Introduction
Preeclampsia (PE) is a pregnancy-specific disorder
that manifests as newly developed maternal hypertension and
proteinuria during the second half of pregnancy, and remains a
major cause of maternal mortality and morbidity worldwide (1,2).
Newly defined features of preeclampsia also include maternal organ
dysfunction, and long-term effects on cardiovascular disease in the
mother later in the life (3,4). At
present, the etiology and pathogenesis of preeclampsia remains
elusive. Defective trophoblastic invasion, anomalous maternal-fetal
inflammation-immune interactions and oxygen dysregulation have been
reported to be associated with the pathogenesis of preeclampsia. A
series of cascading reactions lead to preeclampsia syndrome, which
eventually results in endothelial cell damage, followed by
vasospasm, plasma leakage, ischemia and thrombosis. Delivery of the
fetus and the placenta is the only effective method to eradicate
the clinical manifestations of preeclampsia (5).
In the past decades, great efforts have been made to
explore how microRNAs (miRNAs/miRs) regulate human placental
physiology. miRNAs, a class of small, non-coding RNAs, modulate the
expression of nearly a third of human genes (6). It has been demonstrated that miRNAs
have essential roles in several biological processes, including
proliferation, differentiation, cell growth and development
(7). Equally, it has been
demonstrated that there are numerous differentially expressed
miRNAs in human placenta, which act via targeting of specific genes
with diverse functions, and have an important role in normal and
pathological placental physiology (8–10).
In the present study, the miRNA expression profile
in the placenta of patients with preeclampsia was assessed using
microarray analysis. Based on the results of validation and
literature retrieval, miR-144-3p was detected in the placentas of
patients with preeclampsia and normal placentas using in
situ hybridization (ISH) and immunohistochemical staining. A
dual-luciferase reporter assay was used to investigate the
potential miR-144-3p target gene cyclooxygenase-2 (Cox-2) and
demonstrated that the expression of Cox-2 was negatively regulated
by miR-144-3p. Taken together, it is speculated that miR-144-3p may
be involved in the pathogenesis of preeclampsia by targeting
Cox-2.
Materials and methods
Patients and tissue samples
The placental samples, from 25 women with severe
preeclampsia and 25 with normal pregnancy, were obtained from the
Department of Gynecology and Obstetrics, Yangzhou Women and
Children Hospital (Yangzhou, China) between October 2014 and
November 2016. The clinical data of the pregnant women are
presented in Table I. Chorionic
tissue blocks (~1 cm3) were collected from the mother
surface of the placenta and near the root of the umbilical cord.
Tissues were washed with sterile PBS, then immediately frozen in
liquid nitrogen at the time of surgery and stored at −70°C until
needed. This study was approved by the ethics board of Yangzhou
Women and Children Hospital and informed consent was obtained from
all 50 participants. The present study was approved by the ethics
board of Yangzhou Women and Children Hospital and informed consent
was obtained from all participants. All clinical investigations
were conducted according to the principles of the Declaration of
Helsinki.
 | Table I.Clinical characteristics of normal and
preeclamptic pregnancies. |
Table I.
Clinical characteristics of normal and
preeclamptic pregnancies.
| Characteristics | Normal | Preeclampsia | P-value |
|---|
| Maternal age
(years) | 28.04±3.09 | 29.24±4.05 | 0.245 |
| Systolic blood
pressure (mmHg) | 116.44±8.76 | 160.88±15.29 | <0.001 |
| Diastolic blood
pressure (mmHg) | 73.16±7.37 | 104.08±12.62 | <0.001 |
| Proteinuria | 0 | 2.08±0.76 | <0.001 |
| Gestational age at
delivery (weeks) | 39.16±0.80 | 37.03±1.58 | <0.001 |
| Birthweight
(g) | 3490.80±344.27 | 2844.80±524.34 | <0.001 |
| miR-144-3p
expression | 235.48±105.49 | 163.91±92.33 | 0.014 |
| Cox-2
expression | 1.40±0.65 | 1.96±0.93 | 0.017 |
miRNA microarray and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) and QIAGEN
miRNeasy Mini kit (Qiagen GmbH, Hilden, Germany), quantitated on
the NanoDrop 2000 (Thermo Fisher Scientific, Inc., Wilmington, DE,
USA), labeled using the FlashTag Biotin HSR RNA Labeling kit
(Affymetrix, cat no. 901911; Thermo Fisher Scientific, Inc.) and
hybridized onto the GeneChip2 Hybridization (Affymetrix, cat no.
902413; Thermo Fisher Scientific, Inc.). Following the washing
steps, the slides were scanned using the GeneChip2 Scanner 3000 7G
(Affymetrix; Thermo Fisher Scientific, Inc.). All expressed data
were normalized using the median normalization method, and
significantly differentially expressed miRNAs were identified
through volcano plot filtering. Data were analyzed by GCBI online
software (GCBI, R3.3.1, http://www.gcbi.com.cn; Genminix Informatics Co.,
Ltd., Shanghai, China). RT-qPCR analysis for miR-337-3p,
miR-187-3p, miR-122-5p, miR-26b-5p and miR-144-3p was performed
with a miDETECT A TRACK™ miRNA qRT-PCR Starter kit (RiboBio Co.,
Ltd., Guangzhou, China). The miDETECT A TRACK™ Uni-RT primer was
used for RT (Guangzhou RiboBio Co., Ltd., Guangzhou, China). The
miDETECT A TRACK™ Forward Primer and Uni-Reverse Primer were used
for qPCR (Guangzhou RiboBio Co., Ltd.). The reactions were
incubated in a 96-well plate at 95°C for 10 min, followed by 40
cycles at 95°C for 2 sec, 60°C for 20 sec and 70°C for 10 sec. The
miRNA Primers used in RT-qPCR were synthesized by RiboBio, Co.,
Ltd., and the primer sequences are commercially restricted. The
expression of miRNA was normalized to the small nuclear RNA U6
expression as an endogenous control. The fold changes of mRNA
expression were calculated by the 2−ΔΔCq method.
Cell culture
The HTR-8/SVneo cells were maintained in the
laboratory of Department of Gynecology and Obstetrics, the
Affiliated Drum Tower Hospital of Nanjing University Medical School
(Nanjing, China). The cells were cultured in RPMI-1640 (HyClone; GE
Healthcare Life Sciences, Logan, UT, USA), supplemented with 10%
heat-inactivated fetal bovine serum (Wisent, Inc., St. Bruno,
Quebec, Canada), 100 U/ml penicillin and 100 mg/ml streptomycin, at
37°C in a humidified 5% CO2 incubator.
Luciferase reporter assays
The bioinformatics software TargetScan (version 7.0;
http://www.targetscan.org/) (11) was used to predict the target genes
of miR-144-3p. The Cox-2 3′untranslated region (3′UTR)-hRLuc
reporters (PTGS2 WT) were created by ligation of the Cox-2 3′UTR
PCR product into the XhoI and NotI site of the
dual-luciferase reporter vector (pmiR-RB-REPORT™; RiboBio Co.,
Ltd.). The mutant reporters for the Cox-2 3′UTR (PTGS2 MT;
CAUUUAAUGGGUACUGUAU) were generated
by replacing the binding sites of miR-144-3p. Subsequently,
HTR-8/SVneo cells were cultured in 96-well plates
(1.5×104) and transfected with the wild or mutant
luciferase reporters (100 ng), and miR-144-3p mimics (50 nM) using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). All reactions were run in triplicate. After 48
h, luciferase activities were measured using the dual-luciferase
reporter system (Promega, Madison, WI, USA). Normalized luciferase
activity was reported as Renilla luciferase
activity/luciferase activity.
miRNA ISH analysis
To detect the expression levels of human miR-144-3p
in placenta tissue, ISH was performed using a miRCURY locked
nucleic acid (LNA) detection probe for miR-144-3p
(hsa-miR-144-3p/DigN/AGTACATCATCTATACTGTA) (Exiqon; Qiagen GmbH;
probe concentration 25 µM). The digoxigenin double-labeled
LNA-modified probe, at a final concentration of 500 nmol/l, was
added to the hybridization solution and hybridized according to the
manufacturer's protocol. The sample images were captured using a
light microscope at 100× magnification after the incubation of
slides with 4-nitroblue-tetrazolium for 30 min at 25°C and with
nuclear fast red for 5 min at room temperature (Roche Applied
Science, Indianapolis, IN, USA). The staining intensity and the
proportion of miR-144-3p-positive cells were detected in 50
placenta samples (25 from preeclampsia women and 25 from women
without preeclampsia). A total of 10 fields of view in each slide
was randomly selected for analysis. The 4 intensity grades of
staining cells were negative, weak, intermediate and strong, with
corresponding scores of 0, 1, 2 and 3 respectively. Another score
was given for the proportion of positive cells with the scoring
rule as follows: No positive cells (0 point), 1–24% positive cells
(1 point), 25–49% positive cells (2 points), 50–74% positive cells
(3 points), 75–100% positive cells (4 points). The staining index
(SI) was calculated by multiplying the staining intensity and the
proportion of positive cells. According to the definition of SI
scores, low expression of miR-144-3p (1.0) was described as SI
score ≤4, high expression of miR-144-3p (3.0) was described as SI
score >8 and intermediate (2.0) >4 and ≤8.
Immunohistochemical staining
Immunohistochemical staining was performed using an
immunohistochemistry detection kit (cat no. PV-6000; OriGene
Technologies, Inc., Beijing, China) according to the manufacturer's
instructions. A rabbit polyclonal antibody for Cox-2 (1:1,000;
ab15191, Abcam, Cambridge, UK) was used as primary antibody. A goat
anti-rabbit secondary antibody (1:1,000; A0208; Beyotime Institute
of Biotechnology, Beijing, China) was used as the secondary
antibody. The expression of Cox-2 was evaluated using a scoring
system by the combining the staining intensity score and proportion
of positive cells. Quantification of the staining intensity of
Cox-2 was performed using image analysis as with ISH. A total of 10
fields of view in each slide was randomly selected for
analysis.
Establishment of stably expressing
miRNA-144-3p cell lines by lentivirus
HTR-8/SVneo cells with stable miRNA-144-3p
expression and parental cell lines were established using a
lentiviral expression system. The lentiviruses containing
MiR-144-3p (MIMAT0000436:
GAGCAGGGAGCAGGAAGCTGTGTGTGTCCAGCCCTGACCTGTCCTGTTCTGCCCCCAGCCCCTCACAGTGCTTTTCAAGCCATGCTTCCTGTGCCCCCAGTGGGGCCCTGGCTGGGATATCATCATATACTGTAAGTTTGCGATGAGACACTACAGTATAGATGATGTACTAGTCCGGGCACCCCCAGCTCTGGAGCCTGACAAGGAGGACAGGAGAGATGCTGCAAGCCCAAGAAGCTCTCTGCTCAGCCTGTCACAACCTACTGACTGCCAGGGCACTTGGGA)
or negative control were constructed by GeneChem Co., Ltd.,
(Shanghai, China). All the lentiviral vectors expressed enhanced
green fluorescent protein and puromycin resistance gene, which
allowed the measurement of the infection efficiency and to select
for infected cells. Cells (5×106/15 ml) were infected
with Vector or MiR-144-3p lentivirus (15 µg), followed by culture
in 5 µg/ml puromycin (Clontech Laboratories, Inc., Mountainview,
CA, USA) to select stable lines. The resulting stable lines were
used for further analysis.
Western blot analysis
Cells were collected and homogenized in lysis buffer
containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% SDS, 1%
Nonidet P-40, 0.5% sodium deoxycholate, 0.02% sodium azide, 100
mg/l phenylmethylsulfonyl fluoride and 1 mg/l aprotinin. The
supernatant was collected after centrifuging at 11,000 × g for 15
min at 4°C. Protein concentration was determined using the
bicinchoninic acid method (Pierce; Thermo Fisher Scientific, Inc.).
SDS-PAGE using 10% gels was performed to separate protein (50
µg/lane) and separated proteins were then transferred to
polyvinylidene difluoride membranes. PVDF membranes were blocked
with 5% non-fat milk for 1 h at room temperature and incubated with
primary antibodies against Cox-2 (1:1,000; ab15191; Abcam), GAPDH
as a loading control (1:1,000; sc-32233; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) and Cox-1 as a specific control (1:1,000;
ab109025; Abcam) overnight at 4°C. After washing, the membranes
were incubated with goat anti-rabbit secondary antibody (1:5,000;
cat no. sc-2004; Santa Cruz Biotechnology, Inc.) for 2 h at room
temperature and visualized with enhanced chemiluminescence
(Millipore, Billerica, MA, USA) by Quantity one software (version
62, Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Results were analyzed using the SPSS 22.0
statistical software (IBM Corp., Armonk, NY, USA). Experimental
data are presented as the mean ± standard deviation. The
differences for multiple groups were estimated using one-way
analysis of variance followed by Dunnett's T3 post hoc test, and
the association between miR-144-3p expression and the Cox-2 level
was analyzed by Spearman correlation analysis. The independent
two-tailed Student's t-test was performed for comparisons between
two groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-144-3p is downregulated in
patients with preeclampsia compared with normal placentas
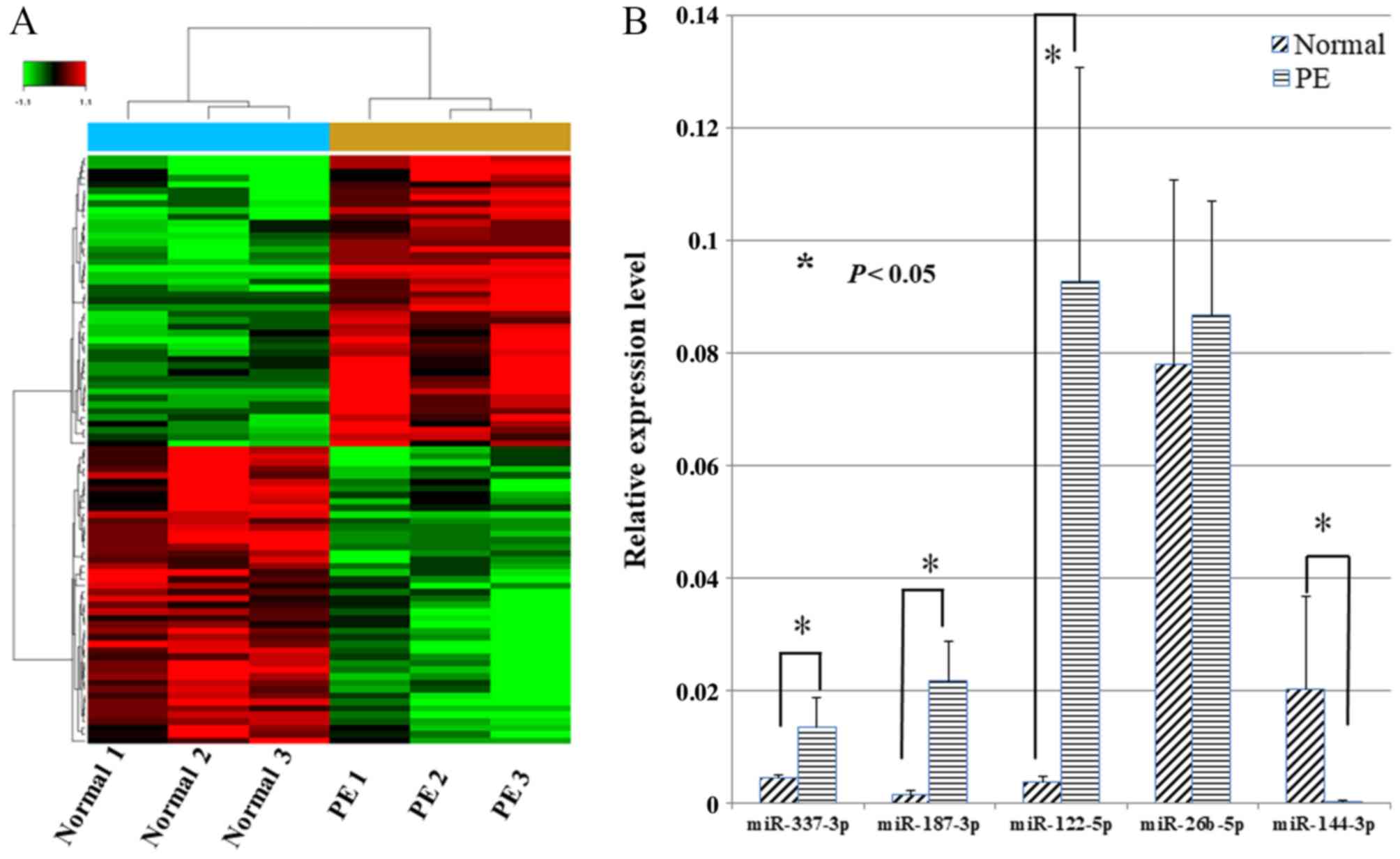
Firstly, miRNA microarray analysis of samples from
six pregnant women (three from normal and three from preeclampsia
cases) and the expression of miRNAs in paired placentas were
compared to identify differential expressed miRNAs. The inclusion
criteria of differential miRNAs was differential expression of ≥2.0
fold changes. In total, 46 miRNAs were downregulated and 45 were
upregulated in the six samples (Fig.
1). Subsequently, the five miRNAs with the highest fold changes
(differential expression of >4.0 fold changes), including
miR-337-3p, miR-187-3p, miR-122-5p, miR-26b-5p and miR-144-3p, were
selected to verify the results of the microarrays. Notably, RT-qPCR
validation demonstrated that the fold changes in the five miRNAs
expression were 2.99, 15.37, 24.91, 0.87 and 0.016, respectively,
in preeclampsia placentas compared with the paired normal placentas
(Fig. 1). Ultimately, miR-144-3p
was selected as the focus of further research as it exhibited the
greatest fold change.
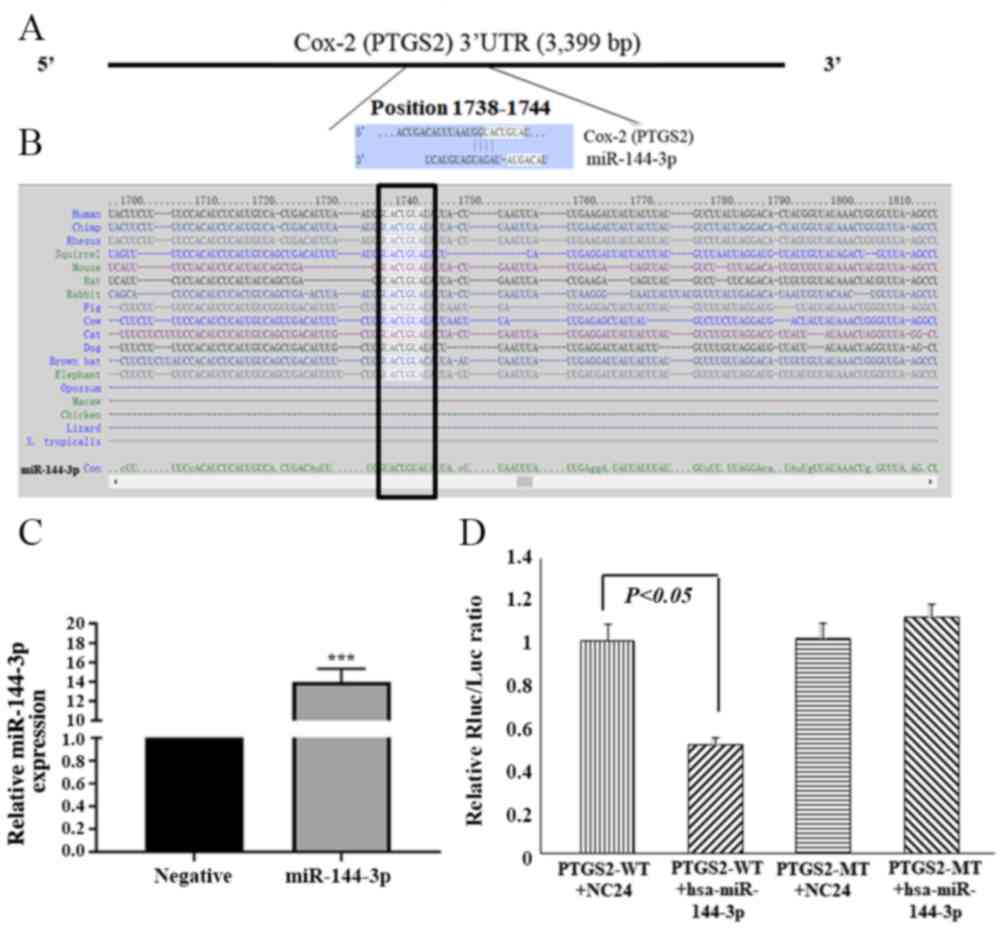
miR-144-3p directly targets Cox-2 by
interacting with the 3′UTR
Cox-2, also known as prostaglandin-endoperoxide
synthase 2 (PTGS2), was selected as a candidate miR-144-3p target
based on bioinformatics analysis (TargetScan) and functional
knowledge. The 3′UTR of Cox-2 has a complementary site for the
miR-144-3p seed sequence at its 3′UTR (Fig. 2A and B), and it is highly conserved
among species. To further determine whether Cox-2 is a direct
target of miR-144-3p, a luciferase reporter construct harboring a
fragment of the Cox-2 3′UTR that contained the miR-144-3p binding
site was prepared (PTGS2 WT). A construct with mutated miR-144-3p
binding site in the Cox-2 3′UTR (PTGS2 MT) was also generated.
Luciferase activity was decreased by 46.5% in HTR-8/SVneo cells
co-transfected with Cox-2 3′UTR (PTGS2 WT) and miR-144-3p (Fig. 2C and D). By contrast, the
luciferase activity was from the Cox-2 3′UTR-mut (PTGS2 MT)
construct was not significantly affected by transfection with
miR-144-3p levels in HTR-8/SVneo cells (Fig. 2D). These results indicate that
miR-144-3p targets the Cox-2 3′UTR directly and downregulates its
expression.
Clinicopathological features of
clinical samples
The basic clinical characteristics of the patients
in the preeclampsia and control group are presented in Table I. Patients in the preeclampsia
group exhibited significant increase in the systolic pressure and
diastolic pressure compared with the control group (P<0.0001).
Notably, patients with preeclampsia exhibited proteinuria.
Furthermore, the gestational age at delivery was significantly
lower for patients with PE than for normal controls (P<0.0001).
miRNA ISH analysis was used to compare the expression of miR-144-3p
in placenta samples of the two groups (Fig. 3A and B). The results revealed that
the expression of miR-144-3p was significantly decreased in
placentas from the PE group compared with the normal group
(P<0.05; Fig. 3C).
Negative correlation between
miR-144-3p and Cox-2 expression was observed in placentas
Tissue assays comprising 25 pairs of placentas with
preeclampsia and normal placentas were examined for miR-144-3p
expression using ISH. ISH revealed differential expression of
miR-144-3p in preeclampsia and normal placentas (Fig. 3). Immunohistochemical staining for
Cox-2 was performed using the same placenta tissues used for
miR-144-3p ISH analysis (Fig. 4A and
B). The expression of Cox-2 was negatively correlated with
miR-144-3p, as demonstrated by Spearman correlation analysis
(r=−0.509, P=0.001; Fig. 4C).
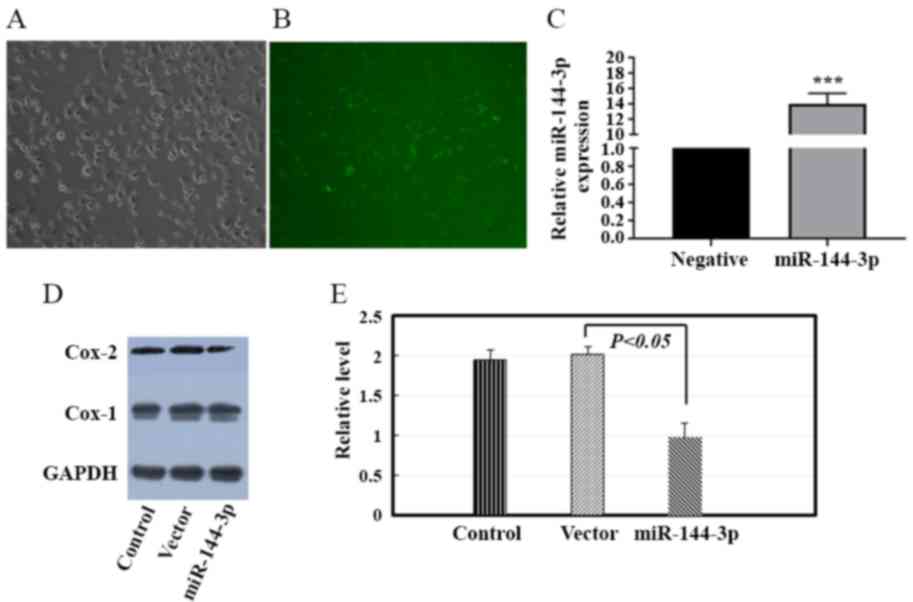
miR-144-3p regulates the expression of
Cox-2 in HTR-8/SVneo cells in vitro
To further investigate the association between
miR-144-3p and Cox-2, HTR-8/SVneo cells were transfected with
miR-144-3p vector or Vector control. The lentiviral transduction
efficiency was assessed (Fig.
5A-C). Western blot analysis was performed to investigate the
effect of miR-144-3p on the expression of Cox-2 in HTR-8/SVneo
cells in vitro. Overexpression of miR-144-3p decreased Cox-2
protein expression by 38.2% in HTR-8/SVneo cells (Fig. 5D and E).
Discussion
Because of the severity of preeclampsia, its
pathogenesis has been the focus of continuous research. Increasing
evidence indicates that miRNAs may be involved in the pathogenesis
of preeclampsia (8–10). In the current study, miR-144-3p was
selected as an RNA of interest based on microarray and RT-qPCR
results. The expression of miR-144-3p and Cox-2 were compared in
placenta samples from patients with preeclampsia and paired normal
placentas, using ISH and immunohistochemical staining. The
expression of miR-144-3p was downregulated in placentas of patients
with preeclampsia compared with paired normal placentas, which was
consistent with previous reports (12,13).
In addition, the current study also demonstrated that the
expression of miR-144-3p was negatively associated with the
expression of Cox-2. A dual-luciferase reporter assay suggested
that Cox-2 was a direct target of miR-144-3p and its expression was
negatively correlated with expression of miR-144-3p.
miRNAs are short, non-coding RNA molecules that
post-transcriptionally regulate gene expression by perfect or
imperfect binding to the 3UTR of target mRNAs, thus causing mRNA
degradation or modification of mRNA translation. miRNAs control
diverse biological possesses, including cell proliferation,
apoptosis, migration and invasion. Pineles et al (8) first reported that miRNAs were
expressed differentially in preeclampsia placentas. Certain miRNAs
have been reported to be involved in the pathogenesis of
preeclampsia. For instance, miR-181a-5p, miR-299, miR-30a-3p and
miR-155 have been reported to be involved in the pathogenesis of
preeclampsia by targeting different genes (14–17).
Xiao et al (12) discovered
that miR-144 contributed to preeclampsia by targeting phosphatase
and tensin homolog in trophoblastic cells. Consistently, the
expression of miR-144-3p was downregulated in placentas of patients
with preeclampsia compared with paired normal placentas in the
current study; however Cox-2 was identified as the miR-144-3p
target in the current study. Several studies have identified the
targets of miR-144-3p in different disease; for instance,
miR-144-3p inhibits cell proliferation and induces apoptosis in
multiple myeloma by targeting c-MET proto-oncogene, receptor
tyrosine kinase (18); miR-144-3p
suppresses proliferation and migration of colorectal cancer cells
via the G1 to S phase transition 1 gene (19); other reported targets of miR-144-3p
also include serum/glucocorticoid regulated kinase 1 (20), TNF superfamily member 11 (21) and runt related transcription factor
1 (22), which have predominantly
been described in cancer research. There is no report on the
interaction between these targets and miRNAs in the pathogenesis of
preeclampsia. Yao et al (23) validated that Cox-2 was a direct
target of miR-144 and that miR-144 negatively regulated the
expression of Cox-2 in gastric cancer. Together, these studies
indicate that each miRNA can have multiple target genes, and
several miRNAs can regulate the same gene. Therefore, miRNAs and
the target genes may form a complex regulatory network during
different pathological and biological possesses. However, the
relationship between miR-144-3p and Cox-2 on preeclampsia has not
been reported previously.
Cox-2, as a rate-limiting enzyme of prostaglandin
synthesis, can regulate the levels of prostaglandins (24). The roles of Cox-2 in preeclampsia
have been discussed in previous studies (25–27).
Goksu et al (26) reported
that the expression of Cox-2 was increased in the placenta of
patients with preeclampsia, presumably due to the decreased levels
of prostaglandin E and prostaglandin F in the placenta.
Inflammation and vascular endothelial damage can lead to vasospasm,
which is one of important physiological manifestations of
preeclampsia (28). Endothelial
cell injury can also activate platelets and clotting factors, which
will cause an aggravation of current hypercoagulable state in
pregnant women. A large number of inflammatory substances can
stimulate and adhere to the white blood cell molecules expressed on
the surface of endothelial cells in a short time. Excessive
prostaglandin production can stimulate the production of
pro-inflammatory cytokines and increase vascular permeability,
promote the adhesion of monocytes and macrophage migration, and
induce macrophage chemotaxis (29–31).
All of these ultimately lead to systemic inflammation and
endothelial cell injury in preeclampsia (32).
In summary, the present investigation demonstrated
that the expression of miR-144-3p was decreased that Cox-2 was
increased in preeclamptic placentas, and the expression of the two
markers had a negative correlation, and the association between
miR-144-3p and Cox-2 was also verified in vitro. However,
the research has some limitations. One limitation is that the
sample size was relatively small, and more samples should be
included to confirm the conclusions, which could be solved through
multi-center cooperation in the future. Another limitation of this
study is that the association between miR-144-3p and Cox-2, and the
mechanisms in preeclampsia should be further explored in
vivo using animal models; for example, in a mouse with a
miR-144-3p conditional deletion or overexpression in the placenta.
Additionally, the microarray analysis also identified other miRNAs,
including miR-337-3p, miR-187-3p, miR-122-5p and miR-26b-5p, which
may be involved in the pathogenesis of preeclampsia. Further work
is required to study the miRNA network associated with the
pathogenesis of preeclampsia. The utility of changing the
expression of miRNAs may provide novel strategies for preeclampsia
therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Jiangsu Province (grant no. BK20140101), the
Fundamental Research Funds for the Central Universities (grant no.
2062140665), the Social development project of Yangzhou (grant no.
YZ2018060), Maternal and child health research project of Jiangsu
Province (grant no. F201672).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors contributions
SH, JL and LM designed the experiments; SH, MT, QL
and YC performed the experiments; HL and YW analyzed the data; SH
and LM wrote the manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics board
of Yangzhou Women and Children Hospital (Yangzhou, China) and
informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kanasaki K and Kalluri R: The biology of
preeclampsia. Kidney Int. 76:831–837. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Redman CW and Sargent IL: Latest advances
in understanding preeclampsia. Science. 308:1592–1594. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mol BWJ, Roberts CT, Thangaratinam S,
Magee LA, de Groot C and Hofmeyr GJ: Pre-eclampsia. Lancet.
387:999–1011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Phipps E, Prasanna D, Brima W and Jim B:
Preeclampsia: Updates in pathogenesis, definitions, and guidelines.
Clin J Am Soc Nephrol. 11:1102–1113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roberts JM and Cooper DW: Pathogenesis and
genetics of pre-eclampsia. Lancet. 357:53–56. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang C: MicroRNomics: A newly emerging
approach for disease biology. Physiol Genomics. 33:139–147. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pineles BL, Romero R, Montenegro D, Tarca
AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P,
et al: Distinct subsets of microRNAs are expressed differentially
in the human placentas of patients with preeclampsia. Am J Obstet
Gynecol. 196:261.e1–e6. 2007. View Article : Google Scholar
|
|
9
|
Hu Y, Li P, Hao S, Liu L, Zhao J and Hou
Y: Differential expression of microRNAs in the placentae of Chinese
patients with severe pre-eclampsia. Clin Chem Lab Med. 47:923–929.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu XM, Han T, Sargent IL, Yin GW and Yao
YQ: Differential expression profile of microRNAs in human placentas
from preeclamptic pregnancies vs normal pregnancies. Am J Obstet
Gynecol. 200:661.e1–e7. 2009. View Article : Google Scholar
|
|
11
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 42015.doi: 10.7554/eLife.05005.
|
|
12
|
Xiao J, Tao T, Yin Y, Zhao L, Yang L and
Hu L: miR-144 may regulate the proliferation, migration and
invasion of trophoblastic cells through targeting PTEN in
preeclampsia. Biomed Pharmacother. 94:341–353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Choi SY, Yun J, Lee OJ, Han HS, Yeo MK,
Lee MA and Suh KS: MicroRNA expression profiles in placenta with
severe preeclampsia using a PNA-based microarray. Placenta.
34:799–804. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu L, Song WY, Xie Y, Hu LL, Hou XM, Wang
R, Gao Y, Zhang JN, Zhang L, Li WW, et al: miR-181a-5p suppresses
invasion and migration of HTR-8/SVneo cells by directly targeting
IGF2BP2. Cell Death Dis. 9:162018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao Y, She R, Wang Q, Li Y and Zhang H:
Up-regulation of miR-299 suppressed the invasion and migration of
HTR-8/SVneo trophoblast cells partly via targeting HDAC2 in
pre-eclampsia. Biomed Pharmacother. 97:1222–1228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Niu ZR, Han T, Sun XL, Luan LX, Gou WL and
Zhu XM: MicroRNA-30a-3p is overexpressed in the placentas of
patients with preeclampsia and affects trophoblast invasion and
apoptosis by its effects on IGF-1. Am J Obstet Gynecol.
218:249.e1–249.e12. 2018. View Article : Google Scholar
|
|
17
|
Zhang Y, Diao Z, Su L, Sun H, Li R, Cui H
and Hu Y: MicroRNA-155 contributes to preeclampsia by
down-regulating CYR61. Am J Obstet Gynecol. 202:466.e1–e7. 2010.
View Article : Google Scholar
|
|
18
|
Zhao Y, Xie Z, Lin J and Liu P: MiR-144-3p
inhibits cell proliferation and induces apoptosis in multiple
myeloma by targeting c-Met. Am J Transl Res. 9:2437–2446.
2017.PubMed/NCBI
|
|
19
|
Xiao R, Li C and Chai B: miRNA-144
suppresses proliferation and migration of colorectal cancer cells
through GSPT1. Biomed Pharmacother. 74:138–144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu M, Huang C, Huang X, Liang R, Feng Y
and Luo X: MicroRNA-144-3p suppresses tumor growth and angiogenesis
by targeting SGK3 in hepatocellular carcinoma. Oncol Rep.
38:2173–2181. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li RD, Shen CH, Tao YF, Zhang XF, Zhang
QB, Ma ZY and Wang ZX: MicroRNA-144 suppresses the expression of
cytokines through targeting RANKL in the matured immune cells.
Cytokine. 108:197–204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han S, Zhu J and Zhang Y: miR-144
potentially suppresses proliferation and migration of ovarian
cancer cells by targeting RUNX1. Med Sci Monit Basic Res. 24:40–46.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yao Q, Gu A, Wang Z and Xue Y:
MicroRNA-144 functions as a tumor suppressor in gastric cancer by
targeting cyclooxygenase-2. Exp Ther Med. 15:3088–3095.
2018.PubMed/NCBI
|
|
24
|
Dubois RN, Abramson SB, Crofford L, Gupta
RA, Simon LS, Van De Putte LB and Lipsky PE: Cyclooxygenase in
biology and disease. FASEB J. 12:1063–1073. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bachawaty T, Washington SL and Walsh SW:
Neutrophil expression of cyclooxygenase 2 in preeclampsia. Reprod
Sci. 17:465–470. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goksu Erol AY, Nazli M and Yildiz SE:
Expression levels of cyclooxygenase-2, tumor necrosis factor-alpha
and inducible NO synthase in placental tissue of normal and
preeclamptic pregnancies. J Matern Fetal Neonatal Med. 25:826–830.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Afroze SH, Kalagiri RR, Reyes M, Zimmerman
JD, Beeram MR, Drever N, Zawieja DC, Kuehl TJ and Uddin MN:
Apoptotic and stress signaling markers are augmented in
preeclamptic placenta and umbilical cord. BBA Clin. 6:25–30. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Redman CW, Sacks GP and Sargent IL:
Preeclampsia: An excessive maternal inflammatory response to
pregnancy. Am J Obstet Gynecol. 180:499–506. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sibai B, Dekker G and Kupferminc M:
Pre-eclampsia. Lancet. 365:785–799. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Akarasereenont P, Techatraisak K,
Chotewuttakorn S and Thaworn A: The expression of cyclooxygenase-2
in human umbilical vein endothelial cell culture from preeclampsia.
J Med Assoc Thai. 82:167–172. 1999.PubMed/NCBI
|
|
31
|
Wetzka B, Nusing R, Charnock-Jones DS,
Schäfer W, Zahradnik HP and Smith SK: Cyclooxygenase-1 and −2 in
human placenta and placental bed after normal and pre-eclamptic
pregnancies. Hum Reprod. 12:2313–2320. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shah TJ and Walsh SW: Activation of
NF-kappaB and expression of COX-2 in association with neutrophil
infiltration in systemic vascular tissue of women with
preeclampsia. Am J Obstet Gynecol. 196:48.e1–e8. 2007. View Article : Google Scholar
|