Introduction
Lung cancer is one of the most prevalent types of
malignant tumors, and has been reported to be the leading cause of
cancer-associated mortality worldwide, particularly in China
(1,2). Lung cancer is divided into two
classes, according to the degree of differentiation and morphologic
characteristics: i) Small cell lung cancer and ii) non-small cell
lung cancer (NSCLC). As the most common subtype of lung cancer,
NSCLC accounts for 85% of lung cancer cases (3). The high rates of mortality for NSCLC
have been associated with smoking; a large number of patients with
NSCLC succumb to metastasis (4).
Advances in treatments for NSCLC have been achieved, including
surgery, radiotherapy and chemotherapy; however, the 5-year
survival rate of patients with NSCLC is <15% and prognosis
remains poor (5,6). Therefore, it is important to
investigate and identify novel biomarkers, and to develop novel
therapeutic strategies for the treatment of NSCLC.
MicroRNAs (miRNAs) are a group of short, regulatory
non-coding RNA molecules of ~22 nucleotides in length (7). The action of miRNAs results in
translational suppression or mRNA degradation by targeting
complementary sequences of the mRNA 3′-untranslated region (UTR)
(7). In addition, miRNAs serve key
roles in a number of processes, including cell proliferation,
apoptosis, motility and metastasis (7–9).
Each miRNA regulates the expression of potentially hundreds of
different genes, and have been reported to exhibit opposing roles
in numerous types of cancer (10,11).
miRNAs may serve as novel therapeutic agents for the treatment of
cancers, as they can function as tumor suppressors or as oncogenes
(12). In particular, miRNA
(miR)-421 has also been reported to exhibit dual roles in various
types of cancer. For example, downregulation of miR-421 may act as
a tumor suppressor; miR-421 was demonstrated to inhibit glucose
metabolism, invasion and angiogenesis, as well as enhance the
radiation sensitivity by targeting myocyte enhancer factor 2D in
glioma (13). Conversely, miR-421
has been observed to be frequently upregulated and function as an
oncogene; miR-421 was reported to promote metastasis, inhibit
apoptosis and induce cisplatin resistance by targeting E-cadherin
and caspase-3 expression in gastric cancer (14). Therefore, the present study aimed
to investigate whether miR-421 may affect the malignant phenotypes
of NSCLC.
In the present study, miR-421 was upregulated in
NSCLC tissue samples and cells. MiR-421 functions as an oncogene in
NSCLC cells. Furthermore, miR-421 could directly target HOPX in
NSCLC cells. The present study confirmed that the homeodomain-only
protein (HOPX) gene may be a direct functional target of miR-421.
miR-421 was observed to suppress HOPX expression, which may
contribute to the development of malignant properties through the
Wnt/β-catenin signaling pathway in NSCLC cells. These results may
provide insight into the mechanisms underlying carcinogenesis and
the identification of potential biomarkers associated with
NSCLC.
Materials and methods
Human NSCLC tissue specimens and cell
lines
Tumor and normal paired tissue samples were
collected from 30 patients with NSCLC from December 2016 to June
2017 at the Department of Thoracic Surgery, The First Hospital of
Qinhuangdao, including 10 females and 20 males whose age from 35 to
67 years old. Patients did not receive therapy at the time of
sample collection. Written informed consent was obtained from all
enrolled patients, and all relevant investigations were performed
according to the principles of The Declaration of Helsinki. The
present study was approved by the Ethical Review Committee of The
First Hospital of Qinhuangdao (ethics approval no. QHD20160235).
The human NSCLC cell lines A549, H358, H460, H2066, SPC-A1 and a
normal human primary lung fibroblast cell line, HLF-a
(ATCC® CCL-199™) were purchased from the Cell Bank of
Type Culture Collection of Chinese Academy of Sciences (Shanghai,
China) and cultured in MEM-a (HyClone; GE Healthcare, Chicago, IL,
USA) medium supplemented with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cells were
incubated in a humidified atmosphere with 5% CO2 at
37°C.
Prediction of miRNA targets
Algorithm programs, including Human TargetScan7.2,
microrna.org and miRDB were used to predict the target
genes of miR-421.
Vector construction
The DNA fragment encoding primary (pri)-miR-421 was
cloned into the mammalian expression vector pcDNA3 (Promega
Corporation, Madison, WI, USA) at BamHI and EcoRI. A
2′-O-methyl-modified antisense oligonucleotide of miR-421
(ASO-miR-421) was commercially synthesized to serve as an inhibitor
of miR-421 (Genewiz, South Plainfield, NJ, USA). The cDNA fragment
containing the HOPX coding sequence (cat. no. BC014225; Tianjin
Saier Corporation, Tianjin, China) was amplified by polymerase
chain reaction (PCR). The subsequent PCR product was cloned into
the pcDNA3 vector at BamHI and EcoRI, thus generating
the HOPX expression plasmid (pHOPX). In addition, enhanced green
fluorescent protein (EGFP) reporter plasmids (pcDNA3/EGFP, Tianjin
Saier Corporation, Tianjin, China) with the wild-type HOPX 3′UTR
(HOPX-WT) containing the target site of miR-421 or the mutant
(HOPX-Mut) were constructed, respectively. The 3′UTR of HOPX that
contain the miR-421 binding sites and mutant 3′UTR fragments with
mutant miR-421 binding sites (site-directed mutagenesis) were
obtained by annealing double-strand DNA and inserting it into the
pcDNA3/EGFP vector. All primer sequences are listed in Table I.
 | Table I.Primers used in the present
study. |
Table I.
Primers used in the present
study.
| A, Plasmid
primers |
|---|
|
|---|
| Primers | Primer sequence
(5′→3′) |
|---|
| pri-miR-421 | F:
CGCGGATCCAGCAGCAACCTGGAGTGG |
|
| R:
CCGGAATTCGAGCTTGGACGTTGTTGG |
| HOPX-3′UTR-WT | F:
GATCCCGCGGGTTAATTACAGACAACTAAAGCTTG |
|
| R:
AATTCAAGCTTTAGTTGTCTGTAATTAACCCGCGG |
| HOPX-3′UTR-Mut | F:
GATCCCGCGGAAGAATTACTGTTAGGGAAAGCTTG |
|
| R:
AATTCAAGCTTTCCCTAACAGTAATTCTTCCGCGG |
| HOPX | F:
CGCGGATCCATGCTCATTTTCCTGGGCTGTTACA |
|
| R:
CCGGAATTCTTAGTCTGTGACGGATCTGCACTCT |
|
| B, RT-qPCR
primers |
|
| Primers | Primer sequence
(5′→3′) |
|
| miR-421-RT |
GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGAGCGCCC |
| miR-421-qPCR-F |
TGCGGATCAACAGACATTAAT |
| U6-RT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTC |
|
|
GCACTGGATACGACAAAATATGGAAC |
| Oligo-dT |
TTTTTTTTTTTTTTTTTT |
| U6-F |
TGCGGGTGCTCGCTTCGGCAGC |
|
miRNA-universal-R |
CCAGTGCAGGGTCCGAGGT |
| HOPX | F:
AGCATTTCCCTTTGAGTC |
|
| R:
GCCCAACAGGCTTCTTTC |
| β-actin | F:
CGTGACATTAAGGAGAAGCTG |
|
| R:
CTAGAAGCATTTGCGGTGGAC |
Cell transfection
A549 cells or SPC-A1 cells (4×105) were
plated in 6-well plates overnight to ensure that cell confluence
could reach 60–80% at the time of transfection. Following 6 h
transfection at 37°C, the medium was replaced with MEM-a media
containing 10% FBS. Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific Inc.) was employed in cell transfection according to the
manufacturer's protocol. They were transiently transfected with 3ug
pri-miR-421 or pcDNA3 and 20 µmol ASO-miR-421 or ASO-NC in a well
of 6-well plate. Cells were collected for reverse
transcription-quantitative PCR (RT-qPCR), proliferation, migration,
invasion, apoptosis and cell cycle assay analyses at 24 h. At 48 h
post-transfection, cells were collected for western blot analysis
and the EGFP fluorescent reporter assay.
RNA extraction and RT-qPCR
Total RNA was extracted for 8×105 cells
using the TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. cDNA
was prepared using the PrimeScript RT Reagent kit (Takara
Biotechnology Co., Ltd., Dalian, China) according to the
manufacturer's protocol. Subsequently, cDNA was used to detect mRNA
expression levels by qPCR using the SYBR® Premix Ex Taq™
II Perfect Real-Time kit (Takara Biotechnology Co., Ltd.) and an
ABI 7500 Real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The thermos cycling conditions for RT-qPCR were
as follows: 95°C for 20 sec, followed by 35 cycles of 95°C for 5
sec, 63°C for 30 sec and 72°C for 5 sec. Gene expression levels of
HOPX and miR-421 were normalized to that of endogenous β-actin (for
HOPX) or U6 (for miR-421), respectively. The relative fold changes
in the transcript levels were calculated using the
2−ΔΔCq method (15).
The primers used for RT-qPCR are presented in Table I.
EGFP fluorescent reporter assay
A total of 2×104 A549 and SPC-A1 cells
were seeded in 48-well plates and co-transfected with 0.5 µg
pri-miR-421, 0.5 µg pcDNA3, 20 nM ASO-miR-421 or 20 nM ASO-NC with
0.5 µg EGFP-HOXP-WT or 0.5 µg EGFP-HOXP-Mut 3′UTR plasmids, as
aforementioned. A pDsRed2-1 vector (0.1 g/well) encoding red
fluorescent protein (RFP; Clontech Laboratories, Inc., Mountain
View, CA, USA) was used for normalization. At 48 h
post-transfection, the fluorescence intensities of EGFP and RFP
were detected at 528 and 488 nm using an F-4500 Fluorescence
Spectrophotometer (Hitachi, Ltd., Tokyo, Japan).
Western blotting
Western blotting was conducted in the present study
as previously described (16).
Briefly, A549 cells (6×108) were washed with 1X PBS
buffer and lysates were prepared with Radio-immunoprecipitation
Assay Buffer (Beyotime Institute of Biotechnology, Shanghai,
China), and protein concentrations were quantified using a
Bicinchoninic Acid Protein Assay kit (Beyotime Institute of
Biotechnology), according to the manufacturer's protocols. Protein
samples (25 µg) were separated by 12% SDS-PAGE and subsequently
transferred to polyvinylidene difluoride membranes. Subsequently,
the membranes were blocked with 5% non-fat milk in 1X Tris-buffered
saline with 0.05% Tween-20 for ~2 h at room temperature, followed
by incubation at 4°C overnight with the following primary
antibodies (unless otherwise noted, all antibodies were from Wanlei
Bio Co., Ltd., Beijing, China; www.wanleibio.cn/): Anti-GAPDH (1:2,000; cat. no.
WL01547); anti-E-cadherin (1:1,000; cat. no. WL01482);
anti-vimentin (1:1,000; cat. no. WL00742); anti-Bcl-2 (1:1,000;
cat. no. WL01556); anti-cleaved caspase-3 (1:1,500; cat. no.
WL01556); anti-cleaved PARP (1:1,000; cat. no. WL01932);
anti-β-catenin (1:1,000; cat. no. WL01160); anti-phosphorylated
β-catenin (1:1,000; cat. no. ab27798; Abcam, Cambridge, UK);
anti-cyclin D1(1:2,000; cat. no. WL01435a); and anti-c-myc
(1:3,000; cat. no. WL01781). Following three extensive washes with
PBS+Tween-20 (0.1%) for 20 min at room temperature, the membranes
were incubated with polyclonal goat anti-rabbit horseradish
peroxidase-conjugated secondary antibody (1:5,000; cat. no.
ab98512; Abcam) for 2 h at room temperature, followed by three
washes with PBS+Tween-20 for 20 min at room temperature. Protein
bands were visualized using an Enhanced Chemiluminescence Detection
kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), and images
were captured using LabWorks™ Image Acquisition (UVP, Inc., Upland,
CA, USA). Analysis Software UVP EC3 v3.0 software (UVP, LLC,
Phoenix, AZ, USA) were used to quantify band intensities and GAPDH
was used to normalize protein expression levels.
Cell Counting Kit-8 (CCK-8) and colony
formation assays
Cell proliferation was determined by a CCK-8 assay
(cat. no. WLa074a; Wanlei Bio, Co., Ltd.). At 24 h
post-transfection, A549 or SPC-A1 cells (5,000 cells/well) were
seeded in 96-well plates and cultured for 24, 48 or 72 h. A total
of 10 µl CCK-8 reagent was added to each well, and the plates were
incubated at 37°C for an additional 6 h. The optical density (OD)
at a wavelength of 450 nm was determined using a microplate reader
(Hitachi, Ltd.). The OD values reflect the relative number of
viable cells. For the colony formation assay, A549 or SPC-A1 cells
(2×103 cells/well) were seeded onto 12-well plates and
incubated for 10–14 days. The colonies were then fixed in 100%
methanol for 30 min, stained with crystal violet for 30 min at room
temperature and the numbers of macroscopically observable colonies
were recorded.
Transwell migration and invasion
assays
Briefly, A549 and SPC-A1 cells were seeded into 8-µm
cell culture inserts and placed in 24-well Transwell cell culture
plates. The upper chamber was coated with 100 µl diluted Matrigel
(2 mg/ml) for invasion assay. The lower chamber was filled with 500
µl of MEM-a medium containing 20% FBS. A549 and SPC-A1 cells
(6×104) in 200 µl of serum-free MEM-a medium were gently
each filter insert (loaded onto upper chamber) and incubated at
37°C for 48 h. The filter inserts were removed from the chambers
and the non-migrative cells were removed using the cotton swab,
fixed with methanol for 10 min and stained with Harris' hematoxylin
for 20 min. The samples were subsequently washed using the 0.9%
saline solution, dried and mounted onto slides. The migratory and
invasive cells were stained blue and six random fields were
examined under an inverted microscope (magnification, ×200) for
statistical analysis.
Cell cycle and apoptosis analysis by
flow cytometry
A549 and SPC-A1 cells were washed with cold 1X PBS.
After washing with PBS, 1×106 cells were fixed in 100%
ethanol for 10 min and incubated with 50 µg/ml propidium iodide
(PI; Nanjing KeyGen Biotech; www.keygentec.com.cn) for 10 min at room temperature
in the dark and analyzed within 20 min using a BD FACSCalibur (BD
Biosciences, Franklin Lakes, NJ, USA). Flow cytometry was used to
detect cell apoptosis and analyzed using FlowJo 7.6.1 (FlowJo LLC,
Ashland, OR, USA). For cell apoptosis, cells were collected for
determination of apoptosis rates using the Annexin V-FLUOS Κit
(Nanjing KeyGen Biotech Co., Ltd., Nanjing, China), according to
the manufacturer's protocols. All samples were assayed in
triplicate.
5-Εthynyl-2′-deoxyuridine (EdU)
staining
EdU staining was performed using the Click-iT EdU
Imaging kit (Invitrogen; Thermo Fisher Scientific, Inc.), according
to the manufacturer's protocols. In brief, A549 and SPC-A1 were
plated in 96-well plates. After the required treatment, EdU was
added into each well and incubated for 3 h. After washing twice
with PBS, the cells were fixed with 4% paraformaldehyde for 30 min
at room temperature. Then, cells were incubated with Apollo
staining reaction liquid for 30 min to detect the positive cells.
The cells were counterstained with 50 nM Hoechst 33342 at room
temperature. Immunofluorescence was observed with a fluorescence
microscope (DMI 3000B; Leica Microsystems GmbH, Wetzlar,
Germany).
Immunofluorescent microscopy
Transfected cells were seeded (2×103
cells/well) into 14-well chambers and incubated for 24 h; the cells
were washed with 1X PBS, fixed with 4% paraformaldehyde,
permeabilized with 0.5% Triton X-100 and incubated with 10% donkey
serum (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China) for 1 h. Subsequently, cells were incubated with
rabbit anti-β-catenin antibody (1:100; cat. no. WL01160; Wanlei Bio
Co., Ltd.) at 4°C overnight. The cells were stained for 1.5 h in
the dark at room temperature with a fluorescein
isothiocyanate-conjugated secondary antibody (1:300; cat. no.
ab150077; Goat Anti-Rabbit IgG H&L Alexa Fluor® 488;
Abcam). Nuclei were counterstained with DAPI (1:1,000;
Sigma-Aldrich; Merck KGaA) for 5 min in the dark at room
temperature. The slides were observed by fluorescent microscopy
(DMI 3000B; Leica Microsystems GmbH) at 488 and 528 nm through the
oil microscope (magnification, ×1,000).
Xenograft tumor formation assay
Animal protocols were approved by The First Hospital
of Qinhuangdao Animal Care and Use Committee (Qinhuangdao, China);
all animals received human care according to the Institutional
Animal Care and Treatment Committee of First Hospital of
Qinhuangdao (2016N185), and experiments were conducted in
accordance with the approved guidelines. Female BALB/c athymic nude
mice [four mice/group, two group (n=8); age, 6 weeks; weight,
>20 g] were purchased from The Institute of Zoology, Chinese
Academy of Sciences (Shanghai, China) were and maintained at
18–29°C, with relative humidity of 50–60%, 10-14-h light/dark cycle
(lights turned on at 8:00 every day and turned off at 18:00), and
free access to clean food and water. A total of 2×106
stably transfected (pcDNA3 or pri-miR-421) A549 cells were
implanted subcutaneously into the armpit of each mouse. All mice
were euthanized with CO2 at 30 days post-implantation.
Tumor weights were measured using an electronic scale.
TOP/FOP flash reporter assays
To determine the transcriptional activity of the Wnt
pathway, 5×106 A549 and SPC-A1 cells treated as
indicated were co-transfected with either the Wnt signaling
reporter TOP Flash or the negative control FOP Flash (EMD
Millipore, Billerica, MA, USA) according to the manufacturer's
protocol. A549 and SPC-A1 cells were transiently transfected with
either 2 µg pTOP flash or pFOP flash plasmids and 0.5 µg
pSV40-Renilla plasmid as an internal control (Promega
Corporation) using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) at room temperature for 48 h. The
dual-luciferase reporter assay system (Dual-Luciferase®
Reporter Assay system; cat. no. E1910; Promega Corporation) was
used to assay the firefly and Renilla luciferase activity
ratio.
Statistical analyses
Each experiment was performed in triplicate. The
differences between two groups were analyzed using a two-tailed
Student's t-tests; multiple group comparisons were performed via
two-way analysis of variance followed by the Bonferroni post hoc
test. All analyses were performed using SPSS 19.0 for Windows (IBM
Corp., Armonk, NY, USA) and GraphPad Prism 5 for Windows (GraphPad
Software Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
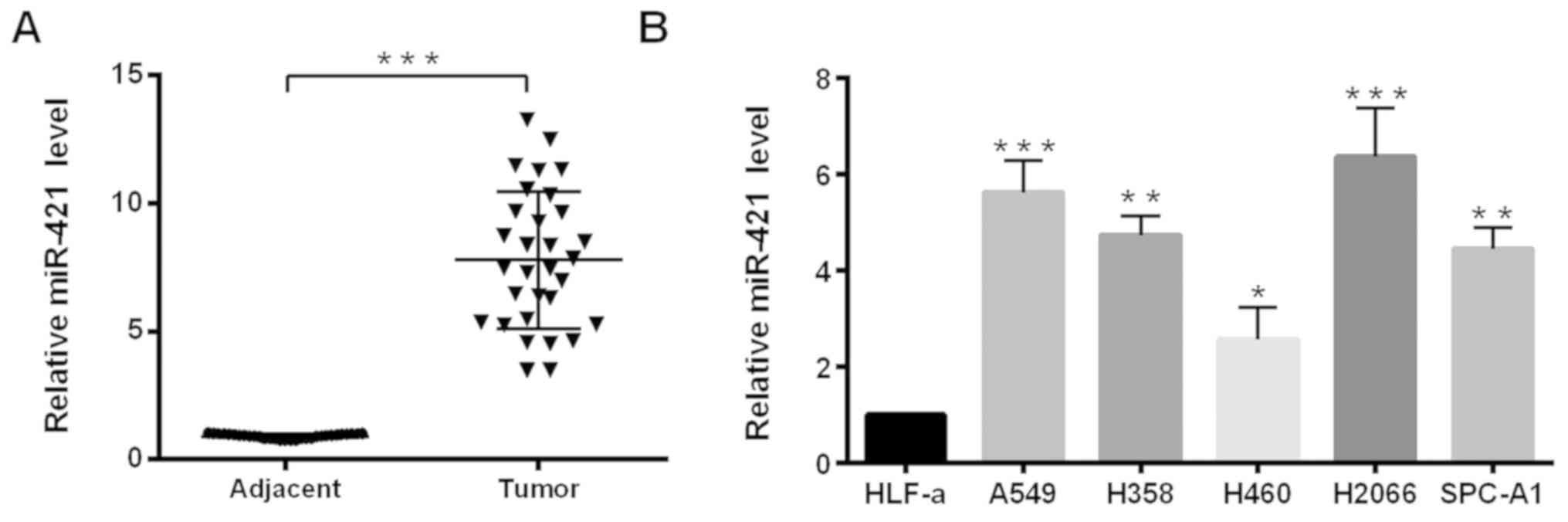
miR-421 is upregulated in NSCLC
tissues and cell lines
RT-qPCR analysis was performed to examine the
expression levels of miR-421 in NSCLC tissue and cell lines. The
results demonstrated that the expression levels of miR-421 were
significantly increased in 30 pairs of NSCLC tissues compared with
expression levels in the adjacent non-cancerous tissues (Fig. 1A). Similarly, compared with in a
normal human primary lung fibroblast cell line, HLF-a, all NSCLC
cell lines (A549, H358, H460, H2066 and SPC-A1) exhibited
significantly higher expression levels of miR-421 (Fig. 1B). A549 and SPC-A1 cell lines were
selected for further study, because they were easier to culture and
transfect compared with in the other NSCLC cell lines.
miR-421 promotes NSCLC cell growth,
migration and invasion in vitro
As miR-421 expression levels in NSCLC tissues and
cell lines were high, it was hypothesized that miR-421 may function
as an oncogene in the progression of NSCLC. First, the miR-421
overexpression plasmid and the synthesized ASO-miR-421 were used,
and the efficiency of the plasmids was confirmed in A549 and SPC-A1
cells by RT-qPCR (Fig. 2A); that
is, miR-421 expression levels were increased and decreased,
respectively, compared with the negative controls. Subsequently,
the effects of miR-421 on the malignant behaviors of NSCLC cells
were also investigated. Results from CCK-8 and colony formation
assays demonstrated that miR-421 overexpression significantly
promoted cell viability and colony formation (Fig. 2B and C, respectively), whereas
viability and colony formation were significantly inhibited in
cells transfected with ASO-miR-421 compared with the respective
pcDNA3 empty vector and ASO-NC groups in the two cells lines. To
investigate how miR-421 may promote A549 and SPC-A1 cell growth,
the effects of miR-421 on the cell cycle were analyzed. The cell
cycle distribution of A549 and SPC-A1 cells was observed to be
affected by alterations in miR-421 expression (Fig. 2D). EdU was used to examine cell
proliferation, the result showed that pri-miR-421 promoted the EdU
concentration and ASO-miR-421 reduced the EdU concentration
compared with the control groups in A549 and SPC-A1 cells (Fig. 2E). To investigate whether miR-421
affected the motility of NSCLC cells, Transwell migration and
invasion assays were performed using transfected A549 and SPC-A1
cells. The migratory and invasive abilities of A549 and SPC-A1
cells transfected with pri-miR-421 were notably increased, and
cells transfected with ASO-miR-421 were notably decreased compared
with the corresponding control-transfected cells (Fig. 2F).
 | Figure 2.miR-421 contributes to the oncogenic
activity of non-small cell lung cancer cells. (A-I) A549 and SPC-A1
cells were transfected with pri-miR-421, pcDNA3, ASO-miR-421 or
ASO-NC. (A) Reverse transcription-quantitative polymerase chain
reaction was used to detect the mRNA expression levels of miR-421.
(B) Viability was examined by Cell Counting Kit-8. (C) Results from
colony formation assays. (D) The cell cycle was analyzed by flow
cytometry following transfection. (E) EdU cell proliferation assay
was performed to assess cell proliferation; Nuclei were stained
with Hoechst 33342. (F) Transwell migration and Matrigel invasion
assays. (G) Protein expression levels of E-cadherin and vimentin
were detected by western blotting; GAPDH was used as a control and
for normalization. (H) Cell apoptosis was analyzed by flow
cytometry. (I) Protein expression levels of Bcl-2, cleaved
caspase-3 and cleaved PARP were examined by western blotting; GAPDH
was used for control normalization. The data are presented as the
mean ± standard deviation; *P<0.05, **P<0.01 and
***P<0.001 vs. the respective Control. ASO, antisense
oligonucleotide; Bcl-2, B cell lymphoma-2; E-cadherin, epithelial
cadherin; EdU, 5-ethynyl-2′-deoxyuridine; miR, microRNA; NC,
negative control; PARP, poly(ADP-ribose) polymerase; pcDNA3, empty
vector; pri, primary. |
Previous studies suggested that EMT serves important
role in cancer metastasis (17).
During the EMT process, a loss of epithelial markers and the
acquisition of mesenchymal markers have been observed (18). Therefore, the present study
evaluated the role of miR-421 in the EMT process. Western blot
analysis revealed that the expression levels of E-cadherin were
significantly decreased and that of vimentin was significantly
increased in miR-421-overexpressing cells compared with the pcDNA3
transfected group; ASO-miR-421 transfection induced a significant
increase in E-cadherin and a significant decrease in vimentin
expression levels within A549 cells compared with the ASO-NC
transfected group (Fig. 2G).
Collectively, these data indicated that miR-421 may promote the EMT
process, as well as the migration and invasion of NSCLC cells. In
addition, the rate of cell apoptosis was examined. The apoptotic
rates of A549 and SPC-A1 cells were significantly decreased in
response to miR-421 overexpression, whereas the downregulation of
miR-421 expression levels by ASO-miR-421 led to increased rates of
cell apoptosis (Fig. 2H).
Additionally, western blotting results indicated that miR-421
overexpression significantly upregulated the expression levels of
Bcl-2, but reduced that of cleaved caspase-3 and cleaved PARP in
A549compared with the pcDNA3vector group (Fig. 2I).
HOPX is a target of miR-421
To investigate the mechanism underlying the effects
of miR-421 on the malignant phenotypes exhibited by NSCLC cells,
the potential targets of miR-421 were predicted using the algorithm
programs TargetScan, microrna.org
and miRDB. Among the 451 predicted target genes, HOPX was selected
or further study as the association between HOPX and NSCLC
progression remains unknown.
To confirm whether HOPX is a target of miR-421,
fluorescent reporter plasmids with the HOPX-WT or HOPX-Mut 3′UTR
fragment were constructed (Fig.
3A). The fluorescence intensity of the HOPX-WT 3′UTR was
significantly suppressed when co-transfected with pri-miR-421
compared with cells that were co-transfected with pcDNA3 empty
vector (Fig. 3B); whereas the
opposite effects were observed when cells were co-transfected with
ASO-miR-421 compared with ASO-NC (Fig.
3B). However, these effects were abolished when the potential
miR-421 binding sites were mutated (Fig. 3B). The results also demonstrated
significantly lower expression of HOPX in cells overexpressing
miR-421 (pri-miR-421) and higher expression of HOPX in cells
knocking down miR-421 (ASO-miR-421) at the mRNA (Fig. 3C) and protein (Fig. 3D) levels in cells compared with the
control group cells. To verify the results obtained from the cell
lines, the collected tissues and the NCBI Gene Expression Omnibus
database were used for further analysis. The results revealed
significantly lower HOPX mRNA expression levels in the NSCLC
tissues and in the GEO database, compared with the adjacent normal
lung tissues (Fig. 3E and F,
respectively). Collectively, these results indicated that HOPX may
be negatively regulated by miR-421 in NSCLC cells.
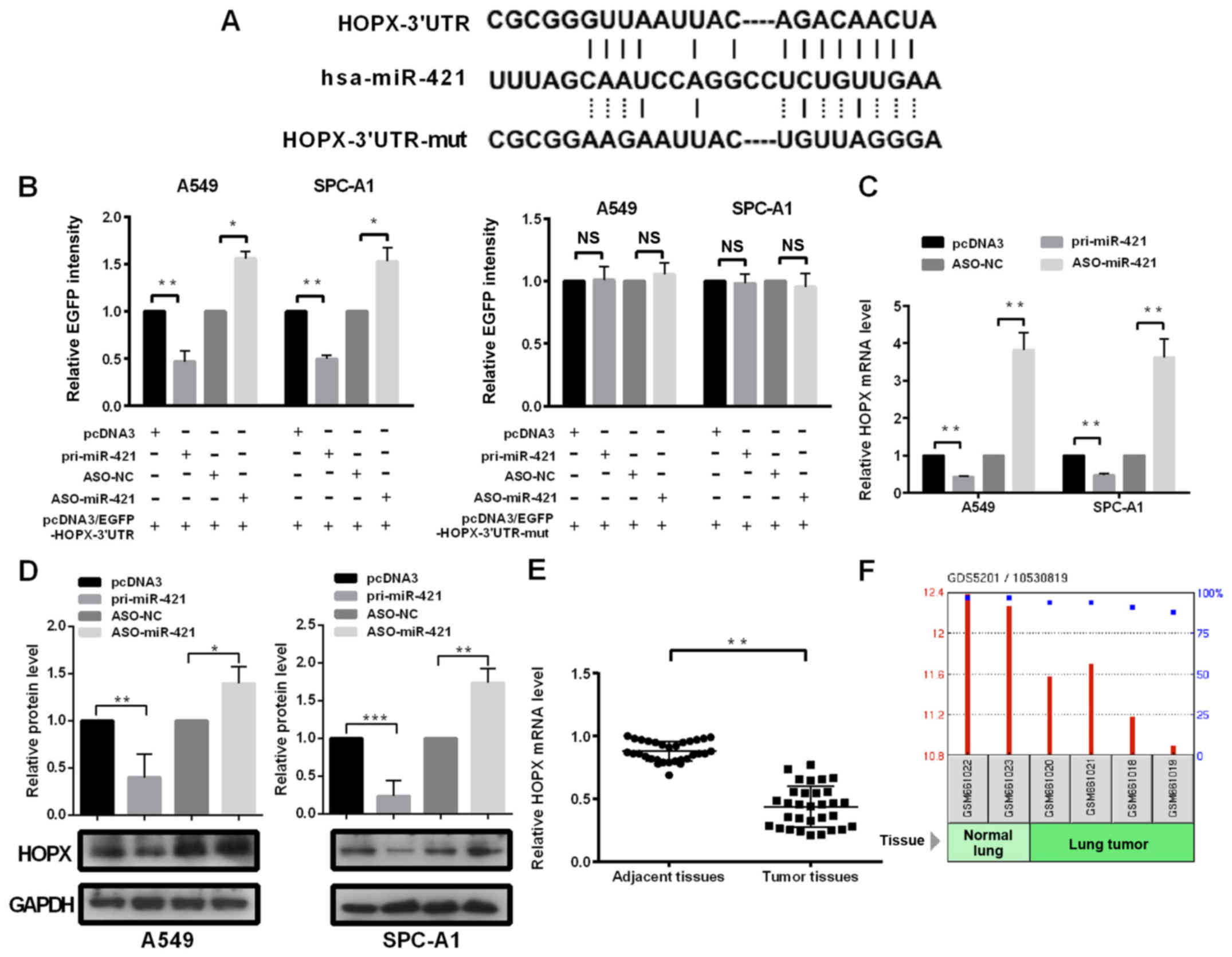 | Figure 3.miR-421 targets the HOPX 3′UTR and
suppresses its expression. (A) Predicted WT and Mut binding sites
for miR-421 in the 3′UTR of HOPX mRNA. (B) A549 and SPC-A1 cells
were co-transfected with the HOPX-WT or HOPX-Mut 3′UTR reporter
plasmids along with pri-miR-421, pcDNA3, ASO-NC or ASO-miR-421;
EGFP fluorescence intensity was measured at 48 h post-transfection
and normalized to pDsRed2-N1 vector. (C and D) A549 and SPC-A1
cells were transfected with pri-miR-421, pcDNA3, ASO-miR-421 or
ASO-NC and the (C) mRNA and (D) protein expression levels of HOPX
were determined by reverse transcription-quantitative polymerase
chain reaction and western blotting, respectively. (E and F)
Relative expression levels of HOPX in (E) paired non-small cell
lung cancer and adjacent non-cancerous tissues, and (F) NCBI Gene
Expression Omnibus database. Normal lung tissues and lung cancer
tissues. Data are presented as the mean ± standard deviation;
*P<0.05, **P<0.01, ***P<0.001. ASO, antisense
oligonucleotide; EGFP, enhanced green fluorescent protein; HOPX,
homeodomain-only protein; miR, microRNA; Mut, mutant; NC, negative
control; NS, not significant; pcDNA3, empty vector; pri, primary;
UTR, untranslated region; WT, wild-type. |
HOPX rescues the effects of miR-421 on
the malignant behaviors of NSCLC cells
As HOPX was predicted to be a direct target of
miR-421 in NSCLC, whether miR-421 may function as an oncogene in a
HOPX-dependent manner was investigated. Prior to performing the
functional studies, a HOPX overexpression vector, pHOPX, was
generated and the efficiency of transfection of A549 and SPC-A1
cells was analyzed by RT-qPCR (Fig.
4A); the results confirmed that HOPX mRNA expression levels
were increased compared with the Control-transfected cells.
Subsequently, the effects of HOPX on NSCLC cell viability, growth,
migration and invasion were investigated. Ectopic expression of
HOPX inhibited cell viability, growth, proliferation, migration,
invasion and EMT, induced cell apoptosis compared with the
pcDNA3+pcDNA3 group (Fig. 4B-I).
These results suggested that HOPX may suppress various malignant
phenotypes of NSCLC cells in vitro, and serve an opposing
role to miR-421.
 | Figure 4.HOPX overexpression reverts
miR-421-mediated cell growth and metastasis-associated traits. (A)
Expression levels of HOPX were analyzed by reverse
transcription-quantitative polymerase chain reaction to confirm
successful transfection of the pHOPX overexpression vector in A549
and SPC-A1 cells; β-actin was used for normalization. (B-I) A549
and SPC-A1 cells were co-transfected with pcDNA3+pHOPX or
pri-miR-421+pHOPX compared with the pcDNA3+pcDNA3 group. (B) Cell
viability was determined by Cell Counting Kit-8. (C) Colony
formation assay. (D) Cell cycle analysis was detected by flow
cytometry. (E) EdU assay was performed to assess the proliferative
ability of transfected cells; nuclei were stained with Hoechst
33342. (F) Effects on cell migration and invasion were examined by
Transwell and Matrigel assays, respectively. (G) Protein expression
levels of epithelial-mesenchymal transition-associated markers
E-cadherin and vimentin were examined by western blotting; GAPDH
was used as a loading control and for normalization. (H) Rate of
apoptosis was determined via flow cytometry. (I) Protein expression
levels of Bcl-2, cleaved caspase-3 and cleaved PARP were analyzed
by western blotting. Data are presented as the mean ± standard
deviation; *P<0.05, **P<0.01 and ***P<0.001. ASO-NC,
antisense negative control; ASO, antisense oligonucleotide; Bcl-2,
B cell lymphoma-2; E-cadherin, epithelial cadherin; EdU,
5-ethynyl-2′-deoxyuridine; HOPX, homeodomain-only protein; miR,
microRNA; NS, not significant; PARP, poly(ADP-ribose) polymerase;
pcDNA3, empty vector; pHOPX, HOPX overexpression vector. |
To confirm that the phenotype of miR-421 may arise
from the suppression of HOPX expression, rescue experiments were
performed. HOPX partially suppressed the viability, growth,
proliferation migration, invasion of NSCLC cells, counteracting the
promoting effects on the malignant phenotype induced by miR-421
(Fig. 4B-I). Collectively, these
data demonstrated that HOPX may be an important functional target
gene in NSCLC cells.
miR-421 regulates the Wnt/β-catenin
signaling pathway mediated by HOPX
Chromatin immunoprecipitation-sequencing analyses
have revealed that HOPX binds to Wnt target genes, which indicated
that HOPX may be involved in the Wnt/β-catenin signaling pathway
(19). The overexpression of
miR-421 significantly induced, whereas ectopic expression of HOPX
significantly decreased phosphorylated β-catenin expression
compared with pcDNA3 vector-transfected cells (Fig. 5A); no significant differences were
observed for total β-catenin expression. This result suggested that
miR-421 may induce the Wnt/β-catenin signaling pathway.
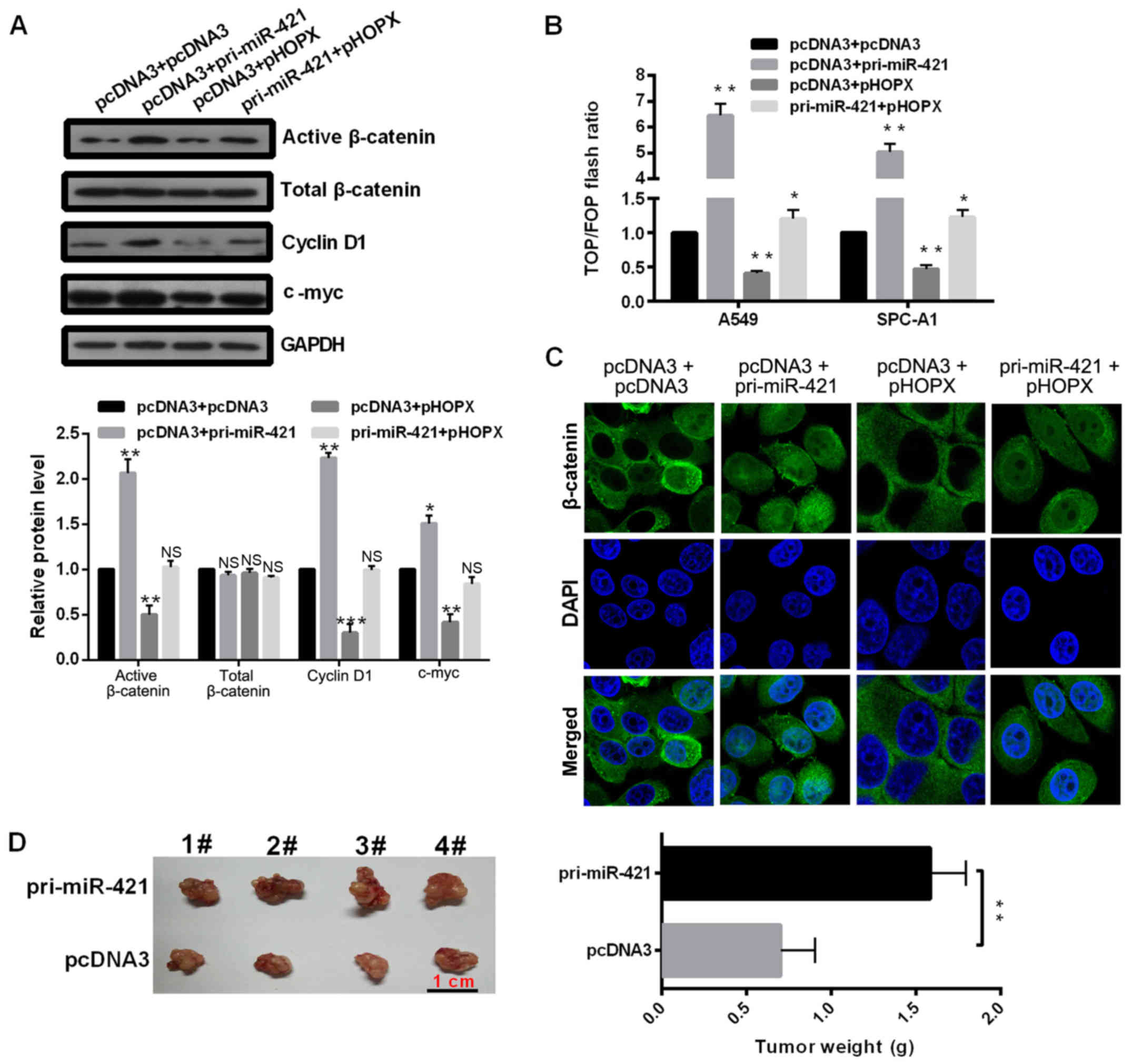 | Figure 5.miR-421 regulates the Wnt/β-catenin
signaling pathway by targeting HOPX. (A) Protein expression levels
of active β-catenin, total β-catenin, cyclin D1 and c-myc were
measured by western blot analysis. (B) TOP/FOP analysis. (C)
β-catenin immunostaining (green) and DAPI staining (blue) were
performed in A549 cells following transfection with the indicated
plasmids. (D) Tumor size and weight were examined from mice
xenografted with A549 cells that were transfected with pri-miR-421
or pcDNA3 negative control. Data are presented as the mean ±
standard deviation; *P<0.05, **P<0.01, ***P<0.001. HOPX,
homeodomain-only protein; pcDNA3, empty vector; pHOPX, HOPX
overexpression vector; miR, microRNA; NS, not significant; pri,
primary. |
Subsequently, the effects of miR-421 and HOPX on the
protein expression levels of cyclin D1 and c-myc were investigated,
which are the downstream target genes of the Wnt signaling pathway.
The results demonstrated that miR-421 overexpression significantly
promoted and HOPX overexpression significantly inhibited the
expression levels of cyclin D1 and c-myc compared with the pcDNA3
group (Fig. 5A). To further
investigate whether the effects of miR-421 on Wnt/β-catenin
signaling pathway are mediated by HOPX, pri-miR-421 and pHOPX
plasmids were co-transfected into the cells. The results
demonstrated that HOPX may attenuate the induction of the
Wnt/β-catenin signaling pathway associated with miR-421. In
addition, TOP/FOP reporter assay was performed and the results
revealed that miR-421 overexpression increased the TOP/FOP flash
ratio, but he ratio was suppressed by HOPX overexpression, compared
with Control-transfected A549 and SPC-A1 cells (Fig. 5B). Immunofluorescence analysis
indicated that miR-421 promoted and HOPX inhibited the nuclear
distribution of β-catenin in A549 cells (Fig. 5C), indicating that miR-421 actives
but HOPX inactivates the Wnt pathway. Finally, to confirm the role
of miR-421 in vivo, a mouse xenograft model was generated
using A549 cells transfected with either pcDNA3 or pri-miR-421
overexpression vector (Fig. 5D).
miR-421 significantly increased tumor weight in nude mice, which
suggested that miR-421 may promote the malignant phenotype of NSCLS
in vivo.
Discussion
Previous studies have demonstrated that numerous
molecules and processes are involved in the onset and development
of NSCLC, including altered epigenetic regulation associated with
miRNAs (20–22). miRNAs are involved in a variety of
cellular processes in numerous human diseases, particularly in the
development and progression of cancer. Several dysregulated miRNAs
have been reported in NSCLC including miR-224, miR-107, miR-484 and
miR-384 (8,23–27).
In addition, evidence has indicated that miRNAs are aberrantly
expressed in human cancers and may function as tumor suppressors or
oncogenes (28). miR-421 has been
demonstrated to be dysregulated in certain cancers and functions
with varying roles. For instance, Li et al (29) reported that upregulated expression
of miR-421 is associated with poor prognosis in non-small-cell lung
cancer. Zhang et al (30)
also reported that MEG3/miR-421/E-cadherin regulatory axis may be a
novel therapeutic target for breast cancer. Chen et al
(31) reported that
miR-421promoted nasopharyngeal carcinoma cell proliferation and
anti-apoptosis and as a novel regulatory mechanism for inactivation
of FOXO4 in nasopharyngeal carcinoma.
In the present study, the expression levels of
miR-421 in NSCLC tissues and cell lines were analyzed and the
results showed that miR-421 was upregulated. In addition, to
investigate the biological roles of miR-421 in NSCLC, a series of
functional experiments were performed. The present study
demonstrated that overexpression of miR-421 promoted NSCLC cell
growth, migration and invasion in vitro. Furthermore, the
results of the present study demonstrated that miR-421 may promote
the progression of the cell cycle. The expression levels of
anti-apoptotic Bcl-2, and cleaved caspase-3 and cleaved PARP were
investigated, and miR-421 was observed to increase the expression
levels of Bcl-2, and inhibit the protein expression levels of
cleaved caspase-3 and cleaved PARP. In addition, the role of
miR-421 on the expression of E-cadherin and vimentin were analyzed
in the present study; these proteins have been reported to serve an
important role in tumor metastasis (32). The results of the present study
indicated that miR-421 inhibited the protein expression of
E-cadherin, but increased that of vimentin, thereby promoting cell
migration and invasion via the EMT process. Furthermore, inhibition
of miR-421 expression had opposing effects. In addition, miR-421
was also observed to promote tumor growth in vivo in the
present study.
The target genes of miRNAs usually mediate their
function. In the present study, the potential targets of miR-421
were predicted and HOPX was selected for further analysis. The HOPX
gene is considered to be necessary for the regulation of cardiac
growth and development (33). The
downregulation of HOPX was reported to lead to cardiac hypertrophy
and heart failure (33). HOPX
usually functions as a transcriptional inhibitor (19,34,35).
It has been reported that HOPX is downregulated in gastric cancer,
esophageal squamous cell carcinoma and uterine endometrial cancer,
due to silencing of the HOPX promoter through hyper methylation
(36–38); however, further investigation into
the function of HOPX in NSCLC is required.
Results from the present study revealed HOPX as a
direct target of miR-421 in NSCLC. Contrary to miR-421, HOPX
overexpression exhibited an inhibitory effect on the malignant
phenotypes of NSCLC cells in the present study. Additionally, the
rescue experiments revealed HOPX rescued the miR-421-mediated role
on cell proliferation and metastasis-associated traits.
Furthermore, the underlying molecular mechanism by which miR-421
affects the progression of NSCLC was investigated in the present
study. miR-421 overexpression increased the protein expression
levels of active β-catenin, cyclin D1 and c-myc, which indicated
induction of the Wnt/β-catenin signaling pathway.
Taken together, the results of the present study
indicated that miR-421 may function as an oncogene in the
progression of NSCLC through the downregulation of HOPX linking it
to the Wnt/β-catenin signaling pathway. The findings of the present
study may provide a novel insight into NSCLC progression and
contribute to the development of novel diagnostic and treatment
approaches for NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL conceived, designed, drafted and performed the
experiments. CW, KG and JL performed the RT-qPCR, western blotting,
EdU and Transwell assays and analyzed the data. RJ provided help in
conceiving and designing the study and revising the manuscript. All
authors approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical Review
Committee of The First Hospital of Qinhuangdao S(ethics approval
no. QHD20160235). Written informed consent was obtained from all
enrolled patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Holdenrieder S and Stieber P: New
challenges for laboratory diagnostics in non-small cell lung
cancer. Cancer Biomark. 6:119–121. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wood SL, Pernemalm M, Crosbie PA and
Whetton AD: Molecular histology of lung cancer: From targets to
treatments. Cancer Treat Rev. 41:361–375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marmarelis M, Thompson JC, Aggarwal C,
Evans TL, Carpenter E, Cohen RB, Langer CJ and Bauml J: Emerging
uses of circulating tumor DNA in advanced stage non-small cell lung
cancer. Ann Transl Med. 5:3802017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Laskin JJ and Sandler AB: State of the art
in therapy for non-small cell lung cancer. Cancer Invest.
23:427–442. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Djuranovic S, Nahvi A and Green R: A
parsimonious model for gene regulation by miRNAs. Science.
331:550–553. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stahlhut C and Slack FJ: Combinatorial
action of MicroRNAs let-7 and miR-34 effectively synergizes with
erlotinib to suppress non-small cell lung cancer cell
proliferation. Cell Cycle. 14:2171–2180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu L, Cui S, Zhang R, Shi Y and Luo L:
MiR-421 inhibits the malignant phenotype in glioma by directly
targeting MEF2D. Am J Cancer Res. 7:857–868. 2017.PubMed/NCBI
|
|
14
|
Ge X, Liu X, Lin F, Li P, Liu K, Geng R,
Dai C, Lin Y, Tang W, Wu Z, et al: MicroRNA-421 regulated by HIF-1α
promotes metastasis, inhibits apoptosis, and induces cisplatin
resistance by targeting E-cadherin and caspase-3 in gastric cancer.
Oncotarget. 7:24466–24482. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Long MJ, Wu FX, Li P, Liu M, Li X and Tang
H: MicroRNA-10a targets CHL1 and promotes cell growth, migration
and invasion in human cervical cancer cells. Cancer Lett.
324:186–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weidenfeld K and Barkan D: EMT and
stemness in tumor dormancy and outgrowth: Are they intertwined
processes? Front Oncol. 8:3812018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jain R, Li D, Gupta M, Manderfield LJ,
Ifkovits JL, Wang Q, Liu F, Liu Y, Poleshko A, Padmanabhan A, et
al: HEART DEVELOPMENT. Integration of Bmp and Wnt signaling by Hopx
specifies commitment of cardiomyoblasts. Science. 348:aaa60712015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Q, Jiang S, Song A, Hou S, Wu Q, Qi L
and Gao X: HOXD-AS1 functions as an oncogenic ceRNA to promote
NSCLC cell progression by sequestering miR-147a. Onco Targets Ther.
10:4753–4763. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pan JY, Zhang F, Sun CC, Li SJ, Li G, Gong
FY, Bo T, He J, Hua RX, Hu WD, et al: miR-134: A human cancer
suppressor? Mol Ther Nucleic Acids. 6:140–149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu X, Xu Y, Pang Z, Guo F, Qin Q, Yin T,
Sang Y, Feng C, Li X, Jiang L, et al: Knockdown of SUMO-activating
enzyme subunit 2 (SAE2) suppresses cancer malignancy and enhances
chemotherapy sensitivity in small cell lung cancer. J Hematol
Oncol. 8:672015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Z, Yang Y and Zhang X: MiR-770
inhibits tumorigenesis and EMT by targeting JMJD6 and regulating
WNT/β-catenin pathway in non-small cell lung cancer. Life Sci.
188:163–171. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang L, Liu W, Zhang YP and Huang XR: The
miR-224 promotes non-small cell lung cancer cell proliferation by
directly targeting RASSF8. Eur Rev Med Pharmacol Sci. 21:3223–3231.
2017.PubMed/NCBI
|
|
25
|
Lu C, Xie Z and Peng Q: MiRNA-107 enhances
chemosensitivity to paclitaxel by targeting antiapoptotic factor
Bcl-w in non small cell lung cancer. Am J Cancer Res. 7:1863–1873.
2017.PubMed/NCBI
|
|
26
|
Li T, Ding ZL, Zheng YL and Wang W:
MiR-484 promotes non-small-cell lung cancer (NSCLC) progression
through inhibiting Apaf-1 associated with the suppression of
apoptosis. Biomed Pharmacother. 96:153–164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan N, Zhang J, Cheng C, Zhang X, Feng J
and Kong R: MicroRNA-384 represses the growth and invasion of
non-small-cell lung cancer by targeting astrocyte elevated
gene-1/Wnt signaling. Biomed Pharmacother. 95:1331–1337. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang L, Huang J, Yang N, Greshock J,
Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR,
et al: microRNAs exhibit high frequency genomic alterations in
human cancer. Proc Natl Acad Sci USA. 103:9136–9141. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Cui X, Li Y, Zhang T and Li S:
Upregulated expression of miR-421 is associated with poor prognosis
in non-small-cell lung cancer. Cancer Manag Res. 10:2627–2633.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang W, Shi S, Jiang J, Li X, Lu H and
Ren F: LncRNA MEG3 inhibits cell epithelial-mesenchymal transition
by sponging miR-421 targeting E-cadherin in breast cancer. Biomed
Pharmacother. 91:312–319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen L, Tang Y, Wang J, Yan Z and Xu R:
miR-421 induces cell proliferation and apoptosis resistance in
human nasopharyngeal carcinoma via downregulation of FOXO4. Biochem
Biophys Res Commun. 435:745–750. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rafael D, Doktorovová S, Florindo HF,
Gener P, Abasolo I, Schwartz S Jr and Videira MA: EMT blockage
strategies: Targeting Akt dependent mechanisms for breast cancer
metastatic behaviour modulation. Curr Gene Ther. 15:300–312. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Trivedi CM, Cappola TP, Margulies KB and
Epstein JA: Homeodomain only protein X is down-regulated in human
heart failure. J Mol Cell Cardiol. 50:1056–1058. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Trivedi CM, Zhu W, Wang Q, Jia C, Kee HJ,
Li L, Hannenhalli S and Epstein JA: Hopx and Hdac2 interact to
modulate Gata4 acetylation and embryonic cardiac myocyte
proliferation. Dev Cell. 19:450–459. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Palpant NJ, Wang Y, Hadland B, Zaunbrecher
RJ, Redd M, Jones D, Pabon L, Jain R, Epstein J, Ruzzo WL, et al:
Chromatin and transcriptional analysis of mesoderm progenitor cells
identifies HOPX as a regulator of primitive hematopoiesis. Cell
Rep. 20:1597–1608. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamashita K, Kim MS, Park HL, Tokumaru Y,
Osada M, Inoue H, Mori M and Sidransky D: HOP/OB1/NECC1 promoter
DNA is frequently hypermethylated and involved in tumorigenic
ability in esophageal squamous cell carcinoma. Mol Cancer Res.
6:31–41. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamaguchi S, Asanoma K, Takao T, Kato K
and Wake N: Homeobox gene HOPX is epigenetically silenced in
human uterine endometrial cancer and suppresses estrogen-stimulated
proliferation of cancer cells by inhibiting serum response factor.
Int J Cancer. 124:2577–2588. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ooki A, Yamashita K, Kikuchi S, Sakuramoto
S, Katada N, Kokubo K, Kobayashi H, Kim MS, Sidransky D and
Watanabe M: Potential utility of HOP homeobox gene promoter
methylation as a marker of tumor aggressiveness in gastric cancer.
Oncogene. 29:3263–3275. 2010. View Article : Google Scholar : PubMed/NCBI
|