Introduction
According to recent estimates from the International
Agency for Research on Cancer of the World Health Organization, the
total number of lung cancer cases represents 20% in all malignant
tumors, and the mortalities account for 25.4% of all
cancer-associated mortalities (1).
In China, with the increases in the size of the aging population,
changes in lifestyle, and economic and environmental factors, the
incidence and mortality rate of lung cancer are increasing
consistently (2). In all patients
with lung cancer, non-small cell lung cancer (NSCLC) represents
>80% cases (3). NSCLC is not
easily diagnosed. The majority of patients are diagnosed with mid-
and late-stage disease and are not suitable for surgical treatment.
Although medical technology has improved in previous years, the
early diagnosis, comprehensive management and prognosis of patients
with NSCLC remain unsatisfactory. The use of effective diagnostic
and therapeutic methods for early detection and early treatment
have become an important issue in the control of NSCLC.
MicroRNAs (miRNAs) are non-coding RNAs measuring
20–22 nucleotides in length and are highly conserved among species.
There are a number of miRNAs (miRNAs) in the human genome involved
in various cellular processes including differentiation,
proliferation, apoptosis, and tumorigenesis and progression
(4). In previous years, the
association between microRNA (miRNA) and lung cancer, in particular
NSCLC, has become a focus of a number of studies: For example,
miR-210 (5), miR-26a (6), and miR-212 (7) in lung cancer have been demonstrated
to serve as oncogenes. By contrast, miR-1 (8), miR-126 (9), and miR-149 (10) serve as tumor suppressors in lung
cancer. miR-512-5p is the 5′terminal of miR-512 precursor. It was
demonstrated that miR-512-5p level was increased in several tumor
types (11,12). A previous study revealed that
miR-512-5p may induce apoptosis and inhibit glycolysis by targeting
cyclin-dependent kinase inhibitor 1 (p21) in NSCLC cells (13). However, its roles in NSCLC have not
yet been fully understood.
E26 transformation specific-1 (ETS1) is a type of
proto-oncogene that is highly expressed in a variety of
malignancies (14). ETS1
participates in tumor invasion, metastasis, angiogenesis,
proliferation, differentiation and anti-apoptosis activities by
regulating the expression of multiple genes (15–17).
Dysregulation of ETS1 is frequent in NSCLC, but its regulation
mechanism in NSCLC tumorigenesis remains unknown.
In the present study, the role of miR-512-5p in the
development of NSCLC was examined; the results revealed that
miR-512-5p was overexpressed in tumor cells and tissues, that it
suppressed cell proliferation and invasion, induced apoptosis, and
decreased ETS1 expression. Furthermore, it was confirmed that the
miR-152-5p may inhibit ETS1 expression via targeting its 3′-UTR,
and then induce the apoptosis pathway and activate the intrinsic
invasion pathway.
Materials and methods
Human tissue samples
The study was known and approved by the Ethics
Committee of the Nanjing Gulou Hospital (Nanjing, China), Jiangsu
Provincial Hospital of Traditional Chinese Medicine (Nanjing,
China) and Affiliated Hospital of Xuzhou Medical College (Xuzhou,
China). Clinical samples were collected from 181 patients with
NSCLC between June 2014 and October 2017, and informed consent was
granted. All patient diagnoses of NSCLC had been finally confirmed,
and none of them patients had received any treatment.
Cell culture and transfection
Human NSCLC A549 and H1299 cell lines and human
normal lung 16HBE cell lines were obtained from the Chinese Academy
of Science Cell Bank (Shanghai, China), which were cultured in
RPMI-1640 medium (Hyclone; GE Healthcare Life Sciences, Logan, UT,
USA) supplemented with 10% fetal bovine serum (Biowest; VWR
International Eurolab S.L., Barcelona, Spain) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) at 37°C in 5% CO2. miR-512-5p mimics, miR-512-5p
inhibitor, negative control (NC) miRNA mimic (AM17110), and NC
miRNA inhibitor (AM17010) were purchased from Ambion; Thermo Fisher
Scientific, Inc. Small interfering (si)RNA against human ETS1 mRNA
and the control siRNA were synthesized by Guangzhou RiboBio Co.,
Ltd. (Guangzhou, China). Transfection was performed with
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. After
12–48 h, the subsequent experiments were performed. Briefly,
3×105 cells were cultured in 6 well plates. Subsequent
to reaching 60–68% confluence, cells were transfected with miRNA
(100 nM) or siRNA (50 nM).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis
In tissue and cell, total RNA was extracted with
TRIzol® (Thermo Fisher Scientific, Inc.), and miRNA was
extracted with miRcute miRNA Isolation kit (Tiangen Biotech, Co.,
Ltd., Beijing, China). Expression of miR-512-5p was analyzed by
RT-qPCR using the TaqMan miR kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.), and expression of ETS1 mRNA was detected using
PrimeScript™ reagent kit (TAKARA, Japan). U6 and GAPDH was used as
the control for miRNA and mRNA, respectively. Data were acquired
using a HT-7900 TaqMan instrument (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The reaction consisted of a hot start (10
min at 95°C), followed with 40 cycles of 15 sec at 95°C and 60 sec
at 60°C; the 2−ΔΔCq method was used (18). Each sample was detected in
triplicate. The PCR primers for miR-512-5p were forward,
5′-CGσGCGGCACTCAGCCTTGAGGG-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′.
The PCR primers for ETS1 were forward, 5′-AGCCGACTCTCACCATCATC-3′
and reverse, 5′-CAAGGCTTGGGACATCATTT-3′. The PCR primers for U6
were forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. The PCR primers for GAPDH were forward,
5′-CTTAGATTTGGTCGTATTGG-3′ and reverse,
5′-GAAGATGGTGATGGGATT-3′.
Cell proliferation assay
Cell proliferation was detected using a Cell
Counting Kit (CCK)-8 (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan), and the assay was performed according to the
protocol of the manufacturer. Briefly, cells were cultured in
96-well plates at 5,000 cells/well, and they were incubated at 37°C
for 48 h following transfection. A total of 10 µl CCK-8 solution
per well was added for 2 h, and then examined. Viable cell numbers
were detected at 450 nm with a Microplate Reader ELx808 (Bio-Tek
Instruments, Inc., Winooski, VT, USA). Each experiment was
performed in sextuplicate.
To confirm the CCK-8 assay results, a
5-ethynyl-2′-deoxyuridine (EdU) assay was also performed to measure
cell proliferation. A total of 1,000 cells/well in a 96-well plate
were serum-starved for 24 h following transfection, and the cells
cultured for a further 24 h. At 2 h prior to cell collection, EdU
was labeled with EdU-labeling reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), which was then detected at a wavelength of 555
nm with a Click-iT assay kit (C10638; Thermo Fisher Scientific,
Inc.) and images of ≥4 fields per treatment condition captured
(Leica Microsystems GmbH, Wetzlar, Germany). The percentage of
cells in S phase was determined using ImageJ software version 1.45
(National Institutes of Health, Bethesda, MD, USA). Each data point
represents the average percentage of labeled cells among the 4
images.
Cell apoptosis assay
Cell apoptosis was detected by the fluorescein
isothiocyanate (FITC)-Annexin V/propidium iodide (PI) Apoptosis
Detection kit (BD Pharmingen; BD Biosciences, San Jose, CA, USA)
according to the manufacturer's protocol. All cells were collected
for apoptosis analysis at 48 h post-transfection. Then, cells were
stained for 5 min at room temperature with Annexin V-FITC (5 ml)
and PI (5 ml). Cell apoptosis was analyzed by a flow cytometer
(Beckman Coulter, Inc., Brea, CA, USA). FlowJo version 7.6 (FlowJo
LLC, Ashland, OR, USA) was used to calculate the apoptosis rate.
All analyses were performed in triplicate.
Wound healing assay
The cells were seeded with 5×105 into
6-well plates, and the cells were lightly scratched using a 10 µl
sterile pipette tip in the central axis of the plate when the
density reached ~70% following transfection. Following culture for
48 h, cells were imaged with a Nikon Ts2 inverted microscope, at
×200 magnification. The extent of cell migration was quantified
using ImageJ version 1.46 (National Institutes of Health). The
percentage of wound closure was calculated as follows: [(wound area
at 0 h-wound area at 24 h)/wound area at 0 h] ×100%. A total of 3
replicates were performed.
Transwell assay
The cells were transfected for 12 h, then placed in
the upper Transwell chamber (Corning Incorporated, Corning, NY,
USA) with an insert pre-coated with Matrigel (BD Biosciences) and
incubated for 12 h at 37°C. The chambers were cultured in 24-well
plates, and RPMI-1640 containing 20% FBS was added to lower
chamber. Following incubation for 24 h at 37°C, the cells that had
penetrated across the membranes were fixed with 90% methanol for 10
min at room temperature, stained with 0.1% crystal violet for 20
min and counted under an inverted light microscope at ×200
magnification. A total of 3 replicates were obtained.
Luciferase assay
The A549 and H1299 cells were seeded onto 24-well
plates and co-transfected with either 25 nM miR-512-5p mimics or
miR-NC and 200 ng pGL3-ETS1-3′UTR or pGL3-ETS1-mut-3′UTR using
Lipofectamine® 2000 reagent (Thermo Fisher Scientific,
Inc.), and the reporter plasmids were obtained from Promega
Corporation (Madison, WI, USA). After 36 h, luciferase activity was
measured using the dual luciferase assay system (Promega
Corporation). Renilla luciferase was used to normalize luciferase
activity. All analyses were performed in triplicate.
Target prediction
For bioinformatics analysis, PicTar (http://pictar.mdc-berlin.de/), miRanda (http://www.microrna.org) and TargetScan (http://www.targetscan.org/) were used.
Western blot analysis
Following transfection, cells of 80% density were
lysed using radioimmunoprecipitation assay buffer (Beyotime
Institute of Biotechnology, Haimen, China) containing a Protease
Inhibitor Cocktail (Promega Corporation) for 30 min. The protein
was collected, and the concentrations were measured using a BCA
assay kit (Beyotime Institute of Biotechnology). Proteins (40 µg)
were then separated with 10% SDS-PAGE, and then transferred onto a
polyvinylidene fluoride membrane. Membranes were blocked at room
temperature in 5% non-fat milk for 1 h, and immunoblotted with
anti-caspase-3 (sc-271759), anti-caspase-7 (sc-365034), anti-B-cell
lymphoma-2 (Bcl-2; sc-130307), anti-Bcl-2-associated X protein
(Bax; sc-23959), anti-matrix metalloproteinase (MMP)-2 (sc-53630),
anti-MMP-9 (sc-12759), anti-phosphorylated (p)-p38
mitogen-activated protein kinase (p-p38 MAPK; sc-7973), anti-p38
MAPK (sc-81621), anti-p-c-Jun N-terminal kinases (p-JNK;
(sc-293138), anti-JNK (sc-137020), anti-p-extracellular
signal-regulated kinase (p-ERK; sc-81492), anti-ERK (sc-135900),
anti-ETS1 (sc-55581) and GAPDH (sc-47724; all Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) antibodies at 4°C overnight;
all antibodies were used at a dilution of 1:1,000. The secondary
antibodies against rabbit or mouse IgG (Beyotime Institute of
Biotechnology) were incubated for 2 h at RT. Following washing of
the membranes three times, signals were visualized with enhanced
chemiluminescence (ECL Wuhan Boster Biological Technology, Ltd.,
Wuhan, China) and analyzed using ImageJ version 1.45 (National
Institutes of Health).
Statistical analysis
Statistical analysis was performed using GraphPad
software 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) and the
results are presented as the mean ± standard deviation. Differences
between two groups were determined by Student's t-test. Comparisons
between multiple group were performed using one-way analysis of
variance with a Scheffe post-hoc test for subsequent individual
group comparisons. P<0.01 was considered to indicate a
statistically significant difference.
Results
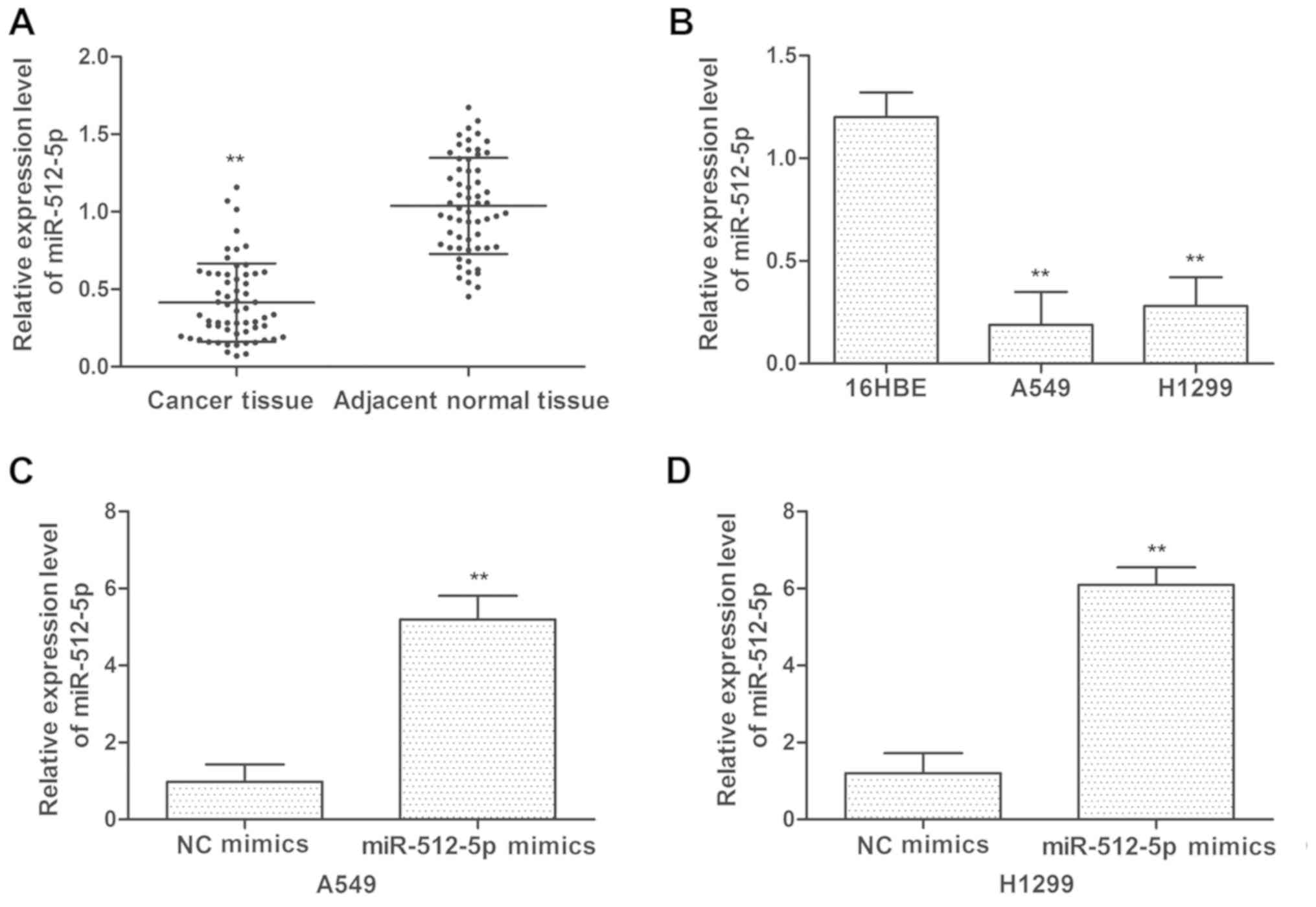
Downregulation of miR-512-5p in NSCLC
tissues and cell lines
To assess the expression levels of miR-512-5p in
NSCLC, the expression level of miR-512-5p was detected in tumor
tissues and adjacent normal tissues from patients with NSCLC by
RT-qPCR. The expression levels of miR-512-5p were decreased in
tumor tissues compared with those in adjacent normal tissues
(Fig. 1A). NSCLC A549 and H1299
cell lines exhibited decreased miR-512-5p expression compared with
the normal lung 16HBE cell line (Fig.
1B). To additionally determine the effect of miR-512-5p on
NSCLC cell lines, miR-512-5p and NC mimics were transfected into
the cells. As indicated in Fig. 1C and
D, the expression levels of miR-512-5p were increased following
transfection with miR-512-5p mimics in A549 and H1299 cells.
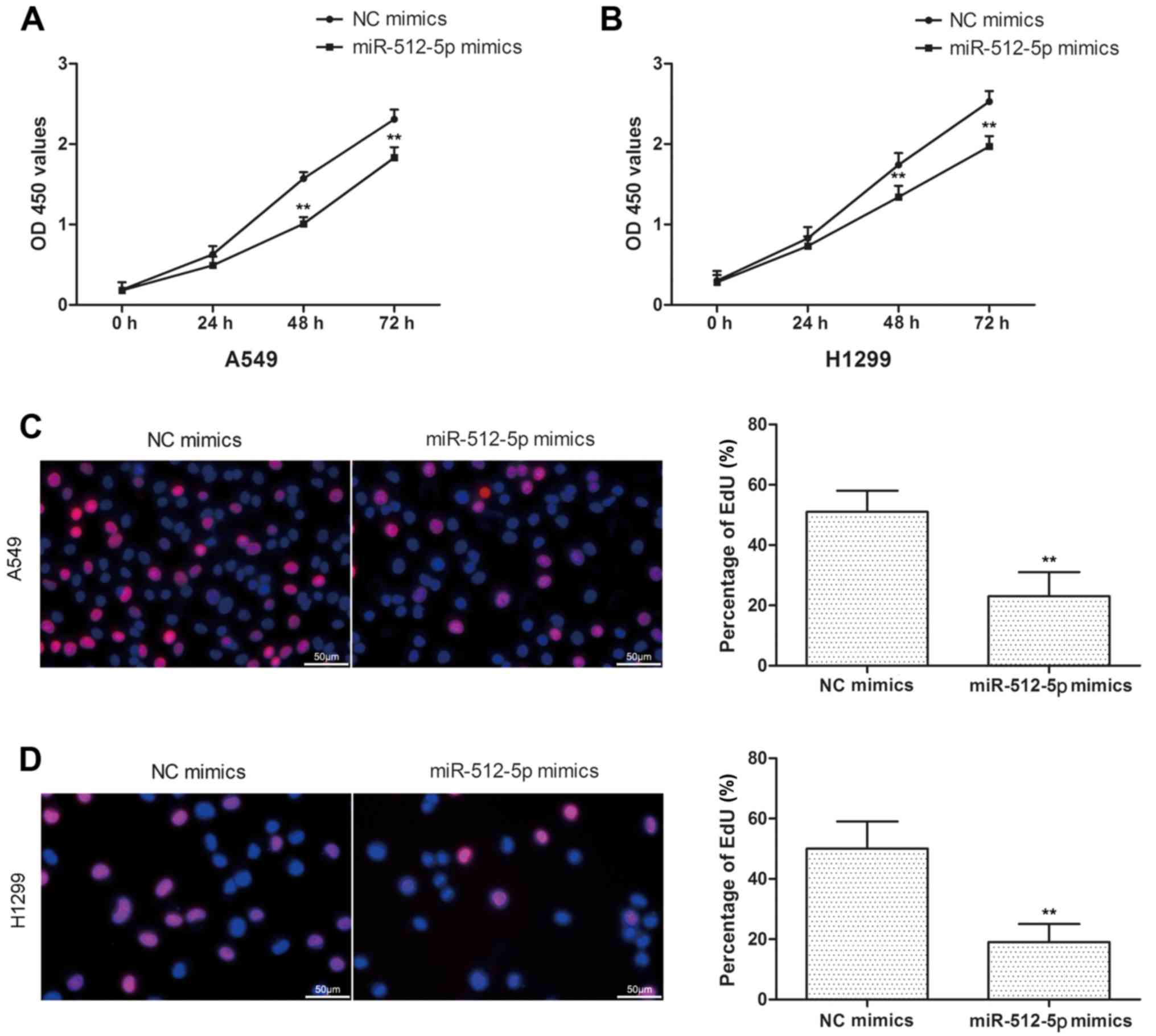
miR-512-5p suppresses NSCLC cell
proliferation and induces apoptosis in vitro
In the present study, the cell proliferation was
assessed by CCK-8 and EdU assays in A549 and H1299 cells. Compared
with the NC mimics, miR-512-5p mimics exhibited a significant
inhibition of cell proliferation in A549 and H1299 cells (Fig. 2A and B), and the results of EdU
assay were coincident with CCK-8 (Fig.
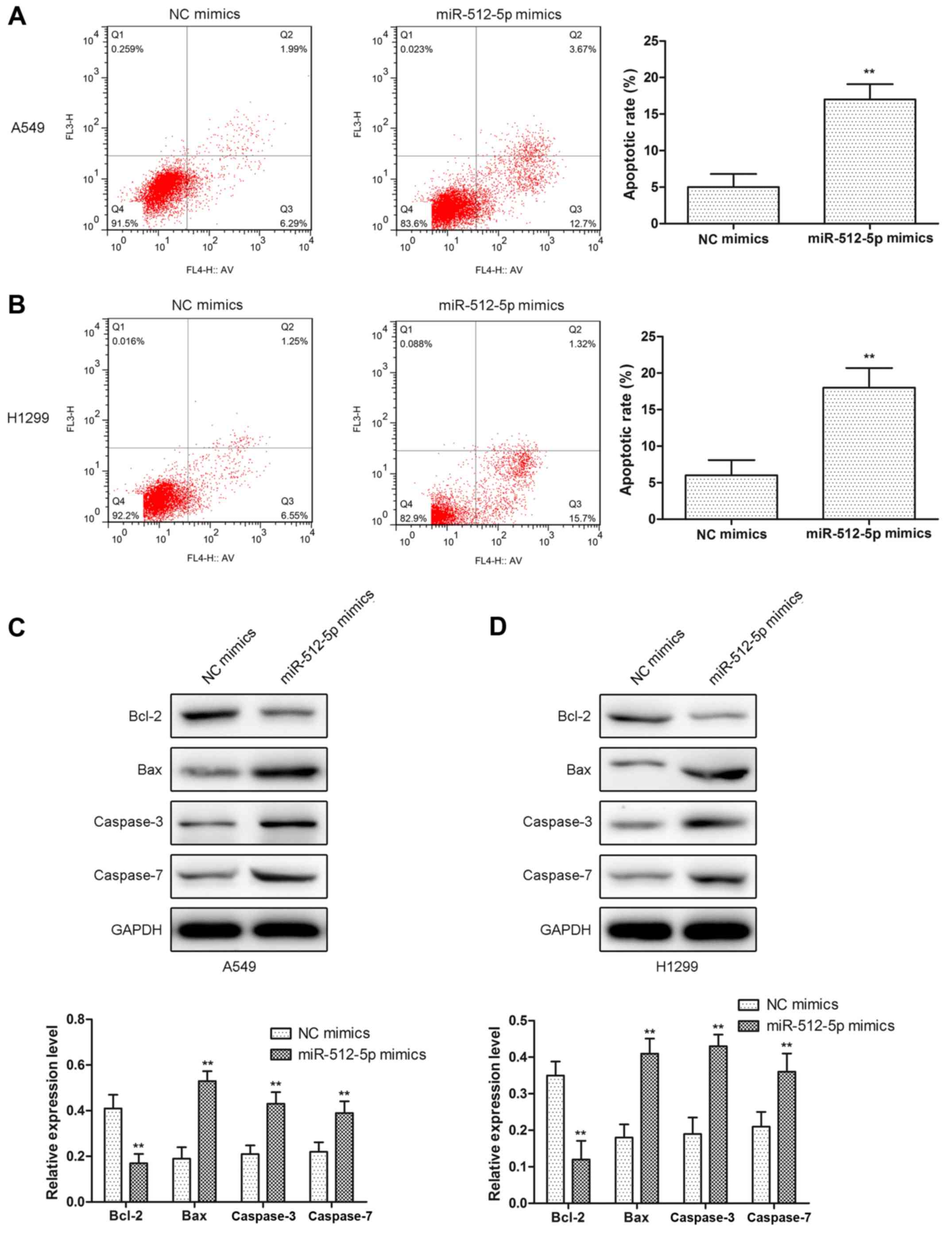
2C and D). In addition, d total apoptosis was measured, and it
was identified that the apoptosis rate was increased in miR-512-5p
mimic-treated cells (Fig. 3A and
B). Furthermore, proapoptotic proteins (Bcl-2, Bax, caspase-3
and caspase-7) were detected by western blot analysis. The results
indicated that the expression levels of Bax, caspase-3 and
caspase-7 were significantly increased, and the expression levels
of Bcl-2 were decreased (Fig. 3C and
D). Taken together, these data revealed that the overexpression
of miR-512-5p may markedly suppress cell proliferation and induced
apoptosis in NSCLC cells.
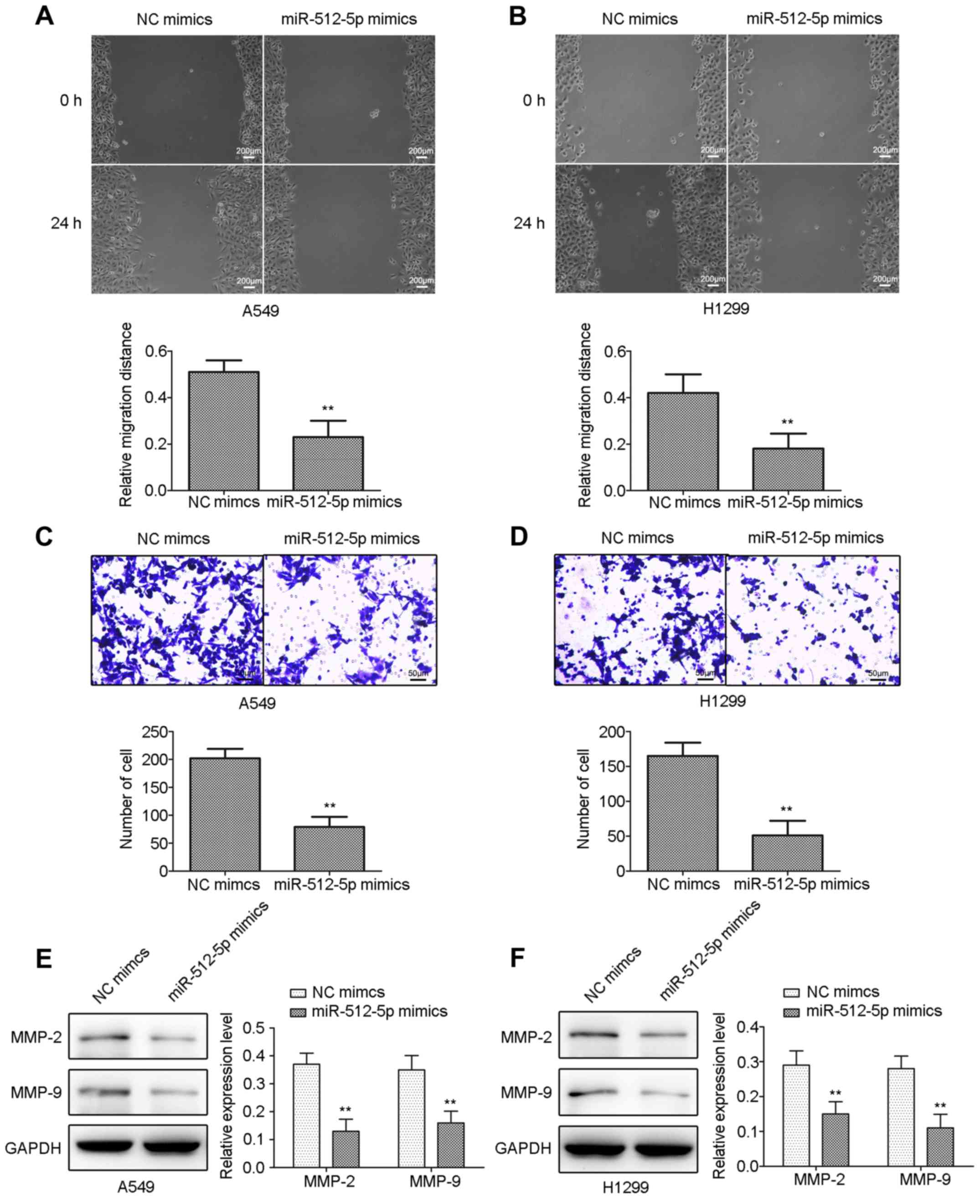
miR-512-5p inhibits the cell migratory
and invasive capacities in A549 and H1299 cells
The effect of miR-512-5p on cell migration and
invasion in A549 and H1299 cells was detected by scratch and
Transwell assays. miR-512-5p mimic-treated cells exhibited a
significant decrease in their wound-closing capacities in the
scratch assay (Fig. 4A and B), and
significantly decreased numbers of invading cells in the Transwell
assay (Fig. 4C and D) compared
with NC mimic-treated cells in A549 and H1299 cells. The results of
the western blot analysis demonstrated that the expression levels
of MMP-2 and MMP-9 were significantly inhibited in A549 and H1299
cells (Fig. 4E and F).
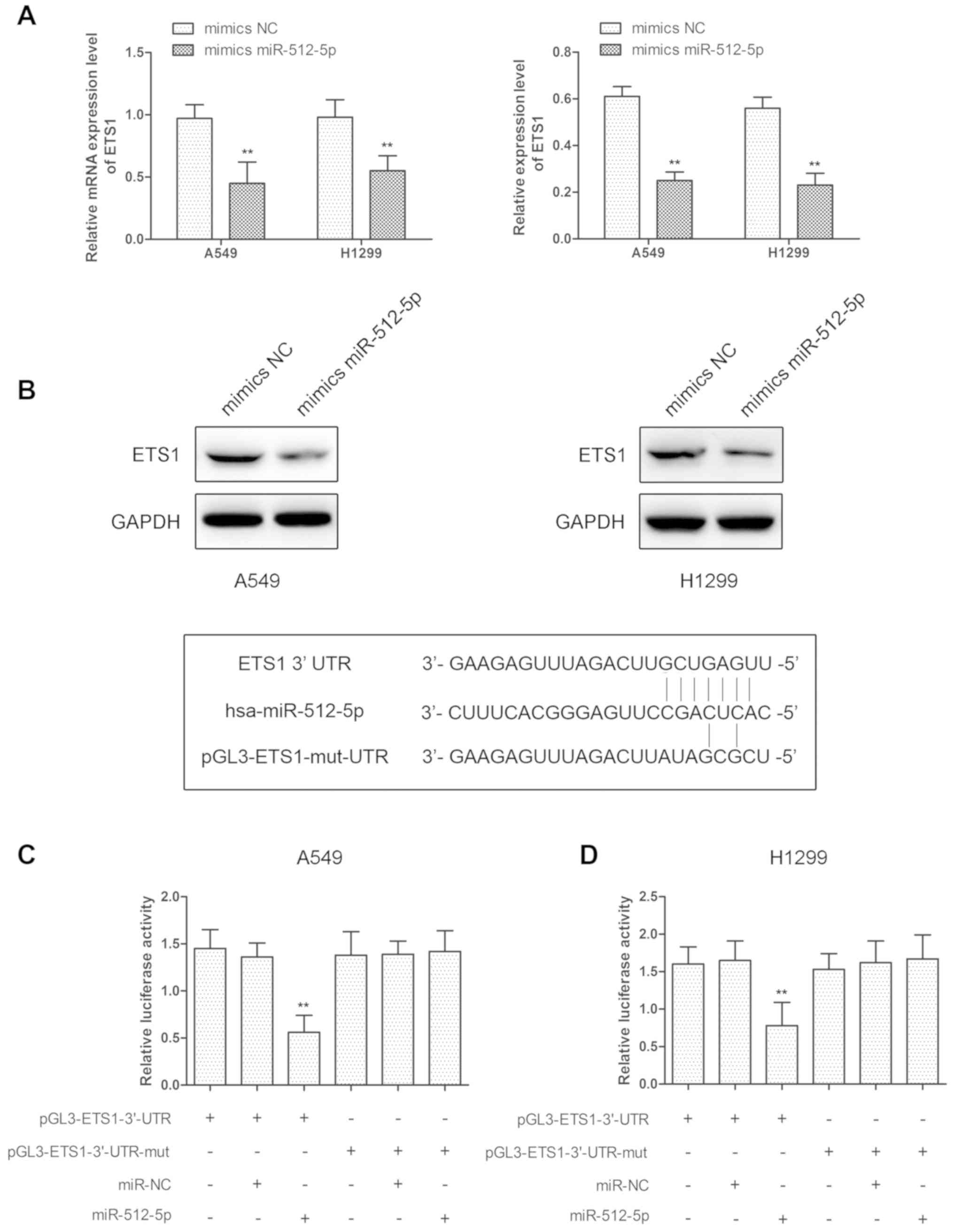
ETS1 is a target of miR-512-5p
ETS1 is important in regulating cell invasion,
metastasis, proliferation and apoptosis, and the present study
identified that ETS1 mRNA and protein levels were decreased
following transfection with miR-512-5p mimics in A549 and H1299
cells (Fig. 5A and B). We
hypothesized that ETS1 was regulated by miR-512-5p in A549 and
H1299 cells, and the ETS1 gene was predicted to have at least one
potential binding site at its 3′-UTR for miR-512-5p. To validate
the interaction between ETS1 and miR-512-5p, luciferase reporter
assays were performed using the miR-512-5p target sequences of
wild-type ETS1 and mutated ETS1 in A549 and H1299 cells. The
results indicated that overexpression of miR-512-5p significantly
decreased luciferase expression in wild-type ETS1-3′-UTR
transfected cells compared with control in A549 and H1299 cells,
and there was no change in mutated ETS1-3′-UTR-transfected cells
(Fig. 5C and D). These data
revealed that miR-512-5p may regulate ETS1 by directly binding to
the 3′-UTR in A549 and H1299
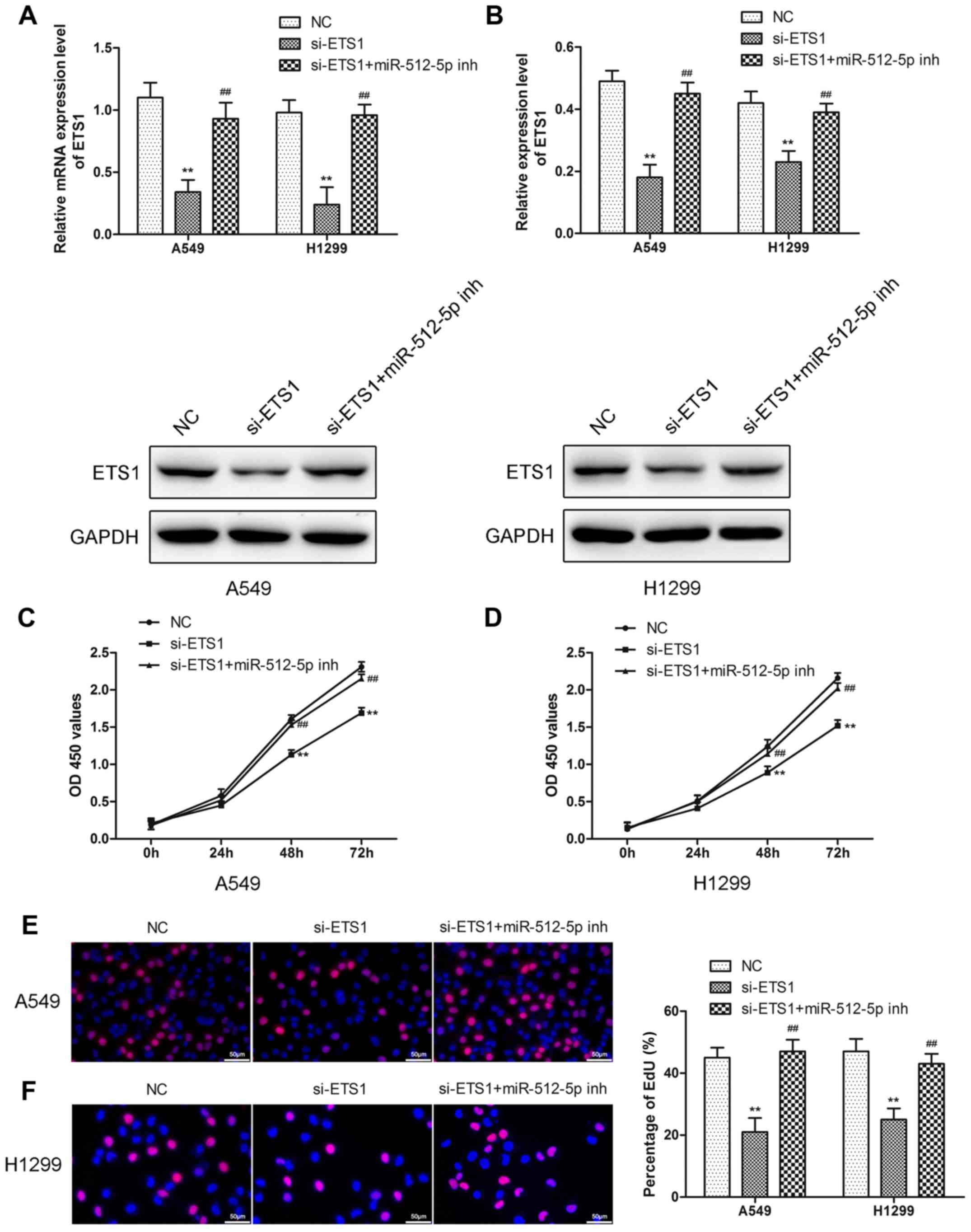
Downregulation of miR-512-5p
attenuates the effect of si-ETS1 on proliferation, invasion,
migration and apoptosis of NSCLC cells
To demonstrate that miR-512-5p affects NSCLC cells
through ETS1, the effect of si-ETS1 and si-ETS1 in the presence of
an miR-512-5p inhibitor in NSCLC cells was investigated. Following
transfection of si-ETS1 into the cells, downregulation of ETS1 was
confirmed by RT-qPCR and western blot analysis (Fig. 6A and B). Downregulation of ETS1
inhibited proliferation (Fig.
6C-F), induced apoptosis (Fig.
7) and migration (Fig. 8) in
the A549 and H1299 cells, increased the expression of Bax,
caspase-3 and caspase-7, and decreased the expression of Bcl-2,
MMP-2 and MMP-9. However, the presence of an miR-512-5p inhibitor
reversed the effect of si-ETS1 on A549 and H1299 cells. These data
demonstrated that the effects of miR-512-5p on proliferation,
invasion, migration and apoptosis of NSCLC cells may be induced by
regulating ETS1.
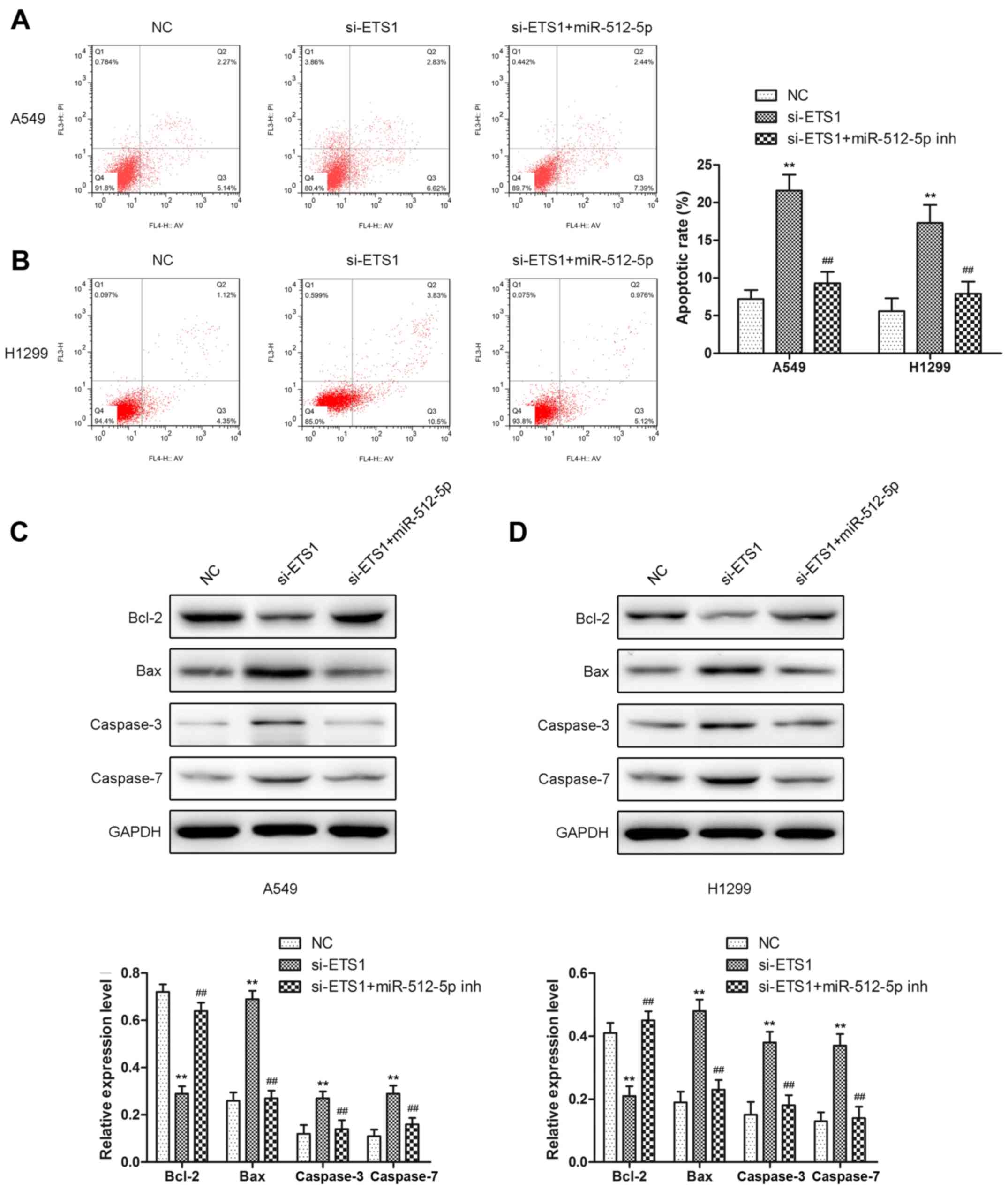 | Figure 7.The effect of knockdown of ETS1 on
cell apoptosis in A549 and H1299 cells. Apoptosis assays were
performed following transfection with si-ETS1 or an miR-512-5p
inhibitor in the presence of si-ETS1 for 48 h in (A) A549 and (B)
H1299 cells. Relative apoptosis proteins (Bcl-2, Bax, caspase-3,
and caspase-7) were detected by western blot analysis following
transfection with si-ETS1 or an miR-512-5p inhibitor in the
presence of si-ETS1 in (C) A549 and (D) H1299 cells. Data are
presented as mean ± standard deviation. **P<0.01 vs. NC;
##P<0.01 vs. si-ETS1. ETS1, E26 transformation
specific-1; si, small interfering; miR, microRNA; NC, negative
control; Bcl-2, B-cell lymphoma-2; Bax, Bcl-2-associated X protein;
inh, inhibitor. |
Discussion
Increasing evidence indicates that the abnormal
expression of target genes mediated by miRNA is involved in a
number of pathological processes (19). The miRNA coding region is often
located in fragile genomic areas, where gene amplification or
deletion easily occurs in tumors (20). Multiple studies have also confirmed
that a large number of abnormally expressed miRNA exist in tumor
cell lines and tumor tissues, and many target genes regulated by
these miRNA are closely associated with the development of tumors
(21).
miR-512-5p is located at chromosome 19q13.42, which
is processed by miR-512-1 or miR-521-2 (12). From previous observations and
studies, profiling of miRNA expression in 357 patients with stage I
NSCLC identified miR-512 as an indicator of good prognosis
(12). miR-152-5p induced
apoptosis and inhibited glycolysis in NSCLC by targeting p21
(13). Overexpression of
miR-512-5p suppressed tumor growth by regulating telomerase reverse
transcriptase in head and neck squamous cell carcinoma in
vitro and in vivo (22).
In the present study, the results indicated that
miR-512-5p was downregulated in NSCLC tissues and cells compared
with normal controls. Furthermore, the effects of miR-512-5p on
NSCLC cell proliferation, apoptosis, migratory and invasive
capabilities was assessed in vitro, and it was identified
that miR-512-5p overexpression decreased the proliferative,
migratory and invasive abilities, and induced apoptosis. Chu et
al (13) demonstrated that
miR-512-5p induced apoptosis in NSCLC cells, similar to the data
from the present study. The present study identified that
miR-512-5p may inhibit cell migratory and invasive abilities in
NSCLC cells, but Chu et al (13) did not investigate these factors.
They demonstrated that miR-512-5p overexpression had no effect on
cell proliferation by CCK-8 assay, conflicting with the data from
the present study. However, the results from the present study
suggested that miR-512-5p overexpression decreased proliferation,
using an EdU assay. The differences between the data from the
present study and those from Chu et al (13) may be due to factors including
detection methods and errors. The expression and regulation of the
Bcl-2 and caspase families are key factors affecting cell apoptosis
(23,24). MMPs promote the invasion of cancer
cells to surrounding tissues by degradation of the extracellular
matrix (25). The results from the
present study indicated that miR-512-5p overexpression
significantly increased expression of Bax, caspase-3 and caspase-9,
and decreased expression of Bcl-2, MMP-2 and MMP-9 in NSCLC cells.
The data from Chu et al (13) indicated that miR-512-5p
overexpression induced NSCLC cells apoptosis by regulating p21.
Those results and the results from the present study indicated that
multiple signaling pathways participate in NSCLC cell apoptosis of
miR-512-5p-regulation. Chu et al (13) also revealed that miR-512-5p
inhibited glycolysis in A549 and H1299 cell lines; this was not
investigated in the present study. The data from the present study
revealed miR-512-5p serves as a tumor suppressor.
ETS1 is highly expressed in a variety of malignant
tumors. It participates in cell invasion, metastasis, proliferation
and apoptosis by regulating the expression of a variety of genes,
including MMPs, Bcl-2 and Bax (26–28).
The results of the RT-qPCR and western blot analysis suggested that
ETS1 was overexpressed in NSCLC cells, and miR-512-5p may decrease
the expression of ETS1. Target prediction analysis additionally
indicated that miR-512-5p may target ETS1. The prediction was
confirmed by luciferase assays, and the results indicated that
miR-512-5p directly targeted ETS1 mRNA and inhibited its
translation. Following transfection of the cells with si-ETS1, it
was identified that the results were concomitant with the
miR-512-5p overexpression data, and in the presence of si-ETS1, an
miR-512-5p inhibitor rescued the effect of si-ETS1 in NSCLC
cells.
Taken together, the present study demonstrated that
miR-512-5p is significantly downregulated in NSCLC tissues and
cells, and may regulate ETS1 expression to affect NSCLC cell
proliferation, migration, invasion and apoptosis. These data
suggest that miR-512-5p may become a potential prognostic marker
and/or therapeutic target in NSCLC.
Acknowledgements
Not applicable
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PS conceived and designed the study. BC and ST
performed the experiments. HT and YC conducted the analysis of
data. PS wrote the manuscript. All authors read and approved the
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Nanjing Gulou Hospital (Nanjing, China), Jiangsu Provincial
Hospital of Traditional Chinese Medicine (Nanjing, China) and
Affiliated Hospital of Xuzhou Medical College (Xuzhou, China).
Patient consent for publication
Informed consent was provided.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yoshimi I and Sobue T: International
comparison in cancer statistics: Eastern Asia (2). Jpn J Clin
Oncol. 34:759–763. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang L: SC17.02 Lung Cancer in China:
Challenges and Perspectives. J Thor Oncol. 12:S113–S114. 2017.
View Article : Google Scholar
|
|
3
|
Higgins KA, O'Connell K, Liu Y, Gillespie
TW, McDonald MW, Pillai RN, Patel KR, Patel PR, Robinson CG, Simone
CB II, et al: National cancer database analysis of proton versus
photon radiotherapy in non-small cell lung cancer (NSCLC). Int J
Radiat Oncol Biol Phys. 97:128–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ludwig N, Leidinger P, Becker K, Backes C,
Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E
and Keller A: Distribution of miRNA expression across human
tissues. Nucleic Acids Res. 44:3865–3877. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Puisségur MP, Mazure NM, Bertero T,
Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K,
Cardinaud B, Hofman V, et al: miR-210 is overexpressed in late
stages of lung cancer and mediates mitochondrial alterations
associated with modulation of HIF-1 activity. Cell Death Differ.
18:465–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu B, Wu X, Liu B, Wang C, Liu Y, Zhou Q
and Xu K: MiR-26a enhances metastasis potential of lung cancer
cells via AKT pathway by targeting PTEN. Biochim Biophys Acta.
1822:1692–1704. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, Zhang D, Chen C, Zhenchao R, Li Y
and Huang Y: MicroRNA-212 displays tumor-promoting properties in
non-small cell lung cancer cells and targets the hedgehog pathway
receptorPTCH1. Mol Biol Cell. 23:14232012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nasser MW, Datta J, Nuovo G, Kutay H,
Motiwala T, Majumder S, Wang B, Suster S, Jacob ST and Ghoshal K:
Down-regulation of Micro-RNA-1 (miR-1) in lung cancer. Suppression
of tumorigenic property of lung cancer cells and their
sensitization to doxorubicin-induced apoptosis by miR-1. J Biol
Chem. 283:33394–33405. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun Y, Bai Y, Zhang F, Wang Y, Guo Y and
Guo L: miR-126 inhibits non-small cell lung cancer cells
proliferation by targeting EGFL7. Biochem Biophys Res Commun.
391:1483–1489. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ke Y, Zhao W, Xiong J and Cao R: miR-149
Inhibits Non-Small-Cell Lung Cancer Cells EMT by Targeting FOXM1.
Biochem Res Int. 2013:5067312013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheung TH, Man KN, Yu MY, Yim SF, Siu NS,
Lo KW, Doran G, Wong RR, Wang VW, Smith DI, et al: Cell Cycle.
11:2876–2884. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Adi Harel S, Bossel Ben-Moshe N, Aylon Y,
Bublik DR, Moskovits N, Toperoff G, Azaiza D, Biagoni F, Fuchs G,
Wilder S, et al: Reactivation of epigenetically silenced miR-512
and miR-373 sensitizes lung cancer cells to cisplatin and restricts
tumor growth. Cell Death Differ. 22:1328–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chu K, Gao G, Yang X, Ren S, Li Y, Wu H,
Huang Y and Zhou C: miR-512-5p induces apoptosis and inhibits
glycolysis by targeting p21 in non-small cell lung cancer cells.
Int J Oncol. 48:577–586. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dittmer J: The biology of the Ets1
proto-oncogene. Mol Cancer. 2:292003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Yan LX, Wu QN, Du ZM, Chen J,
Liao DZ, Huang MY, Hou JH, Wu QL, Zeng MS, et al: miR-125b is
methylated and functions as a tumor suppressor by regulating the
ETS1 proto-oncogene in human invasive breast cancer. Cancer Res.
71:3552–3562. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zheng L, Qi T, Yang D, Qi M, Li D, Xiang
X, Huang K and Tong Q: microRNA-9 suppresses the proliferation,
invasion and metastasis of gastric cancer cells through targeting
cyclin D1 and Ets1. PLoS One. 8:e557192013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pei H, Li C, Adereth Y, Hsu T, Watson DK
and Li R: Caspase-1 is a direct target gene of ETS1 and plays a
role in ETS1-induced apoptosis. Cancer Res. 65:7205–7213. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res. 42:D78–D85.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eulalio A, Huntzinger E and Izaurralde E:
Getting to the root of miRNA-mediated gene silencing. Cell.
132:9–14. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carissimi C, Fulci V and Macino G:
MicroRNAs: Novel regulators of immunity. Autoimmun Rev. 8:520–524.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li J, Lei H, Xu Y and Tao Z: miR-512-5p
suppresses tumor growth by targeting hTERT in telomerase positive
head and neck squamous cell carcinoma in vitro and in vivo. PLoS
One. 10:e01352652015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cory S, Huang DC and Adams JM: The Bcl-2
family: Roles in cell survival and oncogenesis. Oncogene.
22:8590–8607. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ola MS, Nawaz M and Ahsan H: Role of Bcl-2
family proteins and caspases in the regulation of apoptosis. Mol
Cell Biochem. 351:41–58. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nabeshima K, Inoue T, Shimao Y and
Sameshima T: Matrix metalloproteinases in tumor invasion: Role for
cell migration. Pathol Int. 52:255–264. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Puzovic V, Brcic I, Ranogajec I and
Jakicrazumovic J: Prognostic values of ETS-1, MMP-2 and MMP-9
expression and co-expression in breast cancer patients. Neoplasma.
61:439–446. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu Z and Shah DM: Curcumin down-regulates
Ets-1 and Bcl-2 expression in human endometrial carcinoma HEC-1-A
cells. Gynecol Oncol. 106:541–548. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nakazawa Y, Suzuki M, Manabe N, Yamada T,
Kihara-Negishi F, Sakurai T, Tenen DG, Iwama A, Mochizuki M and
Oikawa T: Cooperative interaction between ETS1 and GFI1
transcription factors in the repression of Bax gene expression.
Oncogene. 26:3541–3550. 2007. View Article : Google Scholar : PubMed/NCBI
|