is a critical
oncogene associated with the diagnosis and prognosis of various
types of lung cancer, as well as a potential target for
strengthening the response to chemotherapeutic drugs in lung
cancer.
Epithelial ovarian cancer (EOC)
EOC, characterized by quick disease progression, is
a lethal gynaecological cancer (66). CCAT1 upregulation in EOC was
associated with the International Federation of Gynaecology and
Obstetrics stage, histological grade, lymph node metastasis and
poor survival (93). In addition,
CCAT1 promotes EOC cell migration, invasion and EMT by sponging
miR-152 and miR-130b, which may function as a potential molecular
target for EOC (Fig. 3) (93,94).
Breast cancer (BC)
BC is a prevalent malignancy among women, and it
affects approximately one million women worldwide (95). Among patients with BC, high CCAT1
expression levels are significantly associated with differentiation
grade and TNM stage compared with other clinicopathological factors
(96). Furthermore, reduced CCAT1
may improve breast cancer radiosensitivity by negatively regulating
miR-148b expression (Fig. 3)
(97). These previous studies
provide a crucial basis to identify more effective treatments for
breast cancer.
Other human cancer types
The aberrant upregulation of CCAT1 was detected in
human papillomavirus (HPV)-associated head and neck squamous cell
carcinomas (HNSCC), cervical cancer, endometrial carcinoma,
medulloblastoma, acute myeloid leukaemia (AML), osteosarcoma,
nasopharyngeal carcinoma (NPC), retinoblastoma (RB), melanoma and
glioma cancer (26,98–107) (Tables I and II), and it was associated with
clinicopathological features, in addition to clinical outcomes.
Furthermore, CCAT1 may serve as an intermediate in HPV16 infection
and promote myeloid-derived suppressor cell aggregation in HNSCC
(100,108).
Mechanistically, CCAT1 may promote medulloblastoma
cell proliferation and metastasis through the MAPK signaling
pathway (Fig. 2D), as in GC
(101). In addition, CCAT1
inhibits monocytic differentiation and promotes AML cell growth by
sequestering tumour suppressive miR-155, thereby upregulating c-Myc
(Fig. 3) (102). Additionally, the biological
function of osteosarcoma cells was regulated by the
CCAT1/miR-148a/phosphatidyl inositol 3-kinase interacting protein 1
signaling pathway (Fig. 3)
(103). Wang et al
(104) observed that upregulated
CCAT1 significantly weakened the sensitivity of paclitaxel in NPC
cells through the miR-181a/CPEB2 axis (Fig. 3).
CCAT1 promotes RB and melanoma tumourigenesis and
metastasis through negative modulation of miR-218-5p and miR-33a,
respectively (Fig. 3) (26,105). Furthermore, CCAT1 was able to
promote glioma cell progress by inhibiting miR-410 and miR-181b,
which provides novel insight into the proliferation of glioma
(Fig. 3) (106,107).
Future directions
Overexpression of the oncogenic lncRNA CCAT1 occurs
in numerous cancer types, and positively correlates with clinical
progress and cell biological function via complex molecular
mechanisms. Notably, to date, the mechanism of ceRNA has been
investigated in a number of previous studies. Therefore, further
research is required to investigate the downstream molecular
mechanism of CCAT1 dysregulation. Furthermore, CCAT1 may be
activated by c-Myc and H3K27-acetylation in various tumours, which
suggests that the upstream regulatory mechanisms underlying CCAT1
deregulation in various cancer types may be diverse and remain to
be elucidated. Pertinent to clinical practice, to improve the
clinical utilisation of CCAT1 as a biomarker for the diagnosis and
treatment of cancer, larger cohorts of CCAT1 are required in future
studies. Additionally, identifying sensitive and high-throughput
quantification methods to identify CCAT1 as a specific and early
cancer biomarker, providing clinical benefits in future studies, is
required.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Project of
Standard Diagnosis and Treatment of Key Disease of Jiangsu Province
(grant no. BE2015722), Project of the peak of the six talents of
Jiangsu Province (grant no. WSN-018) and Scientific Research
Foundation for Health of Jiangsu Province (grant no. H201408).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
NW, YY, QL and LM conceived and designed the study.
NW, YY, BX and MZ researched the literature and performed data
analysis. NW drafted the manuscript. QL and LM revised the article.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet 15 Spec No. 1:R17–R29. 2006. View Article : Google Scholar
|
|
5
|
Bonasio R and Shiekhattar R: Regulation of
transcription by long noncoding RNAs. Annu Rev Genet. 48:433–455.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eddy SR: Non-coding RNA genes and the
modern RNA world. Nat Rev Genet. 2:919–929. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
St Laurent G, Wahlestedt C and Kapranov P:
The Landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yildirim E, Kirby JE, Brown DE, Mercier
FE, Sadreyev RI, Scadden DT and Lee JT: Xist RNA is a potent
suppressor of hematologic cancer in mice. Cell. 152:727–742. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hung T, Wang Y, Lin MF, Koegel AK, Kotake
Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al:
Extensive and coordinated transcription of noncoding RNAs within
cell-cycle promoters. Nat Genet. 43:621–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gomez JA, Wapinski OL, Yang YW, Bureau JF,
Gopinath S, Monack DM, Chang HY, Brahic M and Kirkegaard K: The
NeST long ncRNA controls microbial susceptibility and epigenetic
activation of the interferon-γ locus. Cell. 152:743–754. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Engreitz JM, Pandya-Jones A, McDonel P,
Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander
ES, et al: The Xist lncRNA exploits three-dimensional genome
architecture to spread across the X chromosome. Science.
341:12379732013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li W, Notani D, Ma Q, Tanasa B, Nunez E,
Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al: Functional
roles of enhancer RNAs for oestrogen-dependent transcriptional
activation. Nature. 498:516–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Di Ruscio A, Ebralidze AK, Benoukraf T,
Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes
LL, Alberich-Jorda M, Zhang P, et al: DNMT1-interacting RNAs block
gene-specific DNA methylation. Nature. 503:371–376. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nissan A, Stojadinovic A,
Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, Bochem A,
Dayanc BE, Ritter G, Gomceli I, et al: Colon cancer associated
transcript-1: A novel RNA expressed in malignant and pre-malignant
human tissues. Int J Cancer. 130:1598–1606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang
XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al: Human colorectal
cancer-specific CCAT1-L lncRNA regulates long-range chromatin
interactions at the MYC locus. Cell Res. 24:513–531. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu Y, Tan C, Weng WW, Deng Y, Zhang QY,
Yang XQ, Gan HL, Wang T, Zhang PP, Xu MD, et al: Long non-coding
RNA Linc00152 is a positive prognostic factor for and demonstrates
malignant biological behavior in clear cell renal cell carcinoma.
Am J Cancer Res. 6:285–299. 2016.PubMed/NCBI
|
|
25
|
Deng L, Yang SB, Xu FF and Zhang JH: Long
noncoding RNA CCAT1 promotes hepatocellular carcinoma progression
by functioning as let-7 sponge. J Exp Clin Cancer Res. 34:182015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H, Zhong J, Bian Z, Fang X, Peng Y
and Hu Y: Long non-coding RNA CCAT1 promotes human retinoblastoma
SO-RB50 and Y79 cells through negative regulation of miR-218-5p.
Biomed Pharmacother. 87:683–691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zanke BW, Greenwood CM, Rangrej J, Kustra
R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T,
Crowdy E, et al: Genome-wide association scan identifies a
colorectal cancer susceptibility locus on chromosome 8q24. Nat
Genet. 39:989–994. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yeager M, Orr N, Hayes RB, Jacobs KB,
Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee
N, et al: Genome-wide association study of prostate cancer
identifies a second risk locus at 8q24. Nat Genet. 39:645–649.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Younger ST and Rinn JL: ‘Lnc’-ing
enhancers to MYC regulation. Cell Res. 24:643–644. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He X, Tan X, Wang X, Jin H, Liu L, Ma L,
Yu H and Fan Z: C-Myc-activated long noncoding RNA CCAT1 promotes
colon cancer cell proliferation and invasion. Tumour Biol.
35:12181–12188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhuang K, Wu Q, Jiang S, Yuan H, Huang S
and Li H: CCAT1 promotes laryngeal squamous cell carcinoma cell
proliferation and invasion. Am J Transl Res. 8:4338–4345.
2016.PubMed/NCBI
|
|
32
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RG, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ling H, Spizzo R, Atlasi Y, Nicoloso M,
Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al:
CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic
progression and chromosomal instability in colon cancer. Genome
Res. 23:1446–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|
|
36
|
Wang X, Yang Z, Tian H, Li Y, Li M, Zhao
W, Zhang C, Wang T, Liu J, Zhang A, et al: Circulating MIC-1/GDF15
is a complementary screening biomarker with CEA and correlates with
liver metastasis and poor survival in colorectal cancer.
Oncotarget. 8:24892–24901. 2017.PubMed/NCBI
|
|
37
|
Yue B, Cai D, Liu C, Fang C and Yan D:
linc00152 functions as a competing endogenous RNA to confer
oxaliplatin resistance and holds prognostic values in colon cancer.
Mol Ther. 24:2064–2077. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Luo J, Qu J, Wu DK, Lu ZL, Sun YS and Qu
Q: Long non-coding RNAs: A rising biotarget in colorectal cancer.
Oncotarget. 8:22187–22202. 2017.PubMed/NCBI
|
|
39
|
Fang L, Lu W, Choi HH, Yeung SC, Tung JY,
Hsiao CD, Fuentes-Mattei E, Menter D, Chen C, Wang L, et al:
ERK2-dependent phosphorylation of CSN6 is critical in colorectal
cancer development. Cancer Cell. 28:183–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jing F, Jin H, Mao Y, Li Y, Ding Y, Fan C
and Chen K: Genome-wide analysis of long non-coding RNA expression
and function in colorectal cancer. Tumour Biol.
39:10104283177036502017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Alaiyan B, Ilyayev N, Stojadinovic A,
Izadjoo M, Roistacher M, Pavlov V, Tzivin V, Halle D, Pan H, Trink
B, et al: Differential expression of colon cancer associated
transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence.
BMC Cancer. 13:1962013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim T, Cui R, Jeon YJ, Lee JH, Lee JH, Sim
H, Park JK, Fadda P, Tili E, Nakanishi H, et al: Long-range
interaction and correlation between MYC enhancer and oncogenic long
noncoding RNA CARLo-5. Proc Natl Acad Sci USA. 111:4173–4178. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ye Z, Zhou M, Tian B, Wu B and Li J:
Expression of lncRNA-CCAT1, E-cadherin and N-cadherin in colorectal
cancer and its clinical significance. Int J Clin Exp Med.
8:3707–3715. 2015.PubMed/NCBI
|
|
44
|
Ozawa T, Matsuyama T, Toiyama Y, Takahashi
N, Ishikawa T, Uetake H, Yamada Y, Kusunoki M, Calin G and Goel A:
CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21
‘gene desert’, serve as important prognostic biomarkers in
colorectal cancer. Ann Oncol. 28:1882–1888. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao W, Song M, Zhang J, Kuerban M and
Wang H: Combined identification of long non-coding RNA CCAT1 and
HOTAIR in serum as an effective screening for colorectal carcinoma.
Int J Clin Exp Pathol. 8:14131–14140. 2015.PubMed/NCBI
|
|
46
|
Kam Y, Rubinstein A, Naik S, Djavsarov I,
Halle D, Ariel I, Gure AO, Stojadinovic A, Pan H, Tsivin V, et al:
Detection of a long non-coding RNA (CCAT1) in living cells and
human adenocarcinoma of colon tissues using FIT-PNA molecular
beacons. Cancer Lett. 352:90–96. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shi J and Vakoc CR: The mechanisms behind
the therapeutic activity of BET bromodomain inhibition. Mol Cell.
54:728–736. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
McCleland ML, Mesh K, Lorenzana E, Chopra
VS, Segal E, Watanabe C, Haley B, Mayba O, Yaylaoglu M, Gnad F and
Firestein R: CCAT1 is an enhancer-templated RNA that predicts BET
sensitivity in colorectal cancer. J Clin Invest. 126:639–652. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ren J, Zhu D, Liu M, Sun Y and Tian L:
Downregulation of miR-21 modulates Ras expression to promote
apoptosis and suppress invasion of Laryngeal squamous cell
carcinoma. Eur J Cancer. 46:3409–3416. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chu EA and Kim YJ: Laryngeal cancer:
Diagnosis and preoperative work-up. Otolaryngol Clin North Am.
41:673–695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang W, Lin P, Han C, Cai W, Zhao X and
Sun B: Vasculogenic mimicry contributes to lymph node metastasis of
laryngeal squamous cell carcinoma. J Exp Clin Cancer Res.
29:602010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang Y and Hu H: Long non-coding RNA
CCAT1/miR-218/ZFX axis modulates the progression of laryngeal
squamous cell cancer. Tumour Biol.
39:10104283176994172017.PubMed/NCBI
|
|
53
|
Matsushima K, Isomoto H, Yamaguchi N,
Inoue N, Machida H, Nakayama T, Hayashi T, Kunizaki M, Hidaka S,
Nagayasu T, et al: MiRNA-205 modulates cellular invasion and
migration via regulating zinc finger E-box binding homeobox 2
expression in esophageal squamous cell carcinoma cells. J Transl
Med. 9:302011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang E, Han L, Yin D, He X, Hong L, Si X,
Qiu M, Xu T, De W, Xu L, et al: H3K27 acetylation activated-long
non-coding RNA CCAT1 affects cell proliferation and migration by
regulating SPRY4 and HOXB13 expression in esophageal squamous cell
carcinoma. Nucleic Acids Res. 45:3086–3101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang Y, Ma M, Liu W, Ding W and Yu H:
Enhanced expression of long noncoding RNA CARLo-5 is associated
with the development of gastric cancer. Int J Clin Exp Pathol.
7:8471–8479. 2014.PubMed/NCBI
|
|
57
|
Mizrahi I, Mazeh H, Grinbaum R, Beglaibter
N, Wilschanski M, Pavlov V, Adileh M, Stojadinovic A, Avital I,
Gure AO, et al: Colon cancer associated transcript-1 (CCAT1)
expression in adenocarcinoma of the stomach. J Cancer. 6:105–110.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y
and Fang G: Long noncoding RNA CCAT1, which could be activated by
c-Myc, promotes the progression of gastric carcinoma. J Cancer Res
Clin Oncol. 139:437–445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhou B, Wang Y, Jiang J, Jiang H, Song J,
Han T, Shi J and Qiao H: The long noncoding RNA colon
cancer-associated transcript-1/miR-490 axis regulates gastric
cancer cell migration by targeting hnRNPA1. IUBMB Life. 68:201–210.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liu JN and Shangguan YM: Long non-coding
RNA CARLo-5 upregulation associates with poor prognosis in patients
suffering gastric cancer. Eur Rev Med Pharmacol Sci. 21:530–534.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pino I, Pio R, Toledo G, Zabalegui N,
Vicent S, Rey N, Lozano MD, Torre W, Garcia-Foncillas J and
Montuenga LM: Altered patterns of expression of members of the
heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung
cancer. Lung Cancer. 41:131–143. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Buvoli M, Cobianchi F, Biamonti G and Riva
S: Recombinant hnRNP protein A1 and its N-terminal domain show
preferential affinity for oligodeoxynucleotides homologous to
intron/exon acceptor sites. Nucleic Acids Res. 18:6595–6600. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Carpenter B, MacKay C, Alnabulsi A, MacKay
M, Telfer C, Melvin WT and Murray GI: The roles of heterogeneous
nuclear ribonucleoproteins in tumour development and progression.
Biochim Biophys Acta. 1765:85–100. 2006.PubMed/NCBI
|
|
64
|
Boukakis G, Patrinou-Georgoula M,
Lekarakou M, Valavanis C and Guialis A: Deregulated expression of
hnRNP A/B proteins in human non-small cell lung cancer: Parallel
assessment of protein and mRNA levels in paired tumour/non-tumour
tissues. BMC Cancer. 10:4342010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
David CJ, Chen M, Assanah M, Canoll P and
Manley JL: HnRNP proteins controlled by c-Myc deregulate pyruvate
kinase mRNA splicing in cancer. Nature. 463:364–368. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang F, Xie C, Zhao W, Deng Z, Yang H and
Fang Q: Long non-coding RNA CARLo-5 expression is associated with
disease progression and predicts outcome in hepatocellular
carcinoma patients. Clin Exp Med. 17:33–43. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhu H, Zhou X, Chang H, Li H, Liu F, Ma C
and Lu J: CCAT1 promotes hepatocellular carcinoma cell
proliferation and invasion. Int J Clin Exp Pathol. 8:5427–5434.
2015.PubMed/NCBI
|
|
70
|
Zhu HQ, Zhou X, Chang H, Li HG, Liu FF, Ma
CQ and Lu J: Aberrant expression of CCAT1 regulated by c-Myc
predicts the prognosis of hepatocellular carcinoma. Asian Pac J
Cancer Prev. 16:5181–5185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Dou C, Sun L, Jin X, Han M, Zhang B, Jiang
X, Lv J and Li T: Long non-coding RNA CARLo-5 promotes tumor
progression in hepatocellular carcinoma via suppressing miR-200b
expression. Oncotarget. 8:70172–70182. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Dou C, Sun L, Jin X, Han M, Zhang B and Li
T: Long non-coding RNA colon cancer-associated transcript 1
functions as a competing endogenous RNA to regulate
cyclin-dependent kinase 1 expression by sponging miR-490-3p in
hepatocellular carcinoma progression. Tumour Biol.
39:10104283176975722017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhu AX, Hong TS, Hezel AF and Kooby DA:
Current management of gallbladder carcinoma. Oncologist.
15:168–181. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Jiao Y, Pawlik TM, Anders RA, Selaru FM,
Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P,
et al: Exome sequencing identifies frequent inactivating mutations
in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat
Genet. 45:1470–1473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY,
Gong W and Quan ZW: Long non-coding RNA CCAT1 promotes gallbladder
cancer development via negative modulation of miRNA-218-5p. Cell
Death Dis. 6:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Khan SA, Thomas HC, Davidson BR and
Taylor-Robinson SD: Cholangiocarcinoma. Lancet. 366:1303–1314.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Jiang XM, Li ZL, Li JL, Zheng WY, Li XH,
Cui YF and Sun DJ: LncRNA CCAT1 as the unfavorable prognostic
biomarker for cholangiocarcinoma. Eur Rev Med Pharmacol Sci.
21:1242–1247. 2017.PubMed/NCBI
|
|
78
|
Zhang S, Xiao J, Chai Y, Du YY, Liu Z,
Huang K, Zhou X and Zhou W: LncRNA-CCAT1 promotes migration,
invasion, and EMT in intrahepatic cholangiocarcinoma through
suppressing miR-152. Dig Dis Sci. 62:3050–3058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Li C, Heidt DG, Dalerba P, Burant CF,
Zhang L, Adsay V, Wicha M, Clarke MF and Simeone DM: Identification
of pancreatic cancer stem cells. Cancer Res. 67:1030–1037. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Von Hoff DD, Ervin T, Arena FP, Chiorean
EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, et
al: Increased survival in pancreatic cancer with nab-paclitaxel
plus gemcitabine. N Engl J Med. 369:1691–1703. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yu Q, Zhou X, Xia Q, Shen J, Yan J, Zhu J,
Li X and Shu M: Long non-coding RNA CCAT1 that can be activated by
c-Myc promotes pancreatic cancer cell proliferation and migration.
Am J Transl Res. 8:5444–5454. 2016.PubMed/NCBI
|
|
82
|
Bartek J and Lukas J: DNA repair: Cyclin
D1 multitasks. Nature. 474:171–172. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Bray F, Jemal A, Grey N, Ferlay J and
Forman D: Global cancer transitions according to the Human
Development Index (2008–2030): A population-based study. Lancet
Oncol. 13:790–801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Roth A and Diederichs S: Long noncoding
RNAs in lung cancer. Curr Top Microbiol Immunol. 394:57–110.
2016.PubMed/NCBI
|
|
86
|
White NM, Cabanski CR, Silva-Fisher JM,
Dang HX, Govindan R and Maher CA: Transcriptome sequencing reveals
altered long intergenic non-coding RNAs in lung cancer. Genome
Biol. 15:4292014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Luo J, Tang L, Zhang J, Ni J, Zhang HP,
Zhang L, Xu JF and Zheng D: Long non-coding RNA CARLo-5 is a
negative prognostic factor and exhibits tumor pro-oncogenic
activity in non-small cell lung cancer. Tumour Biol.
35:11541–11549. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Cabanski CR, White NM, Dang HX,
Silva-Fisher JM, Rauck CE, Cicka D and Maher CA: Pan-cancer
transcriptome analysis reveals long noncoding RNAs with conserved
function. RNA Biol. 12:628–642. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hu B, Zhang H, Wang Z, Zhang F, Wei H and
Li L: LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance
in non-small-cell lung cancer cell line by targeting SOX4. Cancer
Biol Ther. 18:974–983. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lu L, Xu H, Luo F, Liu X, Lu X, Yang Q,
Xue J, Chen C, Shi L and Liu Q: Epigenetic silencing of miR-218 by
the lncRNA CCAT1, acting via BMI1, promotes an altered cell cycle
transition in the malignant transformation of HBE cells induced by
cigarette smoke extract. Toxicol Appl Pharmacol. 304:30–41. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lu L, Qi H, Luo F, Xu H, Ling M, Qin Y,
Yang P, Liu X, Yang Q, Xue J, et al: Feedback circuitry via let-7c
between lncRNA CCAT1 and c-Myc is involved in cigarette smoke
extract-induced malignant transformation of HBE cells. Oncotarget.
8:19285–19297. 2017.PubMed/NCBI
|
|
92
|
Chen J, Zhang K, Song H, Wang R, Chu X and
Chen L: Long noncoding RNA CCAT1 acts as an oncogene and promotes
chemoresistance in docetaxel-resistant lung adenocarcinoma cells.
Oncotarget. 7:62474–62489. 2016.PubMed/NCBI
|
|
93
|
Cao Y, Shi H, Ren F, Jia Y and Zhang R:
Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in
epithelial ovarian cancer. Exp Cell Res. 359:185–194. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Liu SP, Yang JX, Cao DY and Shen K:
Identification of differentially expressed long non-coding RNAs in
human ovarian cancer cells with different metastatic potentials.
Cancer Biol Med. 10:138–141. 2013.PubMed/NCBI
|
|
95
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhang XF, Liu T, Li Y and Li S:
Overexpression of long non-coding RNA CCAT1 is a novel biomarker of
poor prognosis in patients with breast cancer. Int J Clin Exp
Pathol. 8:9440–9445. 2015.PubMed/NCBI
|
|
97
|
Lai Y, Chen Y, Lin Y and Ye L:
Down-regulation of LncRNA CCAT1 enhances radiosensitivity via
regulating miR-148b in breast cancer. Cell Biol Int. 42:227–236.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Jia L, Zhang Y, Tian F, Chu Z and Xin H:
Long noncoding RNA colon cancer associated transcript-1 promotes
the proliferation, migration and invasion of cervical cancer. Mol
Med Rep. 16:5587–5591. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhao X, Wei X, Zhao L, Shi L, Cheng J,
Kang S, Zhang H, Zhang J, Li L, Zhang H and Zhao W: The rs6983267
SNP and long non-coding RNA CARLo-5 are associated with endometrial
carcinoma. Environ Mol Mutagen. 57:508–515. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Ma X, Sheng S, Wu J, Jiang Y, Gao X, Cen
X, Wu J, Wang S, Tang Y, Tang Y and Liang X: LncRNAs as an
intermediate in HPV16 promoting myeloid-derived suppressor cell
recruitment of head and neck squamous cell carcinoma. Oncotarget.
8:42061–42075. 2017.PubMed/NCBI
|
|
101
|
Gao R, Zhang R, Zhang C, Zhao L and Zhang
Y: Long noncoding RNA CCAT1 promotes cell proliferation and
metastasis in human medulloblastoma via MAPK pathway. Tumori.
104:43–50. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Chen L, Wang W, Cao L, Li Z and Wang X:
Long non-coding RNA CCAT1 acts as a competing endogenous RNA to
regulate cell growth and differentiation in acute myeloid leukemia.
Mol Cell. 39:330–336. 2016. View Article : Google Scholar
|
|
103
|
Zhao J and Cheng L: Long non-coding RNA
CCAT1/miR-148a axis promotes osteosarcoma proliferation and
migration through regulating PIK3IP1. Acta Biochim Biophys Sin
(Shanghai). 49:503–512. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang Q, Zhang W and Hao S: LncRNA CCAT1
modulates the sensitivity of paclitaxel in nasopharynx cancers
cells via miR-181a/CPEB2 axis. Cell Cycle. 16:795–801. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Lv L, Jia JQ and Chen J: LncRNA CCAT1
Upregulates Proliferation and Invasion in Melanoma Cells Via
Suppressing miR-33a. Oncol Res. 26:201–208. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wang ZH, Guo XQ, Zhang QS, Zhang JL, Duan
YL, Li GF and Zheng DL: Long non-coding RNA CCAT1 promotes glioma
cell proliferation via inhibiting microRNA-410. Biochem Biophys Res
Commun. 480:715–720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Cui B, Li B, Liu Q and Cui Y: lncRNA CCAT1
promotes glioma tumorigenesis by sponging miR-181b. J Cell Biochem.
118:4548–4557. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Gabrilovich DI and Nagaraj S:
Myeloid-derived suppressor cells as regulators of the immune
system. Nat Rev Immunol. 9:162–174. 2009. View Article : Google Scholar : PubMed/NCBI
|

















