Introduction
Breast cancer is a major human malignancy and is the
second leading cause of cancer-associated mortality in women
worldwide, following lung cancer (1). As estimated by the American Cancer
Society, there will be >266,000 new cases of breast cancer and
>40,000 cases of breast cancer-associated mortality in the next
10 years in the United States, thus accounting for 30 and 14% of
total cancer cases and cases of cancer-associated mortality,
respectively (1). Survival rates
of patients with breast cancer is relatively low, particularly in
developing countries; however, a 5-year survival rate of >80%
has been achieved in England and the United States (2). As a complex heterogeneous malignancy,
breast cancer remains a severe public health concern, due to
ambiguities regarding the stepwise pathological processes from
normal breast tissue to metastatic cancer tissue (3,4).
Previous studies have revealed that the pathogenesis of breast
cancer is accompanied and driven by a series of successive
mutations in genetic and epigenetic networks in breast cells,
finally resulting in activation of various hallmarks, malignant
transformation and metastasis (5,6). In
recent years, it has been suggested that noncoding RNAs (ncRNAs)
are responsible for genetic and epigenetic dysregulation, which is
associated with various developmental and pathological processes,
including tumorigenesis (7,8).
However, the specific roles of ncRNAs in breast cancer remain to be
investigated.
Long ncRNAs (lncRNAs) refer to ncRNA transcripts
containing >200 nucleotides, which are involved in
post-transcriptional regulation of gene expression by interfering
with microRNA functioning (9).
Through modulating gene expression at the transcriptional,
post-transcriptional and epigenetic levels, lncRNAs have been
reported to be associated with various physiological and
pathological processes, including stem cell development,
neurogenesis and oncogenesis (8,9). In
various types of human cancer, lncRNAs act as promoters and
maintainers of cancer initiation and progression; therefore, they
have been considered potential biomarkers for clinical diagnosis,
and also as therapeutic targets for the development of novel
treatments (10,11). Based on functional studies using
cellular and animal models, it has been reported that lncRNAs are
involved in various cancer-associated cellular phenotypes,
including cell death inhibition, activation of invasion,
proliferation maintenance, dysregulated cellular energetics,
genomic instability and evasion of growth suppressors (12). LncRNAs have also been identified as
key regulators in breast cancer. For example, the lncRNA HOX
transcript antisense RNA, which promotes cancer metastasis by
modulating chromatin states, has been identified as a potential
prognostic marker of cancer metastasis for estrogen receptor
(ER)-positive breast cancer (13).
Furthermore, the lncRNA urothelial cancer-associated 1 has been
reported to enhance breast tumor growth through suppression of p27
protein levels via competitive inhibition (14). An accumulating number of lncRNAs
have been demonstrated to be associated with breast cancer
initiation and progression, thus suggesting that functional
elucidation of key lncRNAs in breast cancer cells may provide
valuable information for clinical screening and management.
Prostate cancer-associated ncRNA 1 (PRNCR1) is closely associated
with resistance of prostate cancer to castration (15), colorectal cancer cell proliferation
and cell cycle progression (16),
and progression of other types of cancer (17). However, to the best of our
knowledge, the implications of PRNCR1 in breast cancer progression
have not been previously addressed.
In the present study, the expression levels of
PRNCR1 were detected in clinical tissues from patients with breast
cancer, as well as in numerous breast cancer cell lines.
Subsequently, PRNCR1 expression was knocked down in breast cancer
cells and a functional analysis was conducted. The present study
identified an association between PRNCR1 and breast cancer, and
therefore provided a novel candidate for ncRNA-based breast cancer
diagnosis and treatment. These findings may also result in future
elucidation of the pathological mechanisms underlying breast cancer
initiation and progression.
Materials and methods
Cancer tissues and cell lines
Breast cancer tissues and paired adjacent normal
tissues were collected from 20 patients at the Department of Breast
Surgery, Foshan First People's Hospital (Foshan, China) between
June 2015 and July 2017. Breast cancer was confirmed by
pathological diagnosis post-operation, and the clinicopathological
data of patients are displayed in Table I. The experimental procedures were
approved by the Ethics Committee of Foshan First People's Hospital,
and written informed consent was obtained from each participant
prior to surgery. The breast cancer cell lines HS-578T (cat. no.
HTB-126), MCF-7 (cat. no. HTB-22), MDA-MB-468 (cat. no. HTB-132),
MDA-MB-231 (cat. no. HTB-26) and BT-549 (cat. no. HTB-122), and the
mammary epithelial cell line MCF10A (cat. no. CRL-10317) were
obtained from the American Type Culture Collection (Manassas, VA,
USA). Breast cancer cells and the mammary epithelial cell line were
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal
bovine serum (Biowest, Nuaillé, France) in a humidified atmosphere
supplied with 5% CO2 at 37°C.
 | Table I.Clinicopathological data of patients
with breast cancer. |
Table I.
Clinicopathological data of patients
with breast cancer.
| Characteristic | Number |
|---|
| Age (years) |
|
|
<55 | 12 |
|
>55 | 8 |
| Sex |
|
|
Male | 0 |
|
Female | 20 |
| Distant
metastasis |
|
|
Absent | 10 |
|
Present | 10 |
| Histological
grade |
|
| 1 | 3 |
| 2 | 11 |
| 3 | 6 |
| Tumor size
(cm) |
|
| ≤2 | 6 |
|
2-5 | 13 |
|
>5 | 1 |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Relative lncRNA expression levels were measured in
total RNA samples extracted from breast cancer tissues and cell
lines by RT-qPCR. Prior to RNA extraction, breast tissues were
homogenized in liquid nitrogen and breast cell lines were collected
by centrifugation at 800 × g for 10 min at room temperature,
followed by three washes with PBS. Total RNA samples were extracted
using TRIzol® solution (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. RNA
concentrations were determined by spectrophotometer (NanoDrop™
1000; NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE,
USA). Subsequently, 2.0 µg RNA was used for cDNA synthesis by
Moloney Murine Leukemia Virus reverse transcriptase (Promega
Corporation, Madison, WI, USA) according to the manufacturer's
protocol, and PCR analysis was performed using the 2X EasyTaq PCR
SuperMix kit (cat. no. AS111-03; Beijing Transgen Biotech Co.,
Ltd., Beijing, China) according to the manufacturer's protocol. The
thermocycling conditions for qPCR were: An initial denaturation at
95°C for 120 sec, followed by 40 cycles of 95°C for 15 sec and 60°C
for 30 sec for primer annealing and elongation, ending with a
melting curve step of 65°C to 95°C with an increment of 0.5°C/5
sec. β-actin was used as an internal standard for quantification.
The relative expression levels of genes were calculated using the
2−ΔΔCq method (18).
For statistical analysis, at least three biological and technical
replicates were performed. The following primers were used for
detection of PRNCR1-2 expression: Forward, 5-CCTTTCCCTCATGACCCAGT-3
and reverse, ATTGGTGTGAGGGGAGTCTG. β-Actin: Forward,
CATGTACGTTGCTATCCAGGC and reverse, CTCCTTAATGTCACGCACGAT.
Cell transfection
For knockdown of PRNCR1-2 expression, breast cancer
cells were digested with trypsin solution, seeded into 6-well
plates and cultured overnight at 37°C in an atmosphere containing
5% CO2. Once cells reached 80% confluence, they were
washed twice with PBS and added to 1.5 ml fresh basal DMEM.
Subsequently, the cells were mixed with 10 µl 20 µM small
interfering RNA (siRNA; sequence: 5′-CCATTAAGCTTGAGGCAAT-3′;
5′-ATTGCCTCAAGCTTAATGG-3′), or negative control siRNA (sequence:
5′-UUCUCCGAACGUGUCACGUTT-3′; 5′-ACGUGACACGUUCGGAGAATT-3′),
synthesized by Guangzhou Forevergen Biosciences Co., Ltd.,
Guangzhou, China and dissolved in 250 µl Gibco™ Opti-MEM (Gibco;
Thermo Fisher Scientific, Inc.). The cells were then incubated with
5 µl Lipofectamine® 2000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), which was pre-mixed
with 250 µl Opti-MEM, at 37°C for 4 h in an atmosphere containing
5% CO2. Finally, cells were cultured in fresh DMEM 48 h
prior to the subsequent assays.
Cell proliferation assay
Proliferation rates of breast cancer cells were
determined using the MTS method. A single-cell suspension was
prepared by digesting breast cancer cells with trypsin solution,
followed by cell counting. Cell density was adjusted to
3×104 cells/ml, and cells were seeded into 96-well
plates (100 µl/well). MTS solution (10 µl; cat. no. ab197010;
Abcam, Cambridge, UK) was added to the cultured breast cancer cells
at a 1:10 ratio, and cells were incubated at 37°C for 4 h in an
atmosphere containing 5% CO2. Finally, cell
proliferation rates were measured by detecting the absorbance at
490 nm using a microplate reader. At least three biological repeats
were performed for statistical analysis.
Cell migration and invasion
assays
To evaluate breast cancer cell migration, cultured
cells were collected by trypsin digestion and centrifugation at 100
× g for 1 min at room temperature, diluted in serum-free medium to
obtain 1×105 cells/ml, and a 100-µl cell suspension was
added to the upper chambers of a Transwell system. The lower
chambers of the Transwell system were filled with 600 µl fresh
DMEM. After being cultured under normal conditions for 48 h,
migrated cells were fixed with 4% paraformaldehyde for 15 min,
stained with 1% crystal violet for 10 min both at room temperature,
washed with PBS, and finally observed and counted under an inverted
fluorescence microscope (Leica Microsystems GmbH, Wetzlar,
Germany). To assess cell invasion, Matrigel matrix (Corning
Incorporation, Corning, NY, USA) was mixed with DMEM at a ratio of
1:3, and was added to the Transwell chambers and incubated at 37°C
for 2 h. Cells were then cultured for 48 h at 37°C, fixed with 4%
paraformaldehyde for 15 min, stained with 1% crystal violet for 10
min, both at room temperature, and finally analyzed under a
microscope.
Cell cycle progression analysis
Breast cancer cells (80% confluence) were washed
three times with PBS, and a single-cell suspension was generated
through trypsin digestion. Subsequently, ~1×106 cells
were fixed with 70% ethanol at −20°C overnight. Cells were then
collected by centrifugation at 500 × g for 5 min and were washed
twice with PBS. Cell precipitates were then resuspended in 500 ml
Cell Cycle staining buffer (cat. no. F559763; Guangzhou Forevergen
Biosciences Co., Ltd.), incubated at 37°C for 30 min, and finally
detected by flow cytometry (Sysmex Partec GmbH, Görlitz, Germany).
Three biological replicates were performed for statistical
analysis.
Cell apoptosis assay
Cell apoptosis was determined using the Annexin
V-allophycocyanin/7-aminoactinomycin D (7-AAD) Apoptosis
Detection kit (cat. no. 40309ES20; Yeasen, Shanghai, China),
according to the manufacturer's protocol. Breast cancer cells were
collected by digestion with EDTA-free trypsin solution and were
centrifuged at 800 × g for 5 min. Cell precipitates were then
resuspended in 1X binding buffer, and cell density was adjusted to
1×106 cells/ml. Finally, 100 µl cells were incubated
with 5 µl Annexin V and 5 µl 7-AAD in the dark for 15 min at room
temperature, incubated with 400 µl binding buffer for 1 h, and
analyzed by flow cytometry (Sysmex Partec GmbH). Data from three
biological repeats were used for statistical analysis.
Western blotting
Protein was extracted using RIPA Lysis Buffer
(Beyotime Institute of Biotechnology, Haimen, China). The
concentration was determined using the Bicinchoninic Acid Kit for
Protein Determination (Sigma-Aldrich; Merck KGaA). Total proteins
were extracted from breast tissues and breast cancer cell lines for
immunoblotting. Briefly, 35 µg total proteins were boiled at 100°C
for 5 min, loaded and separated by 12% SDS-PAGE, and electroblotted
onto polyvinylidene fluoride (PVDF) membranes (EMD Millipore,
Billerica, MA, USA). PVDF membranes were then blocked with 5%
lipid-free milk solution for 1 h at room temperature, and incubated
with primary antibodies against phosphorylated (p)-checkpoint
kinase 2 p-CHK2 (cat. no. ab59408; 1:1,000), CHK2 (cat. no.
ab47433; 1:1,000), p-protein kinase B p-AKT (cat. no. ab38449;
1:500) and AKT (cat. no. ab179463; 1:500; all Abcam) at room
temperature for 1 h. The membranes were then washed three times
with PBS, incubated with horseradish peroxidase-conjugated
secondary antibodies (cat. no. ab6940; 1:10,000; Abcam) at room
temperature for 1 h, and finally detected with enhanced
chemiluminescence solutions (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). GAPDH (cat. no. ab181602; 1:10,000; Abcam) was
used as an internal standard, and three biological repeats were
performed. Signals were densitometrically assessed using Quantity
One® software version 4.5 (Bio Rad Laboratories, Inc.,
Hercules, CA, USA).
Statistical analysis
Data were presented as mean ± standard error of the
mean. The present findings were statistically analyzed using SPSS
18.0 software package (SPSS, Inc., Chicago, IL, USA) via Student's
t-test or one way analysis of variance, least significant
difference test was used to analyze the differences among more than
two groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
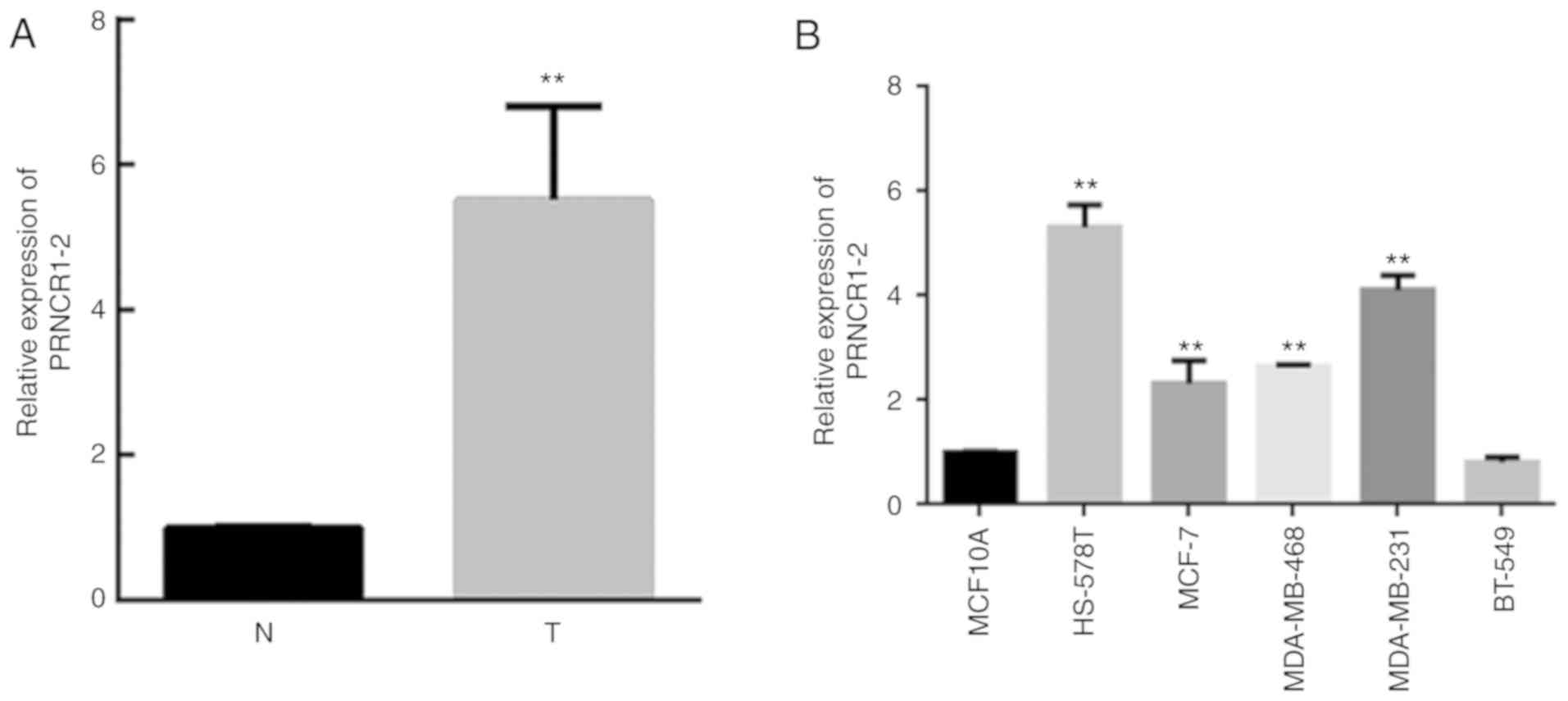
PRNCR1-2 expression is elevated in
breast cancer
To identify a novel lncRNA associated with breast
cancer pathogenesis, the expression levels of PRNCR1-2 were
analyzed in 20 pairs of breast cancer and adjacent normal tissues
(Fig. 1A). Compared with the
normal tissues, the expression levels of PRNCR1-2 were
significantly elevated in breast cancer tissues (P<0.05;
Fig. 1A). For further validation,
PRNCR1-2 expression was confirmed in five breast cancer cell lines
and a mammary epithelial cell line; PRNCR1-2 were successfully
overexpressed in the cancer cell lines HS-587T, MCF-7, MDA-MB-468
and MDA-MB-231 compared with the mammary epithelial cell line
MCF10A, but failed to overexpress in the cancer cell line BT-549
(Fig. 1B). The upregulation of
PRNCR1-2 expression in breast cancer tissues suggested that
PRNCR1-2 may have pathogenic roles in breast cancer. Due to the
high expression level of PRNCR1-2 in HS-587T and MDA-MB-231 cells,
these two cell lines were chosen for the following studies.
PRNCR1-2 depletion in HS-587T and
MDA-MB-231 cells
To investigate the potential roles of the lncRNA
PRNCR1-2 in breast cancer pathogenesis, HS-587T and MDA-MB-231
breast cancer cells were transfected with a specific siRNA against
PRNCR1-2. It was revealed that transfection with siRNA-PRNCR1-2
markedly reduced PRNCR1-2 expression in HS-587T and MDA-MB-231
cells, compared with in the negative control group (Fig. 2A and B). siRNA-PRNCR1-2-transfected
HS-587T and MDA-MB-231 cells were used for subsequent functional
assays.
PRNCR1-2 regulates breast cancer cell
proliferation, migration and invasion
To explore the possible roles of PRNCR1-2 in breast
cancer initiation and progression, the proliferation, migration,
invasion, cell cycle progression and apoptosis of
siRNA-PRNCR1-2-transfected HS-587T and MDA-MB-231 cells were
analyzed. The results of the MTS assay revealed that the
proliferation rates of siRNA-PRNCR1-2-transfected HS-587T cells
were markedly downregulated compared with in the negative control
group (Fig. 3A), thus indicating
the ability of PRNCR1-2 to modulate breast cancer cell
proliferation. Furthermore, the migration and invasion of
PRNCR1-2-depleted HS-587T cells were analyzed using Transwell
assays; the migration and invasion of HS-587T cells were markedly
suppressed by transfection with siRNAs targeting PRNCR1-2 (Fig. 3B). Consistent with PRNCR1-2
depletion-induced alterations in proliferation, the cell cycle
progression of siRNA-PRNCR1-2-transfected HS-587T cells was
markedly altered, with more cells arrested in S phase, thus
indicating the involvement of PRNCR1-2 in promoting S/G2
transition of breast cancer cells (Fig. 3C). The proportion of apoptotic
cells in the siRNA-PRNCR1-2 group was also analyzed by flow
cytometry; however, no marked alterations in cell apoptosis were
observed in siRNA-PRNCR1-2-transfected HS-587T cells (Fig. 3D). The aforementioned experiments
were repeated in MDA-MB-231 cells and similar results were
obtained, as shown in Fig. 4.
Taken together, these cellular assays suggested that PRNCR1-2 may
be involved in breast cancer progression by promoting cell
proliferation, migration and invasion, and by modulating cell cycle
progression, but not via modulation of breast cell apoptosis.
Signaling in HS-587T and MDA-MB-231
cells is modulated by PRNCR1-2 depletion
For more insights into PRNCR1-2-induced regulation
of cell functions during breast cancer progression, key signaling
pathway components associated with cell proliferation, migration,
invasion, cell cycle progression and apoptosis were detected in
HS-587T and MDA-MB-231 cells transfected with siRNA-PRNCR1-2 by
western blotting. The results demonstrated that the protein
expression levels of p-CHK2 were significantly elevated in
siRNA-PRNCR1-2-transfected HS-587T and MDA-MB-231 cells (Fig. 5). Furthermore, depletion of
PRNCR1-2 expression using specific siRNA resulted into significant
inhibition of p-AKT in HS-587T and MDA-MB-231 cells; however,
PRNCR1-2 depletion had no effect on the abundance of total CHK2 and
AKT proteins (Fig. 5). The
selectively altered expression of key signaling components
suggested that regulation of breast cancer cell proliferation,
migration, invasion and cell cycle progression by PRNCR1-2 may be
at least partially mediated by modulating phosphorylation of CHK2
and AKT.
Discussion
Non-coding components that account for large volumes
of the human genome have long been predicted to have important
roles in various physiological and pathological processes (19). Various types of ncRNAs, including
microRNAs, siRNAs, Piwi-interacting RNAs, small nucleolar RNAs and
lncRNAs critically contribute to cancer initiation and progression,
much more than previously expected (8,10–12,20).
Increasing evidence has suggested that lncRNAs may be involved in
tumor pathogenesis, thus promoting trials of their application in
breast cancer diagnosis and prognosis. The lncRNA H19 is
characterized as a highly expressed biomarker in ER-positive MCF-7
breast cancer cells, which is involved in breast cell survival and
estrogen-induced cell proliferation during breast cancer
development (21). In addition,
its diagnostic value as a novel biomarker for breast cancer has
been reported, due to its detectable expression in the urine and
serum of patients with breast cancer (22,23).
Accumulating numbers of such ncRNAs as candidate targets for cancer
diagnosis and treatment highlight the importance of further
characterization of novel ncRNAs in breast cancer pathogenesis.
Aiming to provide a novel candidate for breast
cancer diagnosis and treatment, the present study analyzed the
expression levels of the lncRNA PRNCR1-2 in cancer tissues
collected from patients with breast cancer. PRNCR1-2 expression was
significantly increased in cancer tissues, thus suggesting its
potential association with breast cancer development. Subsequently,
a knockdown assay was conducted to deplete PRNCR1-2 expression in
breast cancer cells. The marked alterations in breast cancer cell
proliferation, migration, invasion and cell cycle progression
following suppression of PRNCR1-2 expression indicated that
PRNCR1-2 may be considered a novel non-coding regulator in breast
cancer pathogenesis. Significantly altered AKT and CHK2
phosphorylation further validated the critical roles served by
PRNCR1-2 in breast cancer cells. In accordance with previous
findings regarding lncRNAs in breast cancer (8), the present results revealed the
cellular functions of PRNCR1-2 in breast cancer cells, which may be
applied as a biomarker for breast cancer diagnosis and therapy.
The molecular mechanisms by which PRNCR1-2 promotes
breast cancer cell proliferation, migration and cell cycle
progression deserve further investigations. Previous studies
reported that interactions of lncRNAs with proteins or RNA partners
may be important for the functioning of lncRNA molecules (10,24).
A recent study also demonstrated that lncRNAs can bind with Janus
kinase 2 (JAK2), thus promoting JAK2 activation and signal
transducer and activation of transcription 3 phosphorylation,
finally mediating breast cancer brain metastases (25). Therefore, it is reasonable to
speculate that PRNCR1-2 may also carry out its role in breast
cancer cell proliferation and migration via its association with
key protein components of these cellular processes. The large-scale
identification of proteins or RNA partners interacting with
PRNCR1-2 during breast cancer development may provide novel
perspectives on pathogenic roles of non-coding molecules in breast
cancer, as well as other types of human cancer. It is also possible
that PRNCR1-2 may be directly involved in the post-translational
modifications of key signaling proteins. Metastasis associated in
lung adenocarcinoma transcript 1 (MALAT1) interacts with both
serine and arginine-rich splicing factor 1 (SRSF1) and SRSF protein
kinase 1. In colorectal cancer cells, the lncRNA MALAT1 regulates
cancer cell proliferation and migration by promoting
phosphorylation of SRSF1 (26). In
the present study, it was revealed that depletion of PRNCR1-2
markedly altered the phosphorylation of AKT and CHK2, whereas total
AKT and CHK2 protein levels were not affected. CHK2 acts as an
important regulator of cell cycle progression and proliferation
(27,28), and CHK2 signaling is activated by
phosphorylation of itself and downstream substrates (29). AKT is another key regulator of
tumor cell proliferation, cell cycle progression, migration and
invasion (30–32), which is also activated by its
phosphorylation (33). These
observations suggested that PRNCR1-2 may modulate the
phosphorylation of CHK2 and AKT by interacting with a specific
kinase or phosphatase. The role of PRNCR1-2 in CHK2 and AKT
phosphorylation and regulation requires further investigation.
In conclusion, this study reported that the lncRNA
PRNCR1-2 is highly expressed in breast cancer tissues, and
depletion of PRNCR1-2 in breast cancer cells results in the
suppression of cell proliferation, migration, invasion and cell
cycle progression. These findings indicated that PRNCR1-2 may be
explored as a breast cancer biomarker for the development of novel
diagnostic or therapeutic methods for patients with breast
cancer.
Acknowledgements
The present study was supported by Guangzhou
Forevergen Company. The authors would also like to thank Dr Kewei
Li of Foshan First People's Hospital for the specimen
collection.
Funding
The present study was supported by grants from the
Specialized Research Fund for the Technology Innovation of Foshan
City (grant no. 2014AG10003) and the Research Fund for the
Guangdong Province Health Bureau (grant no. A2017464).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DP and QH conceived and designed the study, and
drafted and critically revised the manuscript. XL and YXL performed
the experiments and analyzed the data. HD, SC, YDL, LL, FPe and FPa
participated in study design, study implementation and manuscript
revision. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The experimental procedures were approved by the
Ethics Committee of Foshan First People's Hospital, and written
informed consent was obtained from each participant prior to
surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Desantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bombonati A and Sgroi DC: The molecular
pathology of breast cancer progression. J Pathol. 223:307–317.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lopez-Garcia MA, Geyer FC, Lacroix-Triki
M, Marchió C and Reis-Filho JS: Breast cancer precursors revisited:
Molecular features and progression pathways. Histopathology.
57:171–192. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Byler S, Goldgar S, Heerboth S, Leary M,
Housman G, Moulton K and Sarkar S: Genetic and epigenetic aspects
of breast cancer progression and therapy. Anticancer Res.
34:1071–1077. 2014.PubMed/NCBI
|
|
6
|
Welch DR and Wei LL: Genetic and
epigenetic regulation of human breast cancer progression and
metastasis. Endocrine-Related Cancer. 5:155–197. 1998. View Article : Google Scholar
|
|
7
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lo PK, Wolfson B, Zhou X, Duru N,
Gernapudi R and Zhou Q: Noncoding RNAs in breast cancer. Brief
Funct Genomics. 15:200–221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao J: The functional role of long
non-coding RNAs and epigenetics. Biol Proced Online. 16:112014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin C and Yang L: Long noncoding RNA in
cancer: Wiring signaling circuitry. Trends Cell Biol. 28:287–301.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Slaby O, Laga R and Sedlacek O:
Therapeutic targeting of non-coding RNAs in cancer. Biochem J.
474:4219–4251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huarte M: The emerging role of lncRNAs in
cancer. Nature Medicine. 21:1253–1261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sørensen KP, Thomassen M, Tan Q, Bak M,
Cold S, Burton M, Larsen MJ and Kruse TA: Long non-coding RNA
HOTAIR is an independent prognostic marker of metastasis in
estrogen receptor-positive primary breast cancer. Breast Cancer Res
Treat. 142:529–536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu
M and Mo YY: Long non-coding RNA UCA1 promotes breast tumor growth
by suppression of p27 (Kip1). Cell Death Dis. 5:e10082014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Prensner JR, Sahu A, Iyer MK, Malik R,
Chandler B, Asangani IA, Poliakov A, Vergara IA, Alshalalfa M,
Jenkins RB, et al: The lncRNAs PCGEM1 and PRNCR1 are not implicated
in castration resistant prostate cancer. Oncotarget. 5:1434–1438.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang L, Qiu M, Xu Y, Wang J, Zheng Y, Li
M, Xu L and Yin R: Upregulation of long non-coding RNA PRNCR1 in
colorectal cancer promotes cell proliferation and cell cycle
progression. Oncol Rep. 35:318–324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sattarifard H, Hashemi M, Hassanzarei S,
Narouie B and Bahari G: Association between genetic polymorphisms
of long non-coding RNA PRNCR1 and prostate cancer risk in a sample
of the Iranian population. Mol Clin Oncol. 7:1152–1158.
2017.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Esteller M: Non-coding RNAs in human
disease. Nature Reviews Genetics. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jing H, Markowitz GJ and Wang X: Noncoding
RNAs regulating cancer signaling network. Adv Exp Med Biol.
927:297–315. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun H, Wang G, Peng Y, Zeng Y, Zhu QN, Li
TL, Cai JQ, Zhou HH and Zhu YS: H19 lncRNA mediates
17β-estradiol-induced cell proliferation in MCF-7 breast cancer
cells. Oncol Rep. 33:3045–3052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang K, Luo Z, Zhang Y, Zhang L, Wu L,
Liu L, Yang J, Song X and Liu J: Circulating lncRNA H19 in plasma
as a novel biomarker for breast cancer. Cancer Biomark. 17:187–194.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang K, Zhang Y, Luo Z, Lichun WU, Zhang
L and Liu J: Diagnostic value of urinary lncRNA H19 for breast
cancer. Shandong Med J. 56:42–44. 2016.
|
|
24
|
Lin A, Hu Q, Li C, Xing Z, Ma G, Wang C,
Li J, Ye Y, Yao J, Liang K, et al: The LINK-AlncRNA interacts with
PtdIns(3,4,5)P3 to hyperactivate AKT and confer
resistance to AKT inhibitors. Nature Cell Biol. 19:238–251. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang S, Liang K, Hu Q, Li P, Song J, Yang
Y, Yao J, Mangala LS, Li C, Yang W, et al: JAK2-binding long
noncoding RNA promotes breast cancer brain metastasis. J Clin
Invest. 127:4498–4515. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu ZY, Wang XY, Guo WB, Xie LY, Huang YQ,
Liu YP, Xiao LW, Li SN, Zhu HF, Li ZG and Kan H: Long non-coding
RNA MALAT1 increases AKAP-9 expression by promoting SRPK1-catalyzed
SRSF1 phosphorylation in colorectal cancer cells. Oncotarget.
7:11733–11743. 2016.PubMed/NCBI
|
|
27
|
Ingvarsson S, Sigbjornsdottir BI, Chen H,
Hafsteinsdottir SH, Ragnarsson G, Barkardottir RB, Arason A,
Egilsson V and Bergthorsson JT: Mutation analysis of the CHK2 gene
in breast carcinoma and other cancers. Breast Cancer Res. 4:1–6.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sullivan A, Yuille M, Repellin C, Reddy A,
Reelfs O, Bell A, Dunne B, Gusterson BA, Osin P, Farrell PJ, et al:
Concomitant inactivation of p53 and Chk2 in breast cancer.
Oncogene. 21:1316–1324. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu X, Tsvetkov LM and Stern DF: Chk2
activation and phosphorylation-dependent oligomerization. Mol Cell
Biol. 22:4419–4432. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hartmann W, Digonsöntgerath B, Koch A,
Waha A, Endl E, Dani I, Denkhaus D, Goodyer CG, Sörensen N,
Wiestler OD and Pietsch T: Phosphatidylinositol 3′-kinase/AKT
signaling is activated in medulloblastoma cell proliferation and is
associated with reduced expression of PTEN. Clin Cancer Res.
12:3019–3027. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim D, Kim S, Koh H, Yoon SO, Chung AS,
Cho KS and Chung J: Akt/PKB promotes cancer cell invasion via
increased motility and metalloproteinase production. FASEB J.
15:1953–1962. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chang F, Lee JT, Navolanic PM, Steelman
LS, Shelton JG, Blalock WL, Franklin RA and McCubrey JA:
Involvement of PI3K/Akt pathway in cell cycle progression,
apoptosis, and neoplastic transformation: A target for cancer
chemotherapy. Leukemia. 17:590–603. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jacinto E, Facchinetti V, Liu D, Soto N,
Wei S, Jung SY, Huang Q, Qin J and Su B: SIN1/MIP1 maintains
rictor-mTOR complex integrity and regulates Akt phosphorylation and
substrate specificity. Cell. 127:125–137. 2006. View Article : Google Scholar : PubMed/NCBI
|