Introduction
Glaucoma is a widely known, multi-factorial disease,
which may result in apoptosis of retinal ganglion cells. According
to the World Health Organization, glaucoma is the second principal
cause of blindness and the most common cause of irreversible
blindness in the world (1,2). In glaucoma, the anterior and
posterior segments of the eye are affected, and serious damage may
be detected in the trabecular meshwork (3). Oxidative stress is considered to be
responsible for the molecular damage in the anterior chamber.
Primary open-angle glaucoma (POAG) is the most common type of
glaucoma, accounting for 60–70% all glaucoma (4). A candidate protein that may be
associated with POAG is myocilin (MYOC), encoded by the MYOC gene.
MYOC mutations are common in patients with POAG with high levels of
intraocular pressure (IOP) (5,6).
Additionally, mutations in optineurin were identified in patients
with POAG (7). Previous studies
suggested that an abnormal expression of serine/threonine-protein
kinase TBK1 is a cause of normal-tension glaucoma (8–10).
Furthermore, a previous study suggested that the calcium
load-activated calcium channel was involved in glaucoma and that
cyclin-dependent kinase 4 inhibitor B antisense RNA 1 was
upregulated in the retina of a rat model of glaucoma (11).
However, even substantial decreases in IOP are not
able to prevent the development and progression of glaucoma in a
number of clinical cases (12).
Glaucoma-associated cell death is primarily caused by apoptosis,
which is triggered by oxidative stress via mitochondrial damage,
inflammation, endothelial dysregulation and dysfunction, and
hypoxia (13). In general,
glaucoma is not preventable; however, the vast majority of patients
may maintain useful visual function for life if they have early
detection and appropriate treatment (14). Therefore, for the prevention of
glaucoma, emphasis must be placed on early detection, and early
diagnosis and treatment.
The rapid development and application of
high-throughput sequencing technology has provided a comprehensive
and rapid analytical method for the study of the pathogenesis of
glaucoma, and provide novel ideas for the future treatment of
glaucoma (15). The present study
aimed to analyze high-throughput transcriptome data from tissue
samples of patients with glaucoma and a normal control group. The
data was used in bioinformatics analyses to identify key
transcription factors (TFs) associated with glaucoma, to examine
the pathogenesis of glaucoma and provide a basis for the diagnosis
of glaucoma and drug development.
Materials and methods
Microarray expression profiling in
Gene Expression Omnibus (GEO)
The GEO is the largest database of high-throughput
gene expression data that was developed and is maintained by the
National Center for Biotechnology Information (16). The GEO was searched to obtain gene
expression profiling studies of glaucoma subjects. The following
key search terms were used: [‘glaucoma’ (Medical subject headings
Terms) OR ‘glaucoma’ (All Fields)] AND ‘Homo sapiens’
(porgn) AND ‘gse’ (Filter). The selection criteria were as follows:
i) The selected dataset must include genome-wide mRNA transcriptome
data; ii) the data was obtained from the trabecular meshwork tissue
samples of glaucoma and normal control trabecular meshwork tissue
samples; and iii) normalized and raw datasets were considered.
Following selection, two sets of GSE27276 (17) and GSE4316 (18) glaucoma mRNA data were obtained
(19,20).
Identification of differentially
expressed genes (DEGs) in glaucoma compared with normal
controls
Background correction was performed on the raw data.
The normalization was performed using the Linear Models for
Microarray (Limma version 3.30.13) Data package in R (21). Subsequently, two-tailed Student's
t-tests were performed to calculate individual P-values. Stouffer's
test was used to merge individual P-values, and multiple comparison
correction was performed using the Benjamini and Hochberg method to
obtain the false discovery rate (FDR) (22). Genes with FDR<0.001 were
selected as DEGs. Finally, the DEGs in glaucoma vs. normal were
identified.
Functional annotation of DEGs
Gene Ontology (GO) (23) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) (24) pathway
enrichment analysis were performed to detect the biological
functions and potential pathways associated with DEGs using
GeneCoDis3 (http://genecodis.cnb.csic.es/analysis) as previously
described (19). The GO functions
of the DEGs were determined according to the three categories of:
‘Biological process’; ‘molecular functions’; and ‘cellular
component’. Pathway enrichment analysis was based on the KEGG
database, as previously described (25).
Protein-protein interaction (PPI)
network construction
In order to identify candidate genes involved in the
formation of glaucoma, PPI networks of significant DEGs were
constructed, according to the data from Biological General
Repository for Interaction Datasets (BioGRID; http://thebiogrid.org/). Based on the existing data of
protein interaction in the BioGRID database, Cytoscape (www.cytoscape.org/; (version 3.5.0) was used to search
the top 100 upregulated and downregulated mRNAs. The PPI network
interaction map was generated subsequent to the exclusion of genes
that were not differentially expressed.
Screening for TFs of the top 20 DEGs
and construction of TF regulatory network
For the top 20 DEGs, the 2 kb upstream promoter
regions were downloaded from the University of California Santa
Cruz (UCSC) Genome Browser website (genome.ucsc.edu). The TRANScription FACtor (TRANSFAC)
website match tool (gene-regulation.com/pub/databases.html) was
subsequently used to analyze TFs capable of binding to the promoter
region of the DEGs. TFs that exhibited altered expression in
glaucoma with FDR<0.001 were selected. Position Weight Matrix
scanning was used to scan the human genome sequence to obtain the
protein-coding genes that were regulated by the differentially
expressed TFs. Following removal of redundant information, the
glaucoma-specific transcriptional regulatory network was
constructed using Cytoscape software.
In silico validation of DEGs using
GEO
The GEO database (GSE9944) was used to validate the
expression of selected glaucoma DEGs. The expression levels of
these genes were compared between the glaucoma cases and the normal
group. The expression of five genes, cAMP responsive element
binding protein 1 (CREB1), fibronectin 1 (FN1), keratin 19 (KRT19),
lipocalin 2 (LCN2) and paired box 6 (PAX6) was detected, with the
difference of expression levels presented as box-plots.
Results
Differential expression analysis of
genes in glaucoma
The probes corresponding to multiple genes were
removed, and the average gene expression to which multiple probes
corresponded with was calculated. Finally, the intersection of
15,757 genes was obtained.
A total of two gene expression microarray datasets
(GSE27276 and GSE4316) were used for the analysis. Compared with
the normal controls, 1,935 DEGs in glaucoma were obtained
(P<0.05); among these, 951 genes were upregulated and 984 genes
were downregulated. The top 40 most significantly up- or
downregulated genes are summarized in Table I. Among which, PAX6 (26), LCN2 (27), and MAOA (28) were downregulated and were
associated with glaucoma. The DEGs were screened for clustering
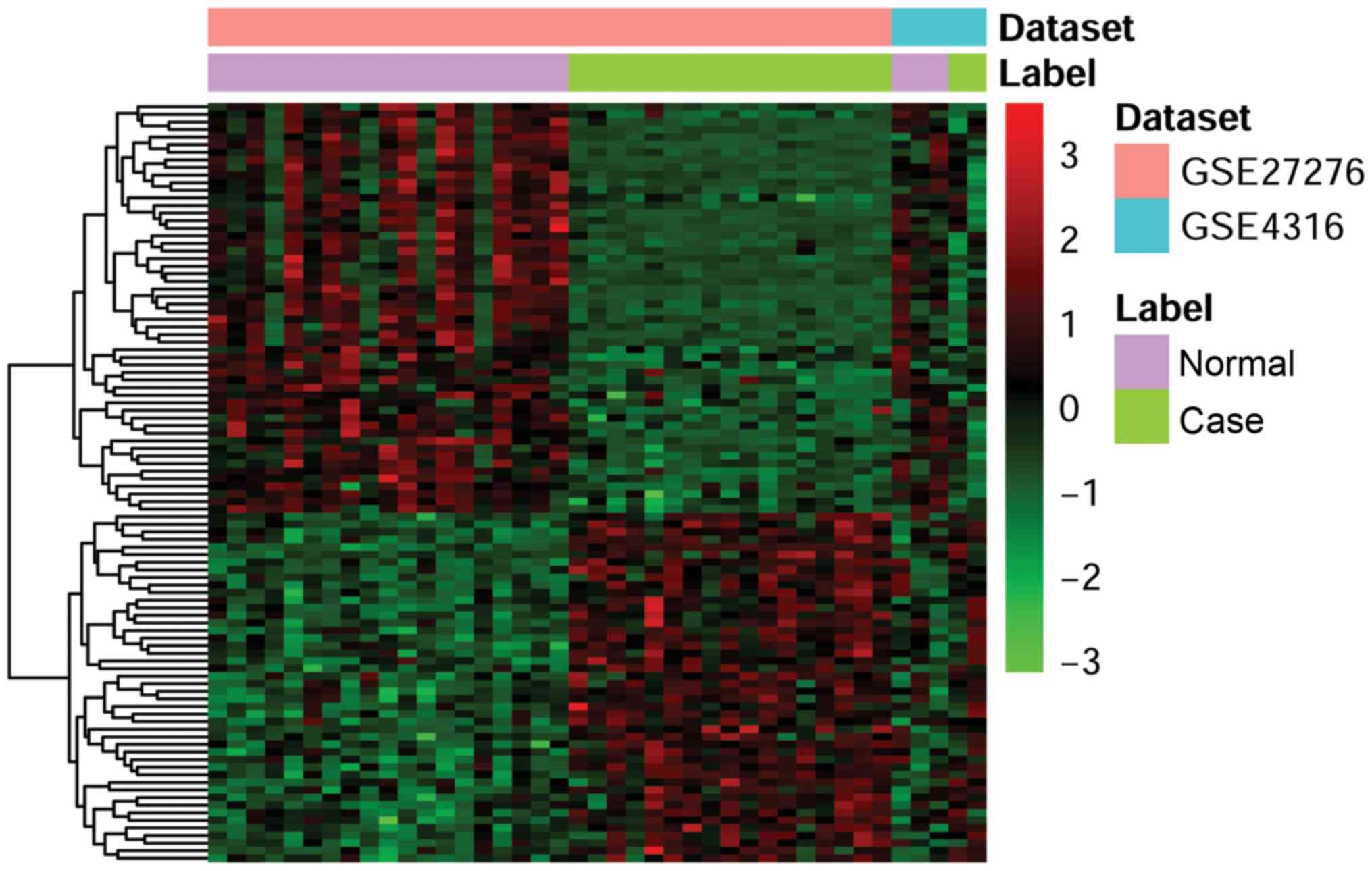
analysis. The heatmap produced by cluster analysis of the two sets
of cDNA microarray data is presented in Fig. 1.
 | Table I.Top 40 differentially expressed
mRNAs. |
Table I.
Top 40 differentially expressed
mRNAs.
| A, Upregulated |
|---|
|
|---|
| ID | Symbol | Combined.ES | P-value | FDR |
|---|
| 3043 | HBB | 5.16018337 | 0 | 0 |
| 9415 | FADS2 | 2.35332790 |
1.40×10−9 |
3.68×10−6 |
| 56172 | ANKH | 2.33974344 |
7.72×10−9 |
1.11×10−5 |
| 3113 | HLA-DPA1 | 2.22895504 |
1.17×10−8 |
1.41×10−5 |
| 5538 | PPT1 | 2.27584322 |
1.72×10−8 |
1.69×10−5 |
| 83643 | CCDC3 | 2.09336726 |
2.56×10−8 |
2.32×10−5 |
| 81618 | ITM2C | 2.10210833 |
5.11×10−8 |
3.81×10−5 |
| 8857 | FCGBP | 2.00251174 |
1.07×10−7 |
6.25×10−5 |
| 4166 | CHST6 | 1.94047822 |
1.48×10−7 |
7.07×10−5 |
| 4753 | NELL2 | 1.97462732 |
2.45×10−7 |
1.07×10−4 |
| 8718 | TNFRSF25 | 1.80276729 |
8.11×10−7 |
2.84×10−4 |
| 81552 | VOPP1 | 1.82118066 |
8.70×10−7 |
2.91×10−4 |
| 2191 | FAP | 1.74786612 |
1.01×10−6 |
3.09×10−4 |
| 187 | APLNR | 1.74800187 |
1.02×10−6 |
3.09×10−4 |
| 51196 | PLCE1 | 1.77349072 |
1.23×10−6 |
3.61×10−4 |
| 23676 | SMPX | 1.76299253 |
1.37×10−6 |
3.78×10−4 |
| 51226 | COPZ2 | 1.72176939 |
1.72×10−6 |
4.24×10−4 |
| 1290 | COL5A2 | 1.71261435 |
1.78×10−6 |
4.31×10−4 |
| 57559 | STAMBPL1 | 1.69907540 |
1.81×10−6 |
4.31×10−4 |
| 2331 | FMOD | 1.75952553 |
1.83×10−6 |
4.31×10−4 |
|
| B,
Downregulated |
|
| ID | Symbol |
Combined.ES | P-value | FDR |
|
| 3880 | KRT19 | −2.54377891 |
3.23×10−10 |
1.70×10−6 |
| 116039 | OSR2 | −2.44873456 |
7.08×10−10 |
2.23×10−6 |
| 5648 | MASP1 | −2.39197514 |
2.01×10−9 |
4.52×10−6 |
| 79845 | RNF122 | −2.31970767 |
3.00×10−9 |
5.91×10−6 |
| 3934 | LCN2 | −2.20821475 |
8.44×10−9 |
1.11×10−6 |
| 57801 | HES4 | −2.22824712 |
1.65×10−8 |
1.69×10−5 |
| 10232 | MSLN | −2.09546485 |
2.65×10−8 |
2.32×10−5 |
| 9245 | GCNT3 | −2.11365461 |
4.07×10−8 |
3.38×10−5 |
| 84525 | HOPX | −2.01200085 |
5.57×10−8 |
3.81×10−5 |
| 4128 | MAOA | −2.03058309 |
7.95×10−8 |
5.22×10−5 |
| 5080 | PAX6 | −2.09500899 |
8.92×10−8 |
5.62×10−5 |
| 64073 | C19orf33 | −2.02417662 |
1.26×10−7 |
6.86×10−5 |
| 3866 | KRT15 | −1.96990194 |
1.40×10−7 |
6.92×10−5 |
| 7148 | TNXB | −1.92147744 |
1.40×10−7 |
6.92×10−5 |
| 163732 | CITED4 | −1.94673475 |
1.94×10−7 |
9.01×10−5 |
| 1675 | CFD | −1.97823285 |
2.68×10−7 |
1.14×10−4 |
| 6510 | SLC1A5 | −1.95192053 |
2.77×10−7 |
1.15×10−4 |
| 1638 | DCT | −1.84682776 |
3.22×10−7 |
1.30×10−4 |
| 3861 | KRT14 | −1.77674842 |
7.12×10−7 |
2.61×10−4 |
| 10053 | AP1M2 | −1.77498212 |
8.52×10−7 |
2.92×10−4 |
Functional annotation
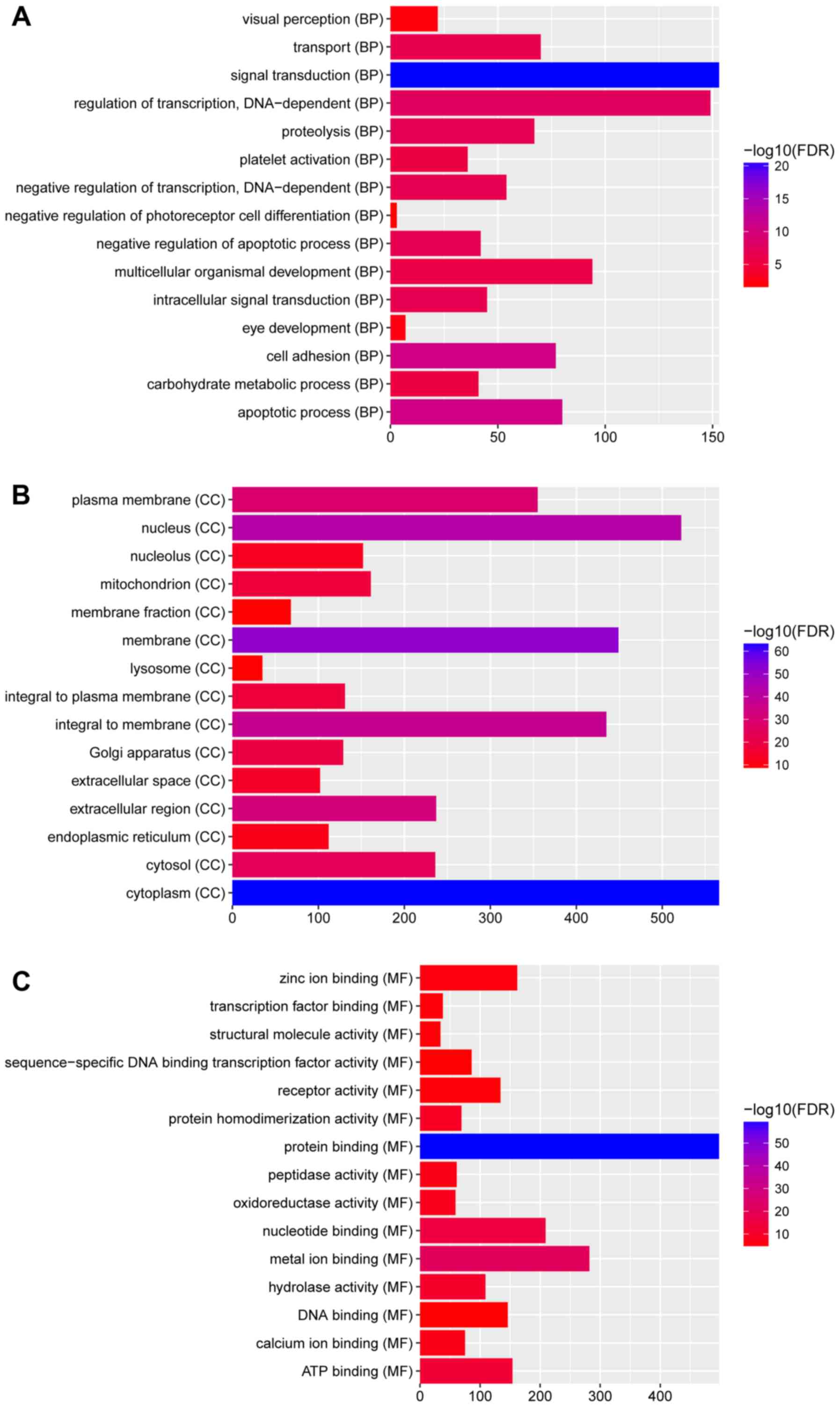
In Fig. 2, GO
enrichment demonstrated that the DEGs were significantly enriched
the ‘Biological processes’ categories: ‘eye development’
(FDR=4.15×10−3); ‘visual perception’
(FDR=7.13×10−3); ‘negative regulation of insulin
receptor signaling pathway’ (FDR=1.47×10−2); the
‘Cellular components’ categories: ‘membrane’
(FDR=3.41×10−52); ‘endoplasmic reticulum’
(FDR=3.76×10−12); ‘cytoplasm’
(FDR=1.73×10−63); and ‘Molecular functions’ categories:
‘nucleotide binding’ (FDR=8.50×10−16); ‘hydrolase
activity’ (FDR=8.50×10−16); and ‘protein binding’
(FDR=6.15×10−60).
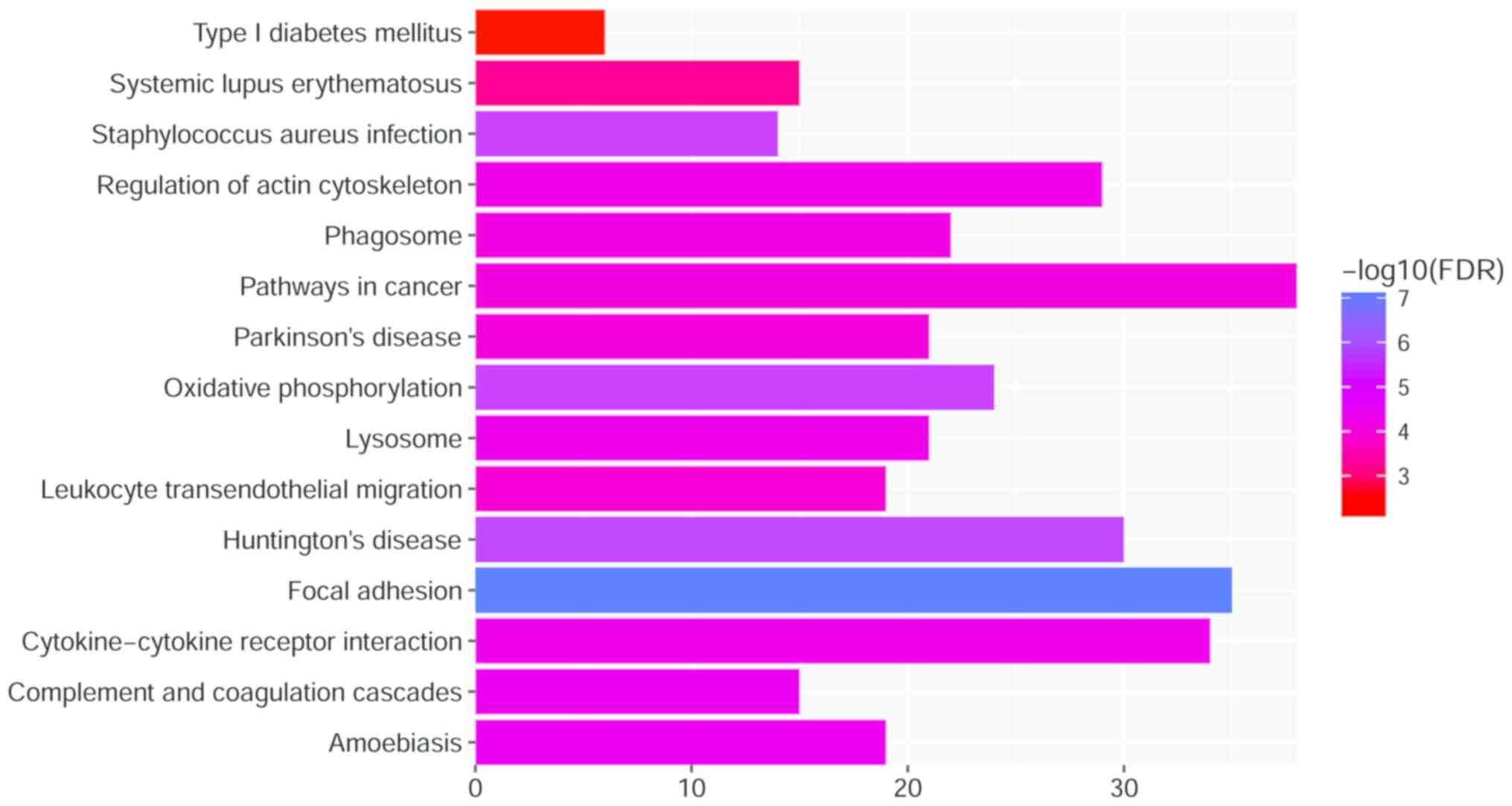
Furthermore, as presented in Fig. 3, the results of the KEGG pathway
enrichment demonstrate that DEGs were enriched in
‘Staphylococcus aureus infection’
(FDR=6.15×10−60); ‘Pathways in cancer’
(FDR=8.66×10−5); ‘Systemic lupus erythematosus’; and
‘Type I diabetes mellitus’ (FDR=0.84×10−3).
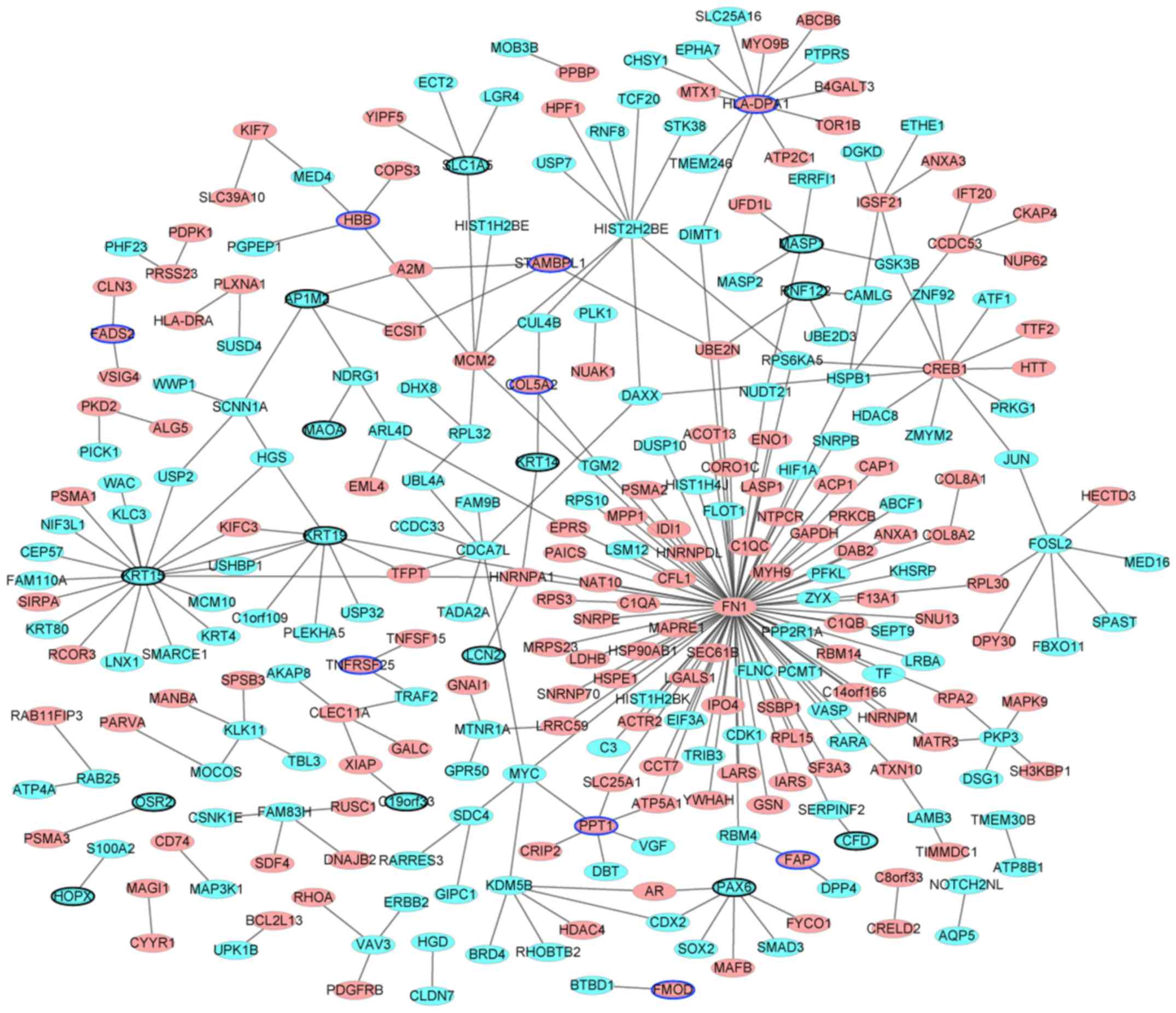
PPI network construction
Following removal of the non-DEGs, the PPI network
was established. The results are presented in Fig. 4. The network consisted of nodes and
edges. The nodes in the network represent the proteins and the
edges represent the interactions between them. There were 290 nodes
and 290 edges identified. Among them, the genes with higher degrees
were: FN1 (degree=92); KRT15 (degree=19); major histocompatibility
complex, class II, DP α 1 (HLA-DPA1; degree=12); CREB1 (degree=11);
KRT19 (degree=9); histone cluster 2 H2B family member e (degree=9);
cell division cycle associated 7 like (degree=7); PAX6 (degree=7);
FOS like 2, AP-1 transcription factor subunit (degree=7);
minichromosome maintenance complex component 2 (degree=6);
palmitoyl-protein thioesterase 1 (degree=6); and lysine demethylase
5B (degree=6). The hub proteins were FN1 (degree=92), KRT15
(degree=19) and HLA-DPA1 (degree=12).
TFs of the top 20 DEGs and TF
regulation network
A total of 250 TF binding associations were
identified, including multiple binding sites of the same TF in a
gene. Among these, 36 TFs were involved. In Table II, the top six TFs with the most
downstream genes (the top 20 differentially expressed genes) were
included: PAX4; solute carrier family 22 member 1 (1-Oct);
hepatocyte nuclear factor 4 α (HNF-4); NK2 homeobox 5 (Nkx2-5);
PAX6; and ELK1, ETS transcription factor (Elk-1).
 | Table II.Top six TFs with the highest number
of downstream regulatory genes and their target genes. |
Table II.
Top six TFs with the highest number
of downstream regulatory genes and their target genes.
| TF name | Number of regulated
genes | Regulated
genes |
|---|
| Pax-4 | 11 | HES4, ITM2C, ANKH,
HLA-DPA1, RNF122, LCN2, CCDC3, NELL2, MSLN, CHST6, KRT19 |
| 1-Oct | 11 | ITM2C, MASP1, HOPX,
ANKH, HLA-DPA1, OSR2, RNF122, CCDC3, HBB, NELL2, MSLN |
| HNF-4 | 11 | ITM2C, MASP1,
HLA-DPA1, LCN2, MAOA, HBB, FADS2, NELL2, MSLN, CHST6, KRT19 |
| Nkx2-5 | 9 | PPT1, HOPX, RNF122,
CCDC3, FADS2, NELL2, MSLN, CHST6, KRT19 |
| Pax-6 | 8 | ITM2C, MASP1, ANKH,
OSR2, CCDC3, NELL2, GCNT3, KRT19 |
| Elk-1 | 5 | ITM2C, MASP1, ANKH,
LCN2, CCDC3, FADS2, CHST6 |
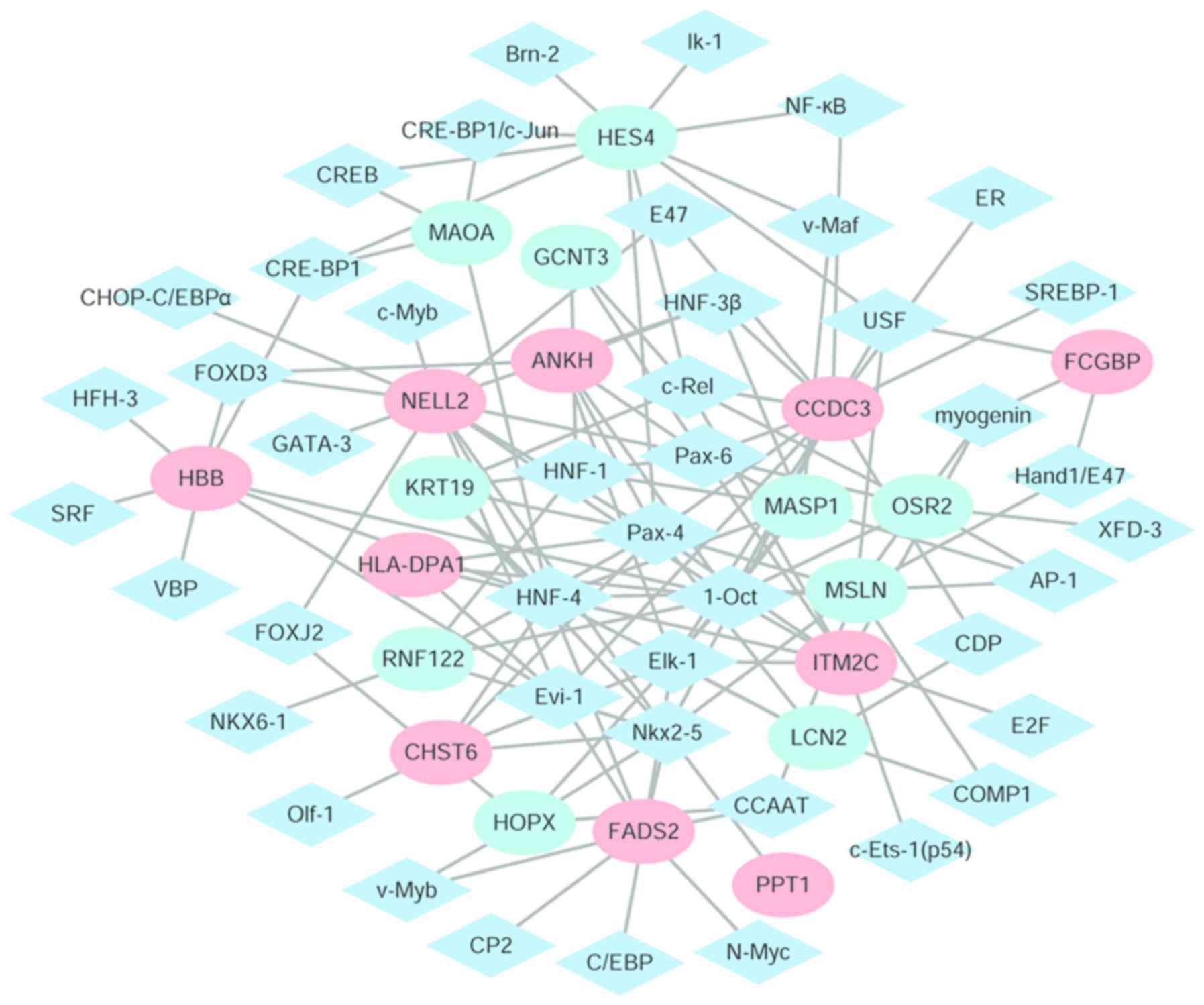
Cytoscape software was used to identify the TFs and
the differentially expressed regulatory network of their target
genes (Fig. 5). There were 61
nodes, including 44 transcription factors and 20 differentially
expressed target genes and 132 edges identified. The eight target
genes with the highest degrees were: Coiled-coil domain containing
3 (degree=14); neural tissue-specific epidermal growth factor-like
repeat domain-containing protein (NELL2; degree=13); hes family
bHLH transcription factor 4 (degree=10); mesothelin (degree=10);
integral membrane protein 2C (degree=9); fatty acid desaturase 2
(degree=9); mannan binding lectin serine peptidase 1 (degree=8);
and hemoglobin subunit b (HBB; degree=8).
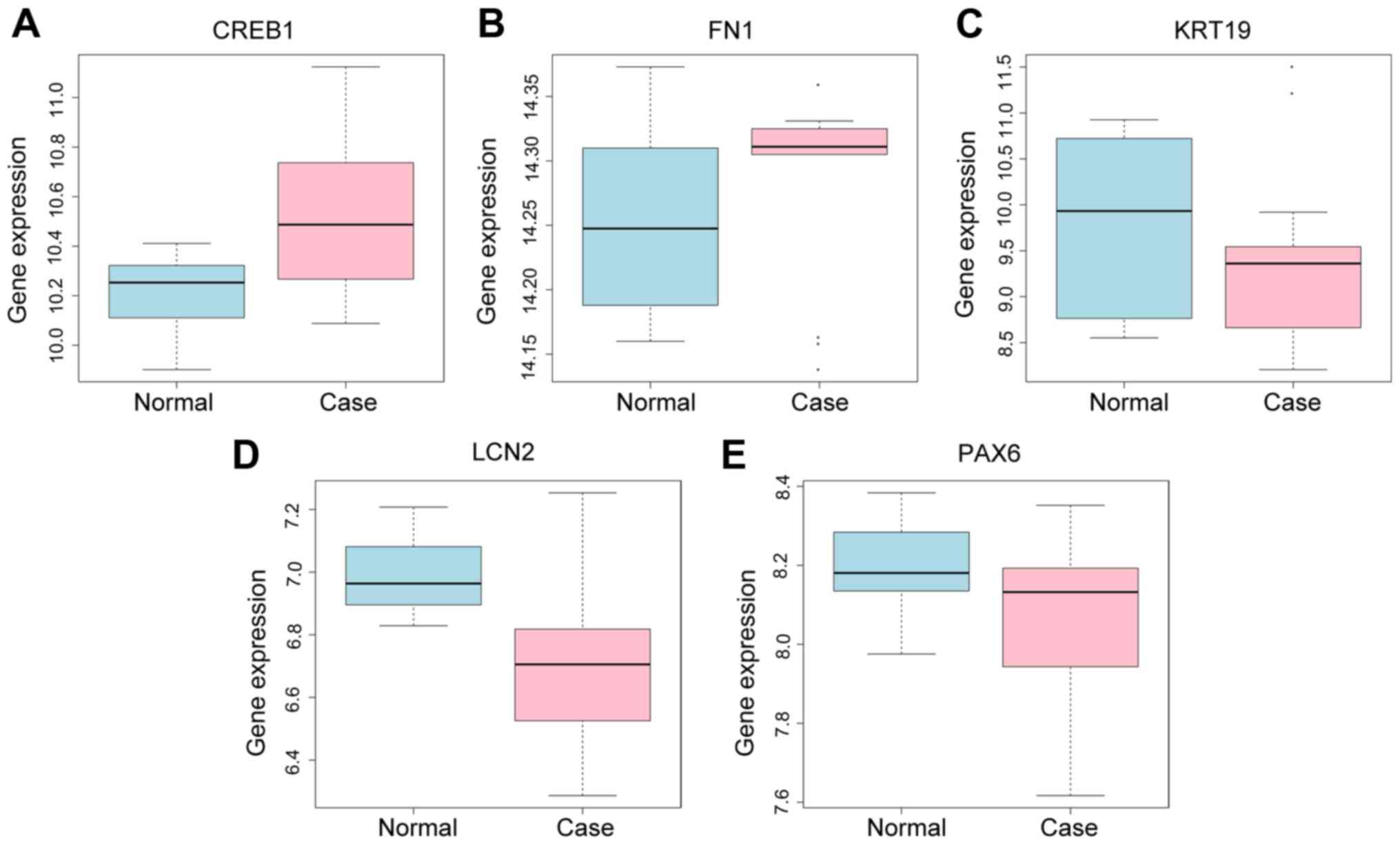
Validation of DEGs in GEO GSE9944
dataset
To define the key genes with key roles in glaucoma,
the GEO GSE9944 repository was searched for high throughput gene
expression data and hybridization arrays, chips and microarrays. As
demonstrated in Fig. 6, the genes
CREB1 and FN1 were upregulated, and the genes KRT19, LCN2 and PAX6
were downregulated. These results were consistent with the
integrated analysis in the GSE27276 and GSE4316 datasets.
Discussion
Glaucoma is the leading cause of irreversible
blindness worldwide and there is no effective treatment at present
(27). In the present study,
integrated analysis was performed using data obtained from the GEO
database. KEGG, GO and other biological information databases, and
R analysis tools were used to analyze the DEGs. A total of 1,935
DEGs in glaucoma (951 genes were upregulated, 984 genes were
downregulated) were obtained. Critical signaling pathways that
affected the pathogenesis of glaucoma, including ‘eye development’
(FDR=0.00415533) and ‘visual perception’ (FDR=0.00713283) were
identified. In addition, based on the promoter sequence of DEGs
obtained from UCSC, a TF regulatory network was constructed using
the match tool of the TRANSFAC website to obtain the corresponding
TFs.
LCN2, encoded by lcn2 gene, is a neutrophil
gelatinase-associated lipocalin. The 25-kD LCN2 protein is able to
bind small lipophilic substances, including bacteria-derived
lipopolysaccharide and formylpeptides (28). The protein additionally functions
as a modulator of inflammation. Khalyfa et al (29) suggested that LCN2 was highly
upregulated in glaucoma. In the results of the present study, LCN2
was downregulated and the validation result in the GSE9944 dataset
was consistent with the analysis. A previous study additionally
demonstrated that the coadministration of a reversible monoamine
oxidase A (MAOA) inhibitor with epinephrine may be useful for
patients with glaucoma (28). In
the results of the present study, MAOA was downregulated.
CREB1 is a transcription regulator activated in
response to harmful stress stimuli, including hypoxia and oxidative
stress, and is involved in the cellular defense against these
stresses (30). High IOP, optic
nerve damage and visual field defects are the primary pathological
features of glaucoma, with high IOP being the most common (27). CREB has been demonstrated to
promote neuronal survival (31),
and a previous study indicated that CREB has a neuroprotective
effect against hydrogen peroxide-induced retinal ganglion cell
death via 2 downstream cell survival genes, including brain-derived
neurotrophic factor and apoptosis regulator Bcl-2 (32). In a study by Yasuda et al
(12), an in silico pathway
analysis conducted in rats with axonal injury induced by
transecting the right optic nerve, CREB1 was the most significant
upstream regulator. In the present study, CREB1 (degree=11) was the
in the top 3 genes with the highest degrees in the PPI network. In
the KEGG analysis, CREB1 was enriched in the function of
Huntington's disease.
FN1 is a glycoprotein within the extracellular
matrix, which has a crucial role in cell adhesion, growth,
migration and differentiation (33,34).
Cellular FN is secreted in a soluble form by a variety of different
cell types, including fibroblasts, chondrocytes, macrophages and
epithelial cells, and is later assembled into an insoluble
extracellular matrix (35). FN has
a multimodular structure organized into functional domains that
interact with multiple binding partners, including integrins,
heparin sulfate proteoglycans, collagen, glycosaminoglycans,
proteoglycans, heparin, fibrin and bacteria (36). A previous study by Anshu et
al (2) identified that FN1
(P=0.0003) was expressed at a significantly increased level in
aqueous humor samples compared with controls. In the present study,
FN1 was the top protein with the highest degree in the PPI network.
Additionally, in the KEGG analysis, FN1 was enriched in the
function of ‘Focal adhesion’.
NELL2 is a secreted glycoprotein that is
predominantly expressed in neural tissues. NELL2 is additionally
involved in promoting the neuronal survival required for the
formation of a sexually dimorphic nucleus of the preoptic area in
male rats (37). In the TF
regulation network in the present study, NELL2 (degree=13) was the
in top 2 genes and was regulated by PAX4, 1-Oct, HNF-4, Nkx2-5 and
PAX6. Additionally, NELL2 was among the top 40 DEGs. These results
indicated that NELL2 may have a key role in the pathogenesis of
glaucoma.
The OMIM database was also utilized in the present
study, and it was identified that PAX6 was key gene. PAX6 is a
member of the paired box gene family, which encodes a
transcriptional regulator involved in oculogenesis and other
developmental processes. Halder et al (38) suggested that the ectopic expression
of Drosophila PAX6 induces ectopic eye development. Wawersik
and Maas (39) reviewed the role
of PAX6 and other genes in vertebrate and fly oculogenesis; the
study demonstrated that regulators of eye development are conserved
across large evolutionary distances. The expression patterns and
conserved functional domains suggested that Pax6 may have important
roles in vertebrate eye formation. Fujimura et al (40) demonstrated that PAX6 appears to
function as a pleiotropic regulator, directing the development of
ocular tissues in concert with the signaling pathway and,
concomitantly, regulating the expression of structural components
of the eye, including shielding pigment. In the present study, PAX6
was downregulated in the patients with glaucoma. In addition, in
the PPI network, PAX6 was one of the top eight genes with the
highest degrees.
In conclusion, six DEGs (LCN2, MAOA, HBB, PAX6, FN1
and CREB1) were identified to be involved in the process of
glaucoma. From the two GEO datasets analyzed, 1,935 DEGs (951
upregulated and 984 downregulated genes) were identified between
glaucoma and normal controls. DEG validation in the GEO GSE9944
dataset was consistent with the integrated analysis of the GSE27276
and GSE4316 datasets. These results may contribute to the
elucidation of novel potential biomarkers, reveal the underlying
pathogenesis and identify novel therapeutic targets for the
treatment of glaucoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by Science and
Technology Development Plan of Jining [grant no. (2016) 56–31] and
Project Medical and Health Technology Development Project of
Shandong (grant no. 2017WS528).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JF and JX conceived the study and wrote the
manuscript. JX performed the data analyses. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tham YC, Li X, Wong TY, Quigley HA, Aung T
and Cheng CY: Global prevalence of glaucoma and projections of
glaucoma burden through 2040: A systematic review and
meta-analysis. Ophthalmology. 121:20812014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anshu A, Price MO, Richardson MR, Segu ZM,
Lai X, Yoder MC and Price FW Jr: Alterations in the aqueous humor
proteome in patients with a glaucoma shunt device. Mol Vis.
17:1891–1900. 2011.PubMed/NCBI
|
|
3
|
Saccà SC, Gandolfi S, Bagnis A, Manni G,
Damonte G, Traverso CE and Izzotti A: From DNA damage to functional
changes of the trabecular meshwork in aging and glaucoma. Ageing
Res Rev. 29:26–41. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kayange PC, Nkume HB, Feyi-Waboso A, Kalua
K, Msukwa G and Schwering Schulze M: Presentation of primary open
angle glaucoma (POAG) at lions sight first eye hospital in
Blantyre, Malawi. Malawi Med J. 26:60–62. 2014.PubMed/NCBI
|
|
5
|
Kwon YH, Fingert JH, Kuehn MH and Alward
WL: Primary open-angle glaucoma. N Engl J Med. 2:1113–1124. 2009.
View Article : Google Scholar
|
|
6
|
Nazir S, Mukhtar M, Shahnawaz M, Farooqi
S, Fatima N, Mehmood R and Sheikh N: A novel single nucleotide
polymorphism in exon 3 of MYOC enhances the risk of glaucoma. PLoS
One. 13:e01951572018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang C, Xie L, Wu Z, Cao Y, Zheng Y, Pang
CP and Zhang M: Detection of mutations in MYOC, OPTN, NTF4, WDR36
and CYP1B1 in Chinese juvenile onset open-angle glaucoma using
exome sequencing. Sci Rep. 8:44982018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fingert JH, Robin AL, Stone JL, Roos BR,
Davis LK, Scheetz TE, Bennett SR, Wassink TH, Kwon YH, Alward WL,
et al: Copy number variations on chromosome 12q14 in patients with
normal tension glaucoma. Hum Mol Genet. 20:2482–2494. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kawase K, Allingham RR, Meguro A, Mizuki
N, Roos B, Solivan-Timpe FM, Robin AL, Ritch R and Fingert JH:
Confirmation of TBK1 duplication in normal tension glaucoma. Exp
Eye Res. 96:178–180. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ritch R, Darbro B, Menon G, Khanna CL,
Solivan-Timpe F, Roos BR, Sarfarzi M, Kawase K, Yamamoto T, Robin
AL, et al: TBK1 gene duplication and normal-tension glaucoma. JAMA
Ophthalmol. 132:544–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burdon KP, Macgregor S, Hewitt AW, Sharma
S, Chidlow G, Mills RA, Danoy P, Casson R, Viswanathan AC, Liu JZ,
et al: Genome-wide association study identifies susceptibility loci
for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet.
43:574–578. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yasuda M, Tanaka Y, Omodaka K, Nishiguchi
KM, Nakamura O, Tsuda S and Nakazawa T: Transcriptome profiling of
the rat retina after optic nerve transection. Sci Rep. 6:287362016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi Z, Rudzinski M, Meerovitch K,
Lebrun-Julien F, Birman E, Di Polo A and Saragovi HU:
Alpha2-macroglobulin is a mediator of retinal ganglion cell death
in glaucoma. J Biol Chem. 283:29156–29165. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Konstas AG, Maskaleris G, Gratsonidis S
and Sardelli C: Compliance and viewpoint of glaucoma patients in
greece. Eye (Lond). 14:752–756. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tajiri-Utagawa E, Hara M, Takahashi K,
Watanabe M and Wakita T: Development of a rapid high-throughput
method for high-resolution melting analysis for routine detection
and genotyping of noroviruses. J Clin Microbiol. 47:435–440. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang WS, Wang YH, Zhu XT and Wu CJ:
Genome-wide profiling of miRNA and mRNA expression in Alzheimer's
disease. Med Sci Monit. 23:2721–2731. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Allingham RR, Qin X, Layfield D,
Dellinger AE, Gibson J, Wheeler J, Ashley-Koch AE, Stamer WD and
Hauser MA: Gene expression profile in human trabecular meshwork
from patients with primary open-angle glaucoma. Invest Ophthalmol
Vis Sci Sep. 54:6382–6389. 2013. View Article : Google Scholar
|
|
18
|
Liton PB, Luna C, Challa P, Epstein DL and
Gonzalez P: Genome-wide expression profile of human trabecular
meshwork cultured cells, nonglaucomatous and primary open angle
glaucoma tissue. Mol Vis. 12:774–790. 2006.PubMed/NCBI
|
|
19
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Rese 41 (Database Issue).
D991–D995. 2013.
|
|
20
|
Li JJ, Wang BQ, Fei Q, Yang Y and Li D:
Identification of candidate genes in osteoporosis by integrated
microarray analysis. Bone Joint Res. 5:594–601. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Klipper-Aurbach Y, Wasserman M,
Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, Karp M,
Benjamini Y, Hochberg Y and Laron Z: Mathematical formulae for the
prediction of the residual beta cell function during the first two
years of disease in children and adolescents with insulin-dependent
diabetes mellitus. Med Hypotheses. 45:486–490. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang F, Wang R, Li Q, Qu X, Hao Y, Yang J,
Zhao H, Wang Q, Li G, Zhang F, et al: A transcriptome profile in
hepatocellular carcinomas based on integrated analysis of
microarray studies. Diagn Pathol. 12:42017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dufier JL, Rozet JM, Kaplan J and Roche O:
From congenital glaucoma to chronic open angle glaucoma in
adulthood: A clinical and genetic continuum. Bull Acad Natl Med.
197:133–141. 2013.PubMed/NCBI
|
|
27
|
Wang X, Huai G, Wang H, Liu Y, Qi P, Shi
W, Peng J, Yang H, Deng S and Wang Y: Mutual regulation of the
Hippo/Wnt/LPA/TGF-β signaling pathways and their roles in glaucoma
(review). Int J Mol Med. 41:1201–1212. 2018.PubMed/NCBI
|
|
28
|
Alpízar-Alpízar W, Laerum OD, Illemann M,
Ramírez JA, Arias A, Malespín-Bendaña W, Ramírez V, Lund LR,
Borregaard N and Nielsen BS: Neutrophil gelatinase-associated
lipocalin (NGAL/Lcn2) is upregulated in gastric mucosa infected
with Helicobacter pylori. Virchows Archiv. 455:225–233.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Khalyfa A, Chlon T, Qiang H, Agarwal N and
Cooper NG: Microarray reveals complement components are regulated
in the serum-deprived rat retinal ganglion cell line. Mol Vis.
13:293–308. 2007.PubMed/NCBI
|
|
30
|
Maeda I, Ueda T, Koide R, Inatomi M,
Fukado Y, Uchida E, Oguchi K and Yasuhara H: Ocular hypotensive
effects of monoamine oxidase-a inhibitors in rabbit. Jpn J
Ophthalmol. 32:211–218. 1988.PubMed/NCBI
|
|
31
|
Ajmone-Cat MA, De Simone R, Nicolini A and
Minghetti L: Effects of phosphatidylserine on p38 mitogen activated
protein kinase, cyclic AMP responding element binding protein and
nuclear factor-kappaB activation in resting and activated
microglial cells. J Neurochem. 84:413–416. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Walton M, Woodgate AM, Muravlev A, Xu R,
During MJ and Dragunow M: CREB phosphorylation promotes nerve cell
survival. J Neurochem. 73:1836–1842. 1999.PubMed/NCBI
|
|
33
|
Shao Y, Yu Y, Zhou Q, Li C, Yang L and Pei
CG: Inhibition of miR-134 protects against hydrogen
peroxide-induced apoptosis in retinal ganglion cells. J Mol
Neurosci. 56:461–471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pankov R and Yamada KM: Fibronectin at a
glance. J Cell Sci. 115:3861–3863. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yi W, Xiao E, Ding R, Luo P and Yang Y:
High expression of fibronectin is associated with poor prognosis,
cell proliferation and malignancy via the NF-κB/p53-apoptosis
signaling pathway in colorectal cancer. Oncol Rep. 36:3145–3153.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jun HK, Jung YJ and Choi BK: Inflammasome
activators induce fibronectin expression and release in
macrophages. Cell Microbiol. 19:2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jeong JK, Ryu BJ, Choi J, Kim DH, Choi EJ,
Park JW, Park JJ and Lee BJ: NELL2 participates in formation of the
sexually dimorphic nucleus of the pre-optic area in rats. J
Neurochem. 106:1604–1613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Halder G, Callaerts P and Gehring WJ:
Induction of ectopic eyes by targeted expression of the eyeless
gene in Drosophila. Science. 267:1788–1792. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wawersik S and Maas RL: Vertebrate eye
development as modeled in Drosophila. Hum Mol Genet. 9:917–925.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fujimura N, Klimova L, Antosova B,
Smolikova J, Machon O and Kozmik Z: Genetic interaction between
Pax6 and β-catenin in the developing retinal pigment epithelium.
Dev Genes Evol. 225:121–128. 2015. View Article : Google Scholar : PubMed/NCBI
|