Introduction
Osteosarcoma (OS) is a highly malignant primary bone
neoplasm, which has a high rate in adolescents (1). In the majority of patients with OS,
survival is significantly prolonged by radiotherapy and
chemotherapy combined with surgery; however, patients with distant
metastasis have poor treatment options, particularly when it has
spread to the lungs (2). With the
advent of high-throughput sequencing technology, interest in long
non-coding RNAs (lncRNAs) is gradually increasing. lncRNAs are a
class of RNA molecules that are >200 nt long and lack an open
reading frame (3,4). Numerous studies have revealed that
lncRNAs are involved in biological developmental regulatory
processes, including cell differentiation, growth and apoptosis
(5,6). Furthermore, several lncRNAs have been
demonstrated to be involved in various human diseases (7). Notably, aberrantly expressed lncRNAs
have been reported to serve roles in inhibiting and promoting
cancer growth in different tumor types (8).
Recently, several studies have confirmed the
dysregulation of lncRNA expression in OS, which is associated with
the progression, metastasis and prognosis of the disease (2,9). In
particular, the dysregulated lncRNAs identified included
lncRNA-activated by Transforming Growth Factor β and lncRNA-taurine
upregulated gene 1. Therefore, lncRNAs may serve as diagnostic
markers for OS. A novel oncogenic lncRNA, forkhead box D2 adjacent
opposite strand RNA1 (FOXD2-AS1), has been shown to serve an
important role in various tumor types, including liver, bladder and
non-small cell lung cancer. For example, high expression of
FOXD2-AS1 regulates the proliferation and metastasis of non-small
cell lung cancer cells via the Wnt/β-catenin signaling pathway;
downregulation of FOXD2-AS1 inhibits the proliferation, metastasis
and invasion of hepatocellular carcinoma cells; and FOXD2-AS1
promotes bladder cancer cell proliferation, migration and invasion
by regulating a feedback loop with Akt and E2F transcription factor
1 (E2F1) (10–12). However, to the best of our
knowledge, the biological function and potential molecular
mechanisms of FOXD2-AS1 in human OS are currently unclear.
The present study aimed to investigate the function
and mechanism of FOXD2-AS1 in OS. In the present study, FOXD2-AS1
expression was significantly increased in OS tissues and cell
lines. In addition, Kaplan-Meier analysis revealed that FOXD2-AS1
expression was closely related to the survival of patients with OS,
thus suggesting that FOXD2-AS1 may be a novel biomarker for the
effective diagnosis of OS. Knockdown of FOXD2-AS1 inhibited the
proliferation of OS cells, and suppressed their migratory and
invasive abilities. In conclusion, the present results revealed
that FOXD2-AS1 may be an effective target for the treatment of
patients with OS.
Materials and methods
Clinical specimens
In the present study, 40 patients with OS (age
range, 6–51; 24 male patients and 16 female patients) were
recruited in the Department of Orthopedic Oncology Institute,
Tangdu Hospital, The Second Affiliated Hospital of Air Force
Medical. The patients underwent resection and the specimens were
collected; the samples were collected between July 2012 and August
2014 at The Second Affiliated Hospital of Air Force Medical
University. None of the patients underwent chemotherapy or
radiotherapy prior to surgery. The collected specimens were
obtained with the written informed consent of the patients or their
families. Informed consent was obtained from the parents of
patients <16 years old. The collected OS tissues and adjacent
tissue controls (>3 cm from the edge of the tumor) were
immediately stored in liquid nitrogen for subsequent use. The use
of human OS tissue was approved by the Air Force Medical University
Ethics Committee.
Relative expression levels of FOXD2-AS1 were
analyzed by reverse transcription-quantitative polymerase chain
reaction (RT-qPCR). Data are presented as log2fold
change (2−ΔΔCq). The samples were divided into two
groups (high and low) based on the median expression level of
FOXD2-AS1. Subsequently, the two groups were examined using the
Kaplan-Meier analysis followed by log-rank test.
Cell culture
The human OS cell line SOSP-9607 was established by
our laboratory and maintained in our laboratory (13). Other cell lines used in this study,
hFOB1.19, U2OS, MG63 and SAOS2, were purchased from the American
Type Culture Collection. SOSP-9607 cell line was cultured in
RPMI-1640 medium (HyClone; GE Healthcare Life Sciences) containing
10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences)
at 37°C in a humidified atmosphere with 5% CO2. U2OS,
SAOS2 and MG63 cells were cultured in DMEM (HyClone; GE Healthcare
Life Sciences) containing 10% FBS at 37°C in a humidified
atmosphere with 5% CO2. The hFOB1.19 cell line was grown
in DMEM containing 10% FBS and 0.3 mg/ml G418 (Sigma-Aldrich; Merck
KGaA) at 33.5°C in a humidified atmosphere containing 5%
CO2.
Construction of lentiviral vectors
containing green fluorescent protein (GFP) and infection
To stably silence FOXD2-AS1, a lentiviral vector
(Hanbio Biotechnology Co., Ltd.) that specifically and stably
expressed a FOXD2-AS1 short hairpin RNA (shRNA). The FOXD2-AS1
shRNA was constructed by Hanbio Biotechnology Co., Ltd. The
FOXD2-AS1 shRNA interference sequence was
5′-GATCCGCGAAGAGTACGTTGCTATTTCAAGAGAATAGCAACGTACTCTTCGCTTTTTTC-3′,
the shRNA control sequence (scramble) was
5′-TTCTCCGAACGTGTCACGTAA-3′. SOSP-9607 and U2OS cells
(2×104 cells/well) were infected with the virus at a
multiplicity of infection of 100 and 50, respectively. After
overnight incubation at 37°C, the virus-containing culture
supernatant was removed and replaced with fresh virus-free medium.
After 48 h of culture, the infection efficiency was determined by
observing the intensity of GFP expression in the cells under
an Olympus IX71 fluorescence microscope (Olympus Corporation).
Cells expressing GFP were selected for further analysis. After
infection, cells were cultured for 96 h prior to subsequent
experiments.
RT-qPCR
Total RNA was extracted from all OS tissues and
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Total
RNA was dissolved in RNase-free water and the concentration was
measured using an Epoch spectrophotometer (ND-1000
spectrophotometer; NanoDrop; Thermo Fisher Scientific, Inc.). cDNA
was synthesized using the 5X-All-In-One RT MasterMix kit (Applied
Biological Materials, Inc.) according to the manufacturer's
protocol, with a Bio-Rad MyCycler system (Bio-Rad Laboratories,
Inc.), with the following conditions: 37°C for 15 min and 85°C for
5 sec, followed by cooling to 4°C. The synthesized cDNA was then
subjected to qPCR using the EvaGreen 2X qPCR MasterMix kit (Applied
Biological Materials, Inc.) on the Rotor-Gene Q 2plex system
(Qiagen GmbH). qPCR thermocycling conditions were as follows:
Denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for
15 sec, 65°C for 10 sec and 72°C for 15 sec. The standard curves
were calculated and the relative quantification of gene expression
was assessed. GAPDH expression was used as a standardized internal
reference and the 2−ΔΔCq method was used for relative
quantification (14). The
sequences of all primers are presented in Table I.
 | Table I.Sequences of primers and probes. |
Table I.
Sequences of primers and probes.
| Primer name | Sequence |
|---|
| GAPDH |
|
|
Forward |
5′CGCTCTCTGCTCCTCCTGTTC3′ |
|
Reverse |
5′CCGTTGACTCCGACCTTCAC3′ |
| FOXD2-AS1 |
|
|
Forward |
5′AGGGACAGCCAAGAATACTC3′ |
|
Reverse |
5′GGGACTCAGAAGGGTTACAC3′ |
| RRM2 |
|
|
Forward |
5′CCACGGAGCCGAAAACTAAG3′ |
|
Reverse |
5′CTCTGCCTTCTTATACATCTGCC3′ |
| PHGDH |
|
|
Forward |
5′ATCTGCGGAAAGTGCTCATC3′ |
|
Reverse |
5′GCAGAGCGAACAATAAGGC3′ |
| COL5A1 |
|
|
Forward |
5′TACCCTGCGTCTGCATTTCC3′ |
|
Reverse |
5′GCTCGTTGTAGATGGAGACCA3′ |
| FZD1 |
|
|
Forward |
5′ATCTTCTTGTCCGGCTGTTACA3′ |
|
Reverse |
5′GTCCTCGGCGAACTTGTCATT3′ |
| EFNA4 |
|
|
Forward |
5′CCCCCTCTGTCTCTTGCTATT3′ |
|
Reverse |
5′TCTTGTCGGTCTGAATTGGCA3′ |
| EPHB2 |
|
|
Forward |
5′GTGTGCAACGTGTTTGAGTCA3′ |
|
Reverse |
5′ACGCACCGAAAACTTCATCTC3′ |
| FOXD2-AS1
probe |
5′-DIG-CGACCCGGACGCCACTGATAGCAAC-DIG-3′ |
Cell Counting kit-8 (CCK8) assay
SOSP-9607 and U2OS cells were seeded into 96-well
tissue culture plates (3×103 cells/well). The CCK8 assay
was performed at 24, 48, 72 and 96 h. Following the addition of 10
µl/well CCK8 reagent (Dojindo Molecular Technologies, Inc.), cells
were incubated for 2 h at 37°C and absorbance was measured at 450
nm using an Infinite M200 Pro Multifunctional microplate reader
(Tecan Group Ltd).
Colony formation assay
SOSP-9607 and U2OS cells infected with the FOXD2-AS1
shRNA were seeded into 6-well plates (500 cells/well) and cultured
at 37°C in a humidified atmosphere containing 5% CO2.
The culture medium was changed every 2 days. After 2 weeks, the
macroscopic observation of cell growth was terminated. The cells
were washed twice with PBS, fixed in methanol for 40 min, stained
with 1% crystal violet dye for 5 min at room temperature and rinsed
twice with water. Finally, images were captured to count the
colonies.
5-Ethynyl-2-deoxyuridine (EdU)
assay
The EdU experiment was performed using the iClick™
EDU Andy Fluor™ 647 Imaging kit (cat. no. A006; GeneCopoeia, Inc.)
according to the manufacturer's protocol. OS cells were treated
with 10 µm EdU solution and were incubated for 2 h at 37°C. Next,
200 µl 1X Hoechst 33342 (5 µg/ml) was added to each well and
incubated at room temperature for 15 min in the dark. Images were
captured under an Olympus IX71 fluorescence microscope (Olympus
Corporation). Andy Fluor 647 azide was detected at 650 nm. Hoechst
33342 was detected at 350 nm.
Cell migration and invasion
assays
Briefly, 24-well Transwell plates (pore size, 8 µm;
Corning, Inc.) were used for cell invasion and migration assays.
For the cell migration assay, 5×104 OS cells were seeded
into the upper chambers of 24-well plates in 200 µl serum-free
DMEM. The lower chamber contained DMEM supplemented with 10% FBS.
After 20 h at 37°C, the non-migrating cells were gently removed
from the upper side of the chamber with a cotton swab, and the
migrated cells were washed twice with PBS, fixed with 95% alcohol
for 10 min and stained with 1% crystal violet for 5 min. Finally,
images were captured and cells were counted under an light inverted
microscope (Olympus Corporation).
For the invasion assay, the wells were pretreated
with Matrigel (BD Biosciences), and OS cells (1×105
cells per chamber) were seeded into the upper chamber. The lower
chamber contained DMEM supplemented with 10% FBS. The subsequent
steps were the same as those conducted for the cell migration
assay.
Flow cytometry
To analyze cell cycle distribution, a sufficient
number (1×106) of stably infected cells was collected,
fixed overnight at 4°C in 500 µl 70% cold ethanol and washed three
times with PBS. Subsequently, the cells were incubated with 450 µl
propidium iodide (PI) and 50 µl RNase A using the Annexin V-APC/PI
apoptosis detection Kit (Nanjing KeyGen Biotech, Co., Ltd.)
according to the manufacturer's protocol in the dark at room
temperature for 1 h. Cell cycle progression was quantified using a
NovoCyte 2040R flow cytometer (ACEA Biosciences, Inc.), the data
were analyzed using the NovoExpress Software (version1.4; ACEA
Biosciences, Inc.).
Nuclear fractionation analysis and
fluorescence in situ hybridization (FISH)
Nuclear and cytoplasmic FOXD2-AS1 was isolated from
1×107 SOSP-9607 and U2OS cells using the PARIS™ kit
(Ambion; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. SOSP-9607 and U2OS cells were used for
RNA-FISH analysis. The relative localization of the transcript was
calculated according to the manufacturer's protocol (PARIS™ kit;
Ambion; Thermo Fisher Scientific, Inc.). The cell suspension
(1×105 cells/ml) was pipetted onto slides, fixed in 4%
paraformaldehyde for 20 min at room temperature and washed with PBS
(pH 7.4) three times (5 min/wash). Following digestion with
proteinase K (20 µg/ml) for 5 min at 37°C, the samples were washed
three times in PBS (5 min/wash). Subsequently, hybridization with
the digoxigenin (DIG)-labeled FOXD2-AS1_lnc probe (8 ng/µl; Wuhan
Servicebio Technology Co., Ltd.) was carried out at 37°C overnight.
The sequence of the probe is presented in Table I. After hybridization, the slides
were washed with 2X SSC for 10 min at 37°C, 1X SSC two times for 5
min at 37°C, and 0.5X SSC for 10 min at 37°C. Subsequently, the
blocking buffer containing 5% BSA (Wuhan Servicebio Technology Co.,
Ltd.) was added and cells were incubated for 30 min at room
temperature. The slides were then incubated with the anti-DIG-488
antibody (1:300; Jackson ImmunoResearch Laboratories, Inc.; cat.
no. 200-482-156) at 37°C for 50 min and washed three times in PBS
(5 min/wash). DAPI solution was then added dropwise and the cells
were incubated in the dark for 8 min at room temperature. After
washing, anti-fade mounting medium (cat. no. G1401; Wuhan
Servicebio Technology Co., Ltd.) was pipetted onto the slides. The
images were acquired using a Nikon direct fluorescence microscope
(Nikon Corporation).
Western blotting
Total cellular protein was extracted using RIPA
buffer (Beijing ComWin Biotech Co., Ltd.) supplemented with 1%
protease inhibitor (100X; Beijing ComWin Biotech Co., Ltd.), and
protein concentration was quantified with a BCA protein assay kit.
Proteins (20–30 µg) were separated by SDS-PAGE, using 6–10%
acrylamide gels, and were then transferred to a PVDF Immobilon-P
membrane (EMD Millipore). After blocking for 2 h at 37°C with 5%
skim milk, membranes were incubated overnight with a primary
antibody anti-phosphoglycerate dehydrogenase (PHGDH; cat. no.
BS71734; 1:1,000; Bioworld Technology, Inc.) and ribonucleotide
reductase regulatory subunit M2 (RRM2; cat. no. BS7520; 1:1,000;
Bioworld Technology, Inc.) at 4°C, followed by incubation with a
horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G
secondary antibody (cat. no. BS10043; 1:2,000; Bioworld Technology,
Inc.) for 2 h at 37°C. The ECL detection reagent (EMD Millipore)
was used to visualize the bands using an ECL detection system
(Thermo Fisher Scientific, Inc.).
Tumor xenograft assay
Nude BALB/c female mice (age, 4 weeks; weight, 16–22
g) were purchased from the experimental animal center of the Air
Force Medical University, and were bred under the following
pathogen-free conditions: 12-h light/dark cycle; temperature,
24–28°C; humidity, 40–60%. Mice had free access to food and water.
The animal feed and heating pad were purchased from the
experimental animal center of the Air Force Medical University, and
were autoclaved and vacuum packed. A total of eight nude mice took
part in the experimental study.
For the tumorigenicity experiment, SOSP-9607 cells
infected with FOXD2-AS1 shRNA or scramble shRNA were suspended in
PBS, and the cell concentration was adjusted to
1×108/ml. Each female BALB/c nude mouse was injected
with a 30-µl cell suspension (n=4 mice/group) into the tibial
plateau, and tumor size was measured every 6 days. After 36 days,
mice were sacrificed by cervical dislocation. The tumor and lung
tissues of each group were removed and fixed for 2 h at 4°C in 4%
paraformaldehyde for subsequent pathological and
immunohistochemical examination, to detect the protein expression
level of Ki-67. The study was approved by the Animal Care and Use
Committee of the Second Affiliated Hospital of the Air Force
Medical University.
Immunohistochemistry
The tumor tissue was fixed in 4% paraformaldehyde at
4°C for 2 h, dehydrated in alcohol and embedded in paraffin. The
sections were incubated in 3% hydrogen peroxide at room temperature
for 25 min in the dark. Subsequently, the sections were washed with
PBS (pH 7.4). Then, the samples were incubated with 3% BSA (cat.
no. G5001; Wuhan Servicebio Technology Co., Ltd.) and blocked at
room temperature for 30 min. After removing the blocking buffer,
the sections were incubated overnight at 4°C with a primary
antibody anti-Ki-67 Rabbit mAb (1:500 cat. no. GB13030-2; Wuhan
Servicebio Technology Co., Ltd.). The slices were incubated in PBS
(pH 7.4) and washed three times with PBS. Next, the sections were
incubated with a secondary antibody goat anti-rabbit immunoglobulin
G (1:1,000; cat. no. BS10043; Bioworld Technology, Inc.) for 50 min
at room temperature. Immunohistochemical staining results were
observed using the Olympus BX51 light microscope (Olympus
Corporation; magnification, ×400).
Statistical analysis
All data were statistically analyzed using SPSS 21.0
software (IBM Corp.). All experiments were repeated at least three
times and the data are expressed as the means ± standard deviation.
Comparisons of data between two groups were performed using a
two-tailed Student's t-test comparisons between multiple groups
were performed using one-way analysis of variance with Fisher's
least significance difference post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
FOXD2-AS1 is upregulated in OS tissues
and cells
In order to investigate the biological function of
FOXD2-AS1 in OS, the expression levels of FOXD2-AS1 were detected
in OS tissues and cells by RT-qPCR. As shown in Fig. 1, the expression levels of FOXD2-AS1
were detected in 40 samples of OS and adjacent tissues. The results
revealed that FOXD2-AS1 expression was upregulated in 80% of OS
tissue samples (32/40) compared with in adjacent tissues (Fig. 1A). Furthermore, the high expression
of FOXD2-AS1 was associated with a significantly lower survival
time compared with relatively low FOXD2-AS1 expression (Fig. 1B). In addition, RT-qPCR
demonstrated that the expression levels of FOXD2-AS1 were increased
in OS cells (9607, U2OS, MG63 and SAOS2) compared with in hFOB1.19
cells (Fig. 1E).
 | Figure 1.Expression of FOXD2-AS1 in OS cell
lines and tissues. (A) Relative expression levels of FOXD2-AS1 in
40 paired specimens of OS and adjacent tissues, as quantified by
RT-qPCR. Data are presented as log2 fold change. (B) Association
between the expression levels of FOXD2-AS1 and survival for
patients with OS, as determined by Kaplan-Meier analysis. (C)
Determination of subcellular localization of FOXD2-AS1 in OS cells
by fluorescence in situ hybridization. (D) Expression of
FOXD2-AS1 in the cytoplasm and nuclei of OS cells; U1 and GAPDH
were used as controls. (E) Expression levels of FOXD2-AS1 in OS
cell lines (9607, U2OS, MG63 and SAOS2) and normal osteoblasts
(hFOB1.19), as quantified by RT-qPCR. *P<0.05, **P<0.01 and
**P<0.01 vs. hFOB1.19. Data are expressed as the means ±
standard deviation. FOXD2-AS1, forkhead box D2 adjacent opposite
strand RNA1; N, normal; OS, osteosarcoma; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; T, tumor. |
Knockdown of FOXD2-AS1 inhibits OS
cell proliferation by inducing cell cycle arrest
To investigate the role of FOXD2-AS1 in the
proliferation of OS cells, lentivirus-mediated FOXD2-AS1 shRNA
infection was conducted in OS cell lines (SOSP-9607 and U2OS), in
order to establish SOSP-9607 and U2OS cell lines with stable low
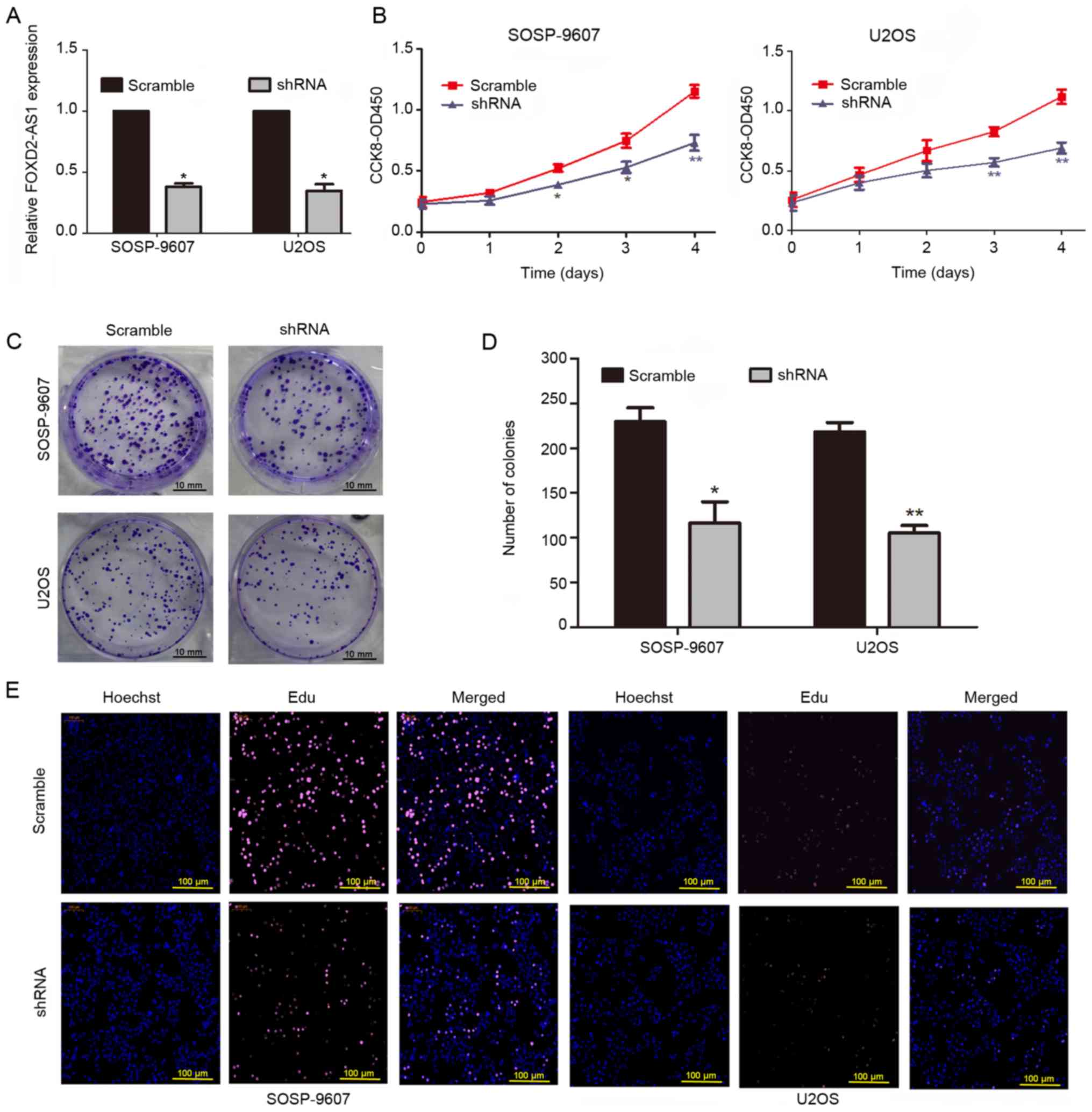
expression of FOXD2-AS1. The knockdown efficiency of FOXD2-AS1 was
verified by RT-qPCR after 72 h of infection (Fig. 2A). The results of a CCK8 assay
revealed that downregulation of FOXD2-AS1 inhibited the
proliferation of OS cells (Fig.
2B). In addition, FOXD2-AS1 knockdown reduced the
colony-forming ability of OS cells (Fig. 2C and D). The EdU assay further
confirmed that knockdown of FOXD2-AS1 inhibited cell proliferation
(Fig. 2E). Finally, flow cytometry
demonstrated that downregulation of FOXD2-AS1 significantly reduced
the abundance of cells in S phase of the cell cycle (Fig. 3C).
Decreased expression of FOXD2-AS1
inhibits the migration and invasion of OS cells
Specific invasion and migration assays were used to
investigate the association between FOXD2-AS1 and the
migratory/invasive abilities of OS cells. The results confirmed
that downregulation of FOXD2-AS1 inhibited SOSP-9607 and U2OS tumor
cell migration and invasion (Fig. 3A
and B). These results revealed that FOXD2-AS1 may act as a
tumor promoter by enhancing cell invasion and migration.
Localization of FOXD2-AS1 in cells and
its potential downstream genes
To investigate the mechanism by which FOXD2-AS1
exerts its effects on OS cells, this study revealed that FOXD2-AS1
expression was predominantly abundant in the cytoplasm using FISH
and RNA isolation experiments (Fig. 1C
and D). lncRNAs located in the cytoplasm can act as competing
endogenous RNAs (ceRNAs). Therefore, FOXD2-AS1 may act as a ceRNA
to sponge microRNAs and thus regulate mRNA expression. To determine
the downstream genes of FOXD2-AS1 in OS cells, transcriptomes of
five pairs of OS tissues and adjacent tissues were sequenced (data
not shown); the results revealed that, when FOXD2-AS1 was
upregulated, the mRNA expression levels of ephrin A4, collagen type
V α1 chain, EPH receptor B2, frizzled class receptor 1, PHGDH and
RRM2 may be upregulated (data not shown). Therefore, it was
hypothesized that FOXD2-AS1 may regulate the expression of these
six mRNAs. To confirm this hypothesis, FOXD2-AS1 was knocked down
and the expression of these six mRNAs was simultaneously detected.
The results of RT-qPCR demonstrated that the downregulation of
FOXD2-AS1 significantly downregulated the expression levels of
PHGDH and RRM2 (Fig. 4A). Western
blotting further confirmed that the expression levels of PHGDH and
RRM2 were reduced compared with in the scramble group (Fig. 4B).
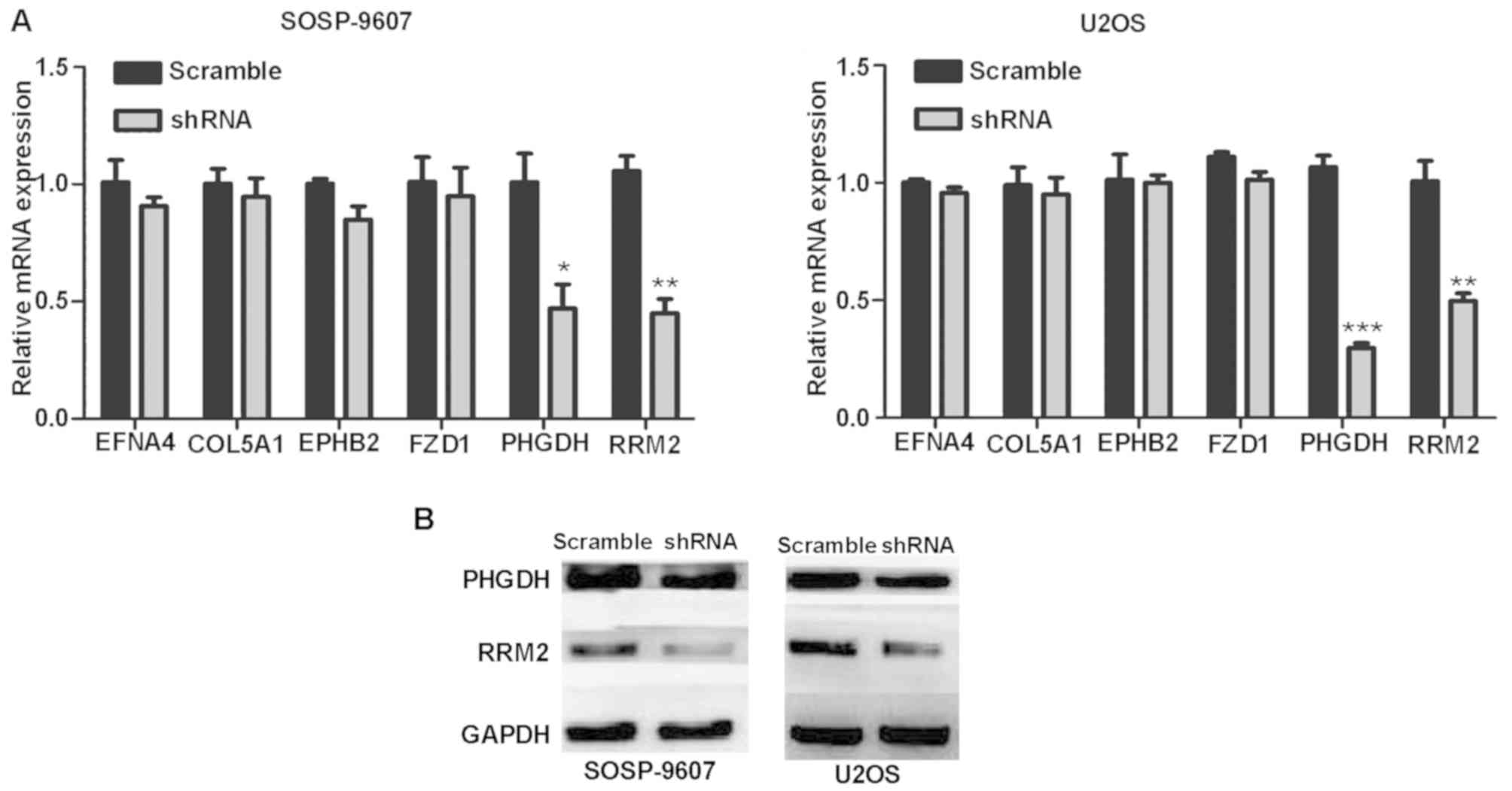 | Figure 4.FOXD2-AS1 affects the expression of
downstream genes and proteins. (A) mRNA expression levels of EFNA4,
COL5A1, EPHB2, FZD1, PHGDH and RRM2 in SOSP-9607 and U2OS
osteosarcoma cells post-infection with FOXD2-AS1 shRNA or scramble
shRNA. (B) Further verification by western blotting revealed that
FOXDA2-AS1 knockdown decreased the expression levels of the
downstream proteins PHGDH and RRM2. *P<0.05, **P<0.01 and
***P<0.001 vs. scramble shRNA. Data are presented as the means ±
standard deviation. COL5A1, collagen type V α1 chain; EFNA4, ephrin
A4; EPHB2, EPH receptor B2; FOXD2-AS1, forkhead box D2 adjacent
opposite strand RNA1; FZD1, frizzled class receptor 1; shRNA, short
hairpin RNA; PHGDH, phosphoglycerate dehydrogenase; RRM2,
ribonucleotide reductase regulatory subunit M2. |
FOXD2-AS1 knockdown inhibits the
growth of SOSP-9607 OS cell line in vivo
To further confirm the effects of FOXD2-AS1 on the
proliferation of SOSP-9607 OS cells in vivo, two groups of
stable cells (scramble shRNA- and FOXD2-AS1 shRNA-infected
SOSP-9607 cells) were injected into the tibial plateau of nude
mice. To evaluate tumor growth, the volume of the in situ
tumor was measured every 6 days post-inoculation. The following
formula was used to measure tumor volume: 1/2 × L × W2,
where L refers to length and W to width. The growth curves revealed
that FOXD2-AS1 knockdown significantly slowed the growth of OS
(Fig. 5A-C) and reduced its
propensity for lung metastasis (Fig.
5D). Histopathological evaluation of orthotopic tumors and lung
metastases in nude mice was performed to compare the shRNA group
and the scramble group (Fig.
5E-G). The present results suggested that downregulation of
FOXD2-AS1 expression inhibited tumor growth and lung metastasis
in vivo.
Discussion
OS is a primary malignant tumor that commonly occurs
in children and adolescents (1),
which is characterized by high lung metastatic potential and a poor
prognosis. In recent years, with the development of neoadjuvant
chemotherapy and combination therapy, the survival rate of patients
with early metastasis-free OS has increased by 55 to 70% (15,16).
Nevertheless, it is considered that developments surrounding the
survival rate and the therapeutic measures available for OS have
entered a relatively stable period. Notably, 15–20% of patients
with OS present with metastases at the time of diagnosis, which are
mostly located in the lungs (17).
Therefore, it is necessary to explore novel clinical targets for
the treatment of patients with OS, and to further elucidate the
pathogenesis of OS.
In recent years, numerous studies have demonstrated
that abnormal expression of lncRNAs is closely associated with the
occurrence and development of cancer (18,19).
Furthermore, aberrantly expressed lncRNAs can promote the growth
and metastasis of tumors; they can act as tumor suppressor genes
and oncogenes (20–22). For example, the lncRNA urothelial
cancer-associated 1 has an oncogenic role in breast cancer and lung
cancer, whereas the lncRNA TCF21 antisense RNA inducing promoter
demethylation suppresses genes through methylation to prevent the
occurrence of cancer (20–22). Accordingly, the abnormal expression
of lncRNAs in OS may be closely related to its biological behavior.
For example, it has been established that lncRNAs are closely
related to the proliferation, metastasis, apoptosis and poor
prognosis of OS (23,24).
Recently, it has been reported that aberrant
expression of FOXD2-AS1 serves a crucial role in carcinogenesis. A
previous study revealed that FOXD2-AS1 mediates chondrocyte
proliferation in osteoarthritis by regulating the expression of
cyclin D1 via sponging microRNA-206 (25). Another study demonstrated that
FOXD2-AS1 promotes bladder cancer cell proliferation, migration and
invasion by regulating a feedback loop with Akt and E2F1 (10). Yang et al (26)revealed that FOXD2-AS1 promotes
colorectal cancer cell proliferation, and that
epithelial-mesenchymal transition and Notch signaling pathway
activity are decreased in cells following knockdown of FOXD2-AS1.
However, few studies have investigated the biological role of
FOXD2-AS1 in OS.
To elucidate the role of FOXD2-AS1 in the
progression of OS, FOXD2-AS1 expression was quantified in 40 pairs
of OS tissues and adjacent tissues using RT-qPCR. To further
confirm the role of FOXD2-AS1 in OS, the expression of FOXD2-AS1
was validated by RT-qPCR in OS cell lines (SOSP-9607, U2OS, SAOS2
and MG63) and normal osteoblasts (hFOB1.19). The results
demonstrated that FOXD2-AS1 expression was increased in OS tissues
and cells compared with in adjacent tissues and normal osteoblasts.
Knockdown of FOXD2-AS1 expression inhibited OS cell proliferation,
migration and invasion. In addition, dysregulation of cell cycle
transitions is an important marker of cell carcinogenesis (27). Therefore, to investigate the
underlying mechanism of FOXD2-AS1 on the proliferation of OS cells,
flow cytometry was conducted; the results demonstrated that
knockdown FOXD2-AS1 expression may inhibit cell proliferation by
preventing entry into the S phase of the cell cycle. The tumor
model in nude mice revealed that downregulation of FOXD2-AS1
significantly slowed the growth of OS and reduced the potential for
lung metastasis. These results indicated that FOXD2-AS1 may be a
tumor-promoting factor.
To further explore the molecular mechanism by which
FOXD2-AS1 exerts its effects on OS cells, the present study
verified the localization of FOXD2-AS1 in the cytoplasm using FISH
and RNA isolation experiments. In addition, the effects of
FOXD2-AS1 knockdown on putative downstream targets were determined
using RT-qPCR and western blotting. Alterations in the protein
expression levels of PHGDH and RRM2 were detected in response to
FOXD2-AS1 knockdown, thus suggesting that FOXD2-AS1 may regulate
the expression of PHGDH and RRM2.
Abnormal expression of PHGDH and RRM2 serves a
crucial role in various cancer behaviors, including tumor cell
proliferation, apoptosis, drug resistance and metastasis (28,29).
It has been reported that high expression of RRM2 has an adverse
effect on the survival and prognosis of patients with OS (30). Previous studies reported that
lncRNAs act as ceRNAs and competitively bind microRNAs, in order to
regulate mRNA expression; the corresponding microRNA response
element is the basis of this interaction (31,32).
However, to the best of our knowledge, which microRNA FOXD2-AS1
binds as a ceRNA, in order to regulate the expression levels of
PHGDH and RRM2, has not been studied. In addition, the mechanism by
which FOXD2-AS1 regulates RRM2 and PHGDH has not been studied in
the present study. Therefore, more experimental studies regarding
the functional role of FOXD2-AS1 in OS are required.
In conclusion, the present findings demonstrated
that FOXD2-AS1 was upregulated in OS cells and OS tissue samples
compared with in normal osteoblasts and adjacent tissue samples. In
addition, knockdown of FOXD2-AS1 expression inhibited OS cell
proliferation, migration and invasion. However, the mechanism by
which FOXD2-AS1 regulates the biological behavior of OS has not
been investigated in this study. We aim to further investigate how
FOXD2-AS1 regulates the expression of RRM2 and PHGDH in subsequent
experiments. In conclusion, these findings suggested that FOXD2-AS1
may be a potential clinical therapeutic target, and further studies
are required to elucidate the molecular mechanism underlying
FOXD2-AS1-mediated OS progression.
Acknowledgements
Not applicable.
Funding
The present research was supported by research
grants from the National Natural Science Foundation of China (grant
nos. 81272441).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ and YL performed the functional in vitro
assays. HZ, TZ and CD performed qPCR and western blot assays. HZ,
JW and XL performed the functional in vivo assays. HZ
analyzed the data and wrote manuscript. XW collected and analyzed
clinical samples. YZ, TY and QM designed the present study. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The use of human OS tissue was approved by the Air
Force Military Medical University Ethics Committee, and the
collected specimens were obtained with the written informed consent
of the patients or their families. The animal study was approved by
the Animal Care and Use Committee of the Second Affiliated Hospital
of the Air Force Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bishop MW, Janeway KA and Gorlick R:
Future directions in the treatment of osteosarcoma. Curr Opin
Pediatr. 28:26–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang Z, Li X, Yang Y, He Z, Qu X and Zhang
Y: Long noncoding RNAs in the progression, metastasis, and
prognosis of osteosarcoma. Cell Death Dis. 7:e23892016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee C and Kikyo N: Strategies to identify
long noncoding RNAs involved in gene regulation. Cell Biosci.
2:372012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mattick JS: Non-coding RNAs: The
architects of eukaryotic complexity. EMBO Rep. 2:986–991. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Di Gesualdo F, Capaccioli S and Lulli M: A
pathophysiological view of the long non-coding RNA world.
Oncotarget. 5:10976–10996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q
and Liu Z: Decreased expression of long non-coding RNA GAS5
indicates a poor prognosisand promotes cell proliferation and
invasion in hepatocellular carcinoma by regulating vimentin. Mol
Med Rep. 13:1541–1550. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lalevee S and Feil R: Long noncoding RNAs
in human disease: Emerging mechanisms and therapeutic strategies.
Epigenomics. 7:877–879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Smolle MA and Pichler M: The role of long
non-coding RNAs in osteosarcoma. Noncoding RNA. 4(pii):
E72018.PubMed/NCBI
|
|
10
|
Su F, He W, Chen C, Liu M, Liu H, Xue F,
Bi J, Xu D, Zhao Y, Huang J, et al: The long non-coding RNA
FOXD2-AS1 promotes bladder cancer progression and recurrence
through a positive feedback loop with Akt and E2F1. Cell Death Dis.
9:2332018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xiong Y, Wang T, Wang M, Zhao J, Li X,
Zhang Z, Zhou Y, Liu J, Jia L and Han Y: Long non-coding RNAs
function as novel predictors and targets of non-small cell lung
cancer: A systematic review and meta-analysis. Oncotarget.
9:11377–11386. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang J, Fan D, Jian Z, Chen GG and Lai
PB: Cancer specific long noncoding RNAs show differential
expression patterns and competing endogenous RNA potential in
hepatocellular carcinoma. PLoS One. 10:e1410422015.
|
|
13
|
Chen X, Yang TT, Wang W, Sun HH, Ma BA, Li
CX, Ma Q, Yu Z and Fan QY: Establishment and characterization of
human osteosarcoma cell lines with different pulmonary metastatic
potentials. Cytotechnology. 61:37–44. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Benjamin RS: Osteosarcoma: Better
treatment through better trial design. Lancet Oncol. 16:12–13.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Durnali A, Alkis N, Cangur S, Yukruk FA,
Inal A, Tokluoglu S, Seker MM, Bal O, Akman T, Inanc M, et al:
Prognostic factorsfor teenage and adult patients with high-grade
osteosarcoma: An analysis of 240 patients. Med Oncol. 30:6242013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miller BJ, Cram P, Lynch CF and Buckwalter
JA: Risk factors for metastatic disease at presentation with
osteosarcoma: An analysis of the SEER database. J Bone Joint Surg
Am. 95:e892013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chan JJ and Tay Y: Noncoding RNA: RNA
regulatory networks in cancer. Int J Mol Sci. 19(pii): E13102018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang YL, Shao J, Wu X, Li T, Xu M and Shi
D: A long non-coding RNA signature for predicting survival in
patients with colorectal cancer. Oncotarget. 9:21687–21695.
2017.PubMed/NCBI
|
|
20
|
Tuo YL, Li XM and Luo J: Long noncoding
RNA UCA1 modulates breast cancer cell growth and apoptosis through
decreasing tumor suppressive miR-143. Eur Rev Med Pharmacol Sci.
19:3403–3411. 2015.PubMed/NCBI
|
|
21
|
Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao
X, Chen WS and Li B: LncRNA-UCA1 exerts oncogenic functions in
non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett.
371:99–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arab K, Park YJ, Lindroth AM, Schäfer A,
Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et
al: Long noncoding RNA TARID directs demethylation and activation
of the tumor suppressor TCF21 via GADD45A. Mol Cell. 55:604–614.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen F, Mo J and Zhang L: Long noncoding
RNA BCAR4 promotes osteosarcoma progression through activating
GLI2-dependent gene transcription. Tumour Biol. 37:13403–13412.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun XH, Yang LB, Geng XL, Wang R and Zhang
ZC: Increased expression of lncRNA HULC indicates a poor prognosis
and promotes cell metastasis in osteosarcoma. Int J Clin Exp
Pathol. 8:2994–3000. 2015.PubMed/NCBI
|
|
25
|
Cao L, Wang Y, Wang Q and Huang J: LncRNA
FOXD2-AS1 regulates chondrocyte proliferation in osteoarthritis by
acting as a sponge of miR-206 to modulate CCND1 expression. Biomed
Pharmacother. 106:1220–1226. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang X, Duan B and Zhou X: Long non-coding
RNA FOXD2-AS1 functions as a tumor promoter in colorectal cancer by
regulating EMT and Notch signaling pathway. Eur Rev Med Pharmacol
Sci. 21:3586–3591. 2017.PubMed/NCBI
|
|
27
|
Hainaut P and Plymoth A: Targeting the
hallmarks of cancer: Towards a rational approach to next-generation
cancer therapy. Curr Opin Oncol. 25:50–51. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ou Y, Wang SJ, Jiang L, Zheng B and Gu W:
p53 Protein-mediated regulation of phosphoglycerate dehydrogenase
(PHGDH) is crucial for the apoptotic response upon serine
starvation. J Biol Chem. 290:457–466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jing Z, Heng W, Xia L, Ning W, Yafei Q,
Yao Z and Shulan Z: Downregulation of phosphoglycerate
dehydrogenase inhibits proliferation and enhances cisplatin
sensitivity in cervical adenocarcinoma cells by regulating Bcl-2
and caspase-3. Cancer Biol Ther. 16:541–548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fellenberg J, Bernd L, Delling G, Witte D
and Zahlten- Hinguranage A: Prognostic significance of
drug-regulated genes in high-grade osteosarcoma. Mod Pathol.
20:1085–1094. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independentregulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language. Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|