Introduction
Non-small-cell lung cancer (NSCLC) accounts for ~80%
of all lung cancer cases according to its pathological
classification (1). Although
advances in diagnostic and therapeutic techniques have improved the
early detection and reduced the mortality rate of lung cancer, it
remains the leading cause of cancer-associated mortality worldwide
(1). Thus, it is imperative to
elucidate the mechanism underlying the development and progression
of NSCLC.
Cancer cells and embryonic stem (ES) cells have long
been known to share common characteristics with respect to
self-renewal, proliferation and indefinite growth (2). Developmental pluripotency-associated
4 (Dppa4) is highly expressed in ES cells. In addition,
self-renewal regulatory factors, such as Oct4, Sox2, Bmi and Nanog,
are highly expressed in tumor cells and serve an important role in
carcinogenesis (3,4). The Dppa gene family is a cluster of
five genes whose developmental expression patterns are similar to
those of Oct4 (5). The structure
and functions of Dppa4 indicate its possible involvement in cancer
progression. Dppa4 has been found to be highly expressed in colon,
prostate and bladder carcinomas, and may be a novel predictor of
prognosis and a potential therapeutic target in colon cancer
(4,6). It has also been reported that Dppa4
functioned as an oncogene in mouse 3T3 cells and immortalized human
dermal fibroblasts, and enhanced cell proliferation by inducing
G1/S arrest (7). However, the
clinicopathological significance of Dppa4, and its possible
mechanism in NSCLC tumorigenesis and progression remain
unclear.
The aim of the present study was to explore the
expression pattern of Dppa4 in NSCLC tissue samples, including
squamous cell carcinoma, adenocarcinoma, large cell carcinoma,
adenosquamous carcinoma and other cell lines in order to determine
whether the overexpression of Dppa4 is associated with unfavorable
clinicopathological variables. In addition, the study assessed
whether Dppa4 may be of value as a novel prognostic marker for
NSCLC. The possible action mechanism of Dppa4 in NSCLC was also
investigated. The results revealed that Dppa4 promoted NSCLC by
regulating glycolysis, possibly through LDHB.
Materials and methods
Human tissue specimens
A total of 100 tumor tissue samples and 100 adjacent
normal tissue samples were collected from the Department of
Cardiothoracic Surgery, Xuzhou Cancer Hospital (Xuzhou, China)
between January 2010 and December 2014. The tissue samples included
squamous cell carcinoma, lung adenocarcinoma, large cell carcinoma
and adenosquamous carcinoma. The mRNA expression of Dppa4 was
evaluated in 20 pairs of fresh NSCLC tissues and adjacent normal
tissues using reverse transcription-quantitative PCR (RT-qPCR),
while a total of 80 paired tissue samples were embedded in paraffin
to examine the Dppa4 protein levels via immunohistochemistry (IHC).
All the patients had been diagnosed with NSCLC, which was confirmed
by histological examination. Patients who had previously received
radiotherapy or chemotherapy were excluded. The study protocol was
approved by the Research Ethics Committee of Xuzhou Cancer
Hospital, and informed consent was obtained from all the
patients.
IHC assay and scoring
Tissues were fixed with 4% paraformaldehyde for at
least 24 h at room temperature. Samples (5 µm sections) were
deparaffinized in xylene and rehydrated in a graded alcohol series.
Next, 3% H2O2 was used to block endogenous
peroxidase activity for 5 min at room temperature, and antigen
retrieval was performed subsequent to heating in citrate buffer.
The samples were blocked by normal 5% goat serum (Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China) for 1 h at
37°C. The sections were incubated with antibodies against Dppa4
(dilution, 1:100; Cell Signaling Technology, Inc., Danvers, MA,
USA; cat. no. 67138), lactate dehydrogenase B (LDHB; dilution
1:500; Abcam, Cambridge, UK; cat. no. ab75167) and β-actin
(dilution 1:5,000; Abcam; cat. no. ab8226) at 4°C overnight. This
was followed by incubation with a horseradish peroxidase-conjugated
secondary antibody (dilution, 1:2,000; GTVision III Detection kit;
GeneTech Co., Ltd., Shanghai, China) at room temperature for 40
min. The 3,3′-diaminobenzidine substrate (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) was added for signal detection for 10 min
at room temperature, and the sections were lightly counterstained
with hematoxylin for 30 sec at room temperature. Images were
acquired using light microscopy (Olympus BX43, magnification
×400).
Double blind scoring was performed independently by
two investigators. Five visual fields from different areas of each
specimen were randomly selected for immunohistochemical evaluation.
Dppa4 expression was subjectively scored by the pathologists
according to the staining intensity (0, no staining; 1, weak
staining; 2, moderate staining; and 3, dark staining) and the
percentage of positive cells (0, no positive cells; 1, ≤10%
positive cells; 2, 10–50% positive cells; and 3, >50% positive
cells). The final score was calculated as follows: Comprehensive
score = staining percentage × intensity. Dppa4 expression was
defined as low in specimens with scores <2 and high in specimens
with scores of ≥2.
Cell culture
The NSCLC cell lines A549, H1299, SPC (BNCC100120)
and LH7, and normal human bronchial epithelial (HBE) cells were
purchased from the American Type Culture Collection (Manassas, VA,
USA). A549 cells were cultured in F-12K medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.)
while other cell lines were cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS. All
cells were incubated at 37°C in a humidified atmosphere with 5%
CO2. A549 cells, which had the highest Dppa4 expression,
were used for Dppa4 knockdown, and H1299 cells, which had the
lowest Dppa4 expression, were used for Dppa4 overexpression. The
remaining cell lines were used solely for Dppa4 expression analysis
in NSCLC cell lines.
Plasmids and transfection
For Dppa4 overexpression, the plasmids
pcDNA3.1-Dppa4 (Dppa4-over) and control vector pcDNA3.1 were
constructed as described previously (8). For expression knockdown, small
interfering RNAs (siRNAs) targeting Dppa4 (namely Dppa4-si1 and
Dppa4-si2; cat. no. SR310582; GenePharma Co., Ltd., Shanghai,
China) were used. NSCLC cells were transfected with 4 µg
overexpression plasmids or 4 µg siRNAs using 8 µl
Lipofectamine® 2000 and 8 µl Lipofectamine RNAiMax
(Invitrogen; Thermo Fisher Scientific, Inc.), respectively.
Transfected cells were subjected to functional assays and western
blotting after 24 or 48 h, respectively. Untreated cells or cells
treated with empty vectors served as the control groups. In
addition, co-transfection with 4 µg LDHB siRNA (cat. no. sc-45899;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and Dppa4-over
plasmid in NSCLC cells was performed with Lipofectamine®
2000 reagent. The transfection efficiency was determined using
western blotting 48 h after transfection.
Cell viability assessment
An MTT assay was used to evaluate cell viability.
Briefly, the transfected 1×103 cells were plated in
96-well culture plates and cultured for 24, 48 and 72 h. Following
incubation with 5 mg/ml MTT (Sigma-Aldrich; Merck KGaA) for 4 h,
the cells were washed with PBS and then solubilized with dimethyl
sulfoxide. The absorbance was measured at a wavelength of 490 nm
using a microplate reader
Western blot analysis
Standard western blotting was performed using
protein (20 µg) lysed by radioimmunoprecipitation assay lysis
buffer (Beyotime Institute of Biotechnology, Haimen, China). The
protein concentration of each group was subsequently determined
using a bicinchoninic acid assay kit (Beyotime Institute of
Biotechnology). The equal amount of proteins were separated via 10%
SDS-PAGE, and the proteins were then transferred onto
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). These were subsequently blocked with 5% bovine serum albumin
(BSA; Thermo Fisher Scientific, Inc.) in TBST for 1 h at room
temperature. The samples were incubated with primary antibodies
against Dppa4 (dilution, 1:1,000; Cell Signaling Technology, Inc.;
cat. no. 67138), LDHB (dilution 1:1,000; Abcam; cat. no. ab75167)
and β-actin (dilution 1:5,000; Abcam; cat. no. ab8226) overnight at
4°C. TBST was used to wash the PVDF membranes three times, which
were then incubated with a secondary antibody anti-mouse (dilution,
1:1,000; Cell Signaling Technology, Inc.; cat. no. 7076) and
anti-rabbit (dilution, 1:1,000; Cell Signaling Technology, Inc.;
cat. no.7074) for 1 h in the room temperature. TBST was used to
wash again three times. Finally, proteins were visualized using an
enhanced chemiluminescence kit (Thermo Fisher Scientific, Inc.),
and images were acquired using ImageQuant LAS 4000 (GE Healthcare
Life Sciences, Little Chalfont, UK) and densitometry analysis was
performed using ImageQuant TL software version 1.1 (GE Healthcare
Life Sciences).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
NSCLC cell lines and tumor samples were used to
detect the Dppa4 expression by RT-qPCR. Furthermore, the relative
levels of glycolytic enzyme expression, namely GLUT-1, GLUT-4,
phosphofructokinase liver type (PFK-L), hexokinase 2 (HK-II),
phosphofructokinase muscle type (PFK-M), pyruvate kinase M-1
(PKM-1), PKM-2, phosphofructokinase platelet (PFK-P), aldolase,
fructose-bisphosphate B (AldoB), phosphoglycerate kinase 1 (PGK-1),
phosphoglycerate mutase (PGAM-1), Enolase, glucose-6-phosphate
isomerase (G6PI), LDHA and LDHB. Total RNA extraction was performed
using TRIzol® reagent (Thermo Fisher Scientific, Inc.)
for cell cultures and the miniBEST universal RNA extraction kit for
fresh tumor and tumor-adjacent tissues (Takara Bio, Inc., Otsu,
Japan). RNA was then reverse transcribed to cDNA (PrimeScript™ RT
reagent Kit with genomic DNA Eraser; Takara Bio, Inc.), which was
then subjected to qPCR analysis (SYBR Green PCR kit, Takara Bio,
Inc.) to evaluate mRNA expression. The RT temperature protocol was:
37°C for 15 min and 85°C for 5 sec. PCR amplification was performed
in a thermal cycler for 40 cycles at the following cycle
conditions: 95°C for 5 sec, 60°C for 30 sec, and 72°C for 30 sec.
The relative mRNA expression of each gene was calculated with the
comparative quantitation cycle (Cq) method, 2−ΔΔCq
(9,10). The specific primers used in qPCR
were as follows: Dppa4 sense, 5′-GACACAGATGGTTGGGTTCA-3′, and
antisense, 5′-GAGGCAGGAAGCAAGAAGAG-3′; GAPDH sense,
5′-AGAAGGCTGGGGCTCATTTG-3′, and antisense,
5′-AGGGGCCATCCACAGTCTTC-3′; GLUT-1 sense,
5′-CTTTGTGGCCTTCTTTGAAGT-3′, and anti-sense,
5′-CCACACAGTTGCTCCACAT-3′; GLUT-4 sense,
5′-CTTCATCATTGGCATGGGTTT-3′, and antisense,
5′-CGGGTTTCAGGCACTTTTAGG-3′; PFK-L sense,
5′-GGACAGGAAAGAGGAAGTGAC-3′, and antisense,
5′-CGTAGATGAGGAAGACTTTGGC-3′; HK-II sense,
5′-GATTTCACCAAGCGTGGACT-3′, and antisense,
5′-CCACACCCACTGTCACTTTG-3′; PFK-M sense, 5′-ATTCGGGCTGTGTTCTGG-3′,
and antisense, 5′-TGGCTAGGATTTTGAGGATGG-3′; PKM-1 sense,
5′-CTATCCTCTGGAGGCTGTGC-3′, and antisense,
5′-CCATGAGGTCTGTGGAGTGA-3′; PKM-2 sense, 5′-GGGTTCGGAGGTTTGATG-3′,
and antisense, 5′-ACGGCGGTGGCTTCTGT-3′; PFK-P sense,
5′-CATCGACAATGATTTCTGCGG-3′, and antisense,
5′-CCATCACCTCCAGAACGAAG-3′; AldoB sense,
5′-ATGCCACTCTCAACCTCAATGCTATC-3′, and antisense,
5′-TTATTTTCTTGGGTGGGTATTCTGG-3′; PGK-1 sense,
5′-CGGTAGTCCTTATGAGCC-3′, and antisense, 5′-CATGAAAGCGGAGGTTCT-3′;
PGAM-1 sense, 5′-CCTGGAGAACCGCTTC-3′, and antisense,
5′-CATGGGCTGCAATCAGTACAC-3′; Enolase sense,
5′-CTGATGCTGGAGTTGGATGG-3′, and antisense,
5′-CCATTGATCACGTTGAAGGC-3′; G6PI sense, 5′-AGGCTGCTGCCACATAAGGT-3′,
and antisense, 5′-AGCGTCGTGAGAGGTCACTTG-3′; LDHA sense,
5′-CAGCTTGGAGTTTGCAGTTAC-3′, and antisense,
5′-TGATGGATCTCCAACATGG-3′; LDHB sense, 5′-CCTAGAGCTCACTAGTCACAG-3′
and antisense, 5′-CTCCTGTGCAAAATGGCAAC-3′.
Lactate dehydrogenase (LDH) activity,
lactate production, glucose utilization assay and intracellular ATP
level
For LDH activity and lactate production assays,
tumor cells were transfected with siRNAs and plasmids, and after 24
h a total of 1×106 cells were examined using the Lactate
Dehydrogenase Activity Assay kit and Lactate Assay kit
(Sigma-Aldrich; Merck KGaA) according to the manufacturer's
protocol. For the glucose utilization assay, tumor cells were
transfected with plasmids and siRNAs for 24 h, and then the media
were replaced with phenol red-free RPMI medium supplemented with 1%
FBS, followed by continuous culture for 3 days. The glucose
utilization was measured using a colorimetric glucose assay kit
(BioVision Inc., Milpitas, CA, USA) and normalized to the cell
number. For intracellular ATP measurement, a firefly
luciferase-based ATP assay kit (Beyotime Institute of
Biotechnology) was used, according to the manufacturer's
instructions.
Statistical analysis
All data are presented as the mean ± standard
deviation. The comparisons between tumor tissue samples and control
tissues were performed using a paired t-test. The mean values of
other two groups were compared with the Student's t-test, while the
mean values of three or more groups were compared by one-way
analysis of variance. Statistical analyses were conducted with SPSS
for Windows, version 17.0 (SPSS, Inc., Chicago, IL, USA). The
least-significant difference test was used in the comparison of two
pairs as a post hoc test. Associations between the expression of
Dppa4 and clinical characteristics were analyzed with χ2
test or Fisher's exact test, as appropriate. The overall survival
was assessed using the Kaplan-Meier method. Univariate and
multivariate Cox regression analyses were also performed.
Parameters with a P-value of <0.05 in the univariate analysis
were included in a Cox multivariate proportional hazards regression
model. In the multivariate analysis, the Backward LR statistic was
used in the conditional logistic regression model. All statistical
analyses were performed using SPSS version 17.0 software. P<0.05
was considered to indicate a statistically significant
difference.
Results
Aberrant overexpression of Dppa4 in
NSCLC tissues and cells
The mRNA expression of Dppa4 was evaluated in 20
pairs of NSCLC tissues and adjacent normal tissues, while a total
of 80 paired tissue samples were used to examine the Dppa4 protein
level. The results demonstrated that the expression of Dppa4 was
higher in NSCLC tissue samples as compared with that in normal
tissues, with a statistically significant difference detected
(P<0.05; Fig. 1A and Table I). Representative images of Dppa4
expression in adenocarcinoma and squamous cell carcinoma are shown
in Fig. 1B. Subsequently, these
results were verified in NSCLC cell lines. Compared with the normal
HBE cells, the tumor A549, SPC and LH7 cell lines exhibited
markedly higher expression of Dppa4 at both the mRNA and protein
levels, with no significant difference detected in H1299 cells
(P<0.05; Fig. 1C and D).
 | Table I.Comparison of Dppa4 expression between
NSCLC and paired adjacent normal tissues. |
Table I.
Comparison of Dppa4 expression between
NSCLC and paired adjacent normal tissues.
|
|
| Dppa4 expression |
|
|---|
|
|
|
|
|
|---|
| Tissues | Cases | Low (%) | High (%) | P-value |
|---|
| NSCLC | 80 | 38 (47.5) | 42 (52.5) |
<0.001a |
| Adjacent
normal | 80 | 65 (81.3) | 15 (18.7) |
|
Correlation between Dppa4 expression
and the clinicopathological characteristics of NSCLC patients
The correlation between Dppa4 expression and the
clinicopathological characteristics of NSCLC patients was next
investigated. As shown in Table
II, Dppa4 was positively correlated with the tumor
differentiation grade (P=0.04), T stage (P=0.006), N stage
(P<0.001) and clinical stage (P<0.001). Other
clinicopathological characteristics, including the patient sex, age
and smoking history, were not found to be significantly correlated
with Dppa4 expression. These data suggest that Dppa4 may be
involved in NSCLC progression.
 | Table II.Correlation between the
clinicopathological characteristics and Dppa4 expression in
non-small cell lung carcinoma patients. |
Table II.
Correlation between the
clinicopathological characteristics and Dppa4 expression in
non-small cell lung carcinoma patients.
|
|
| Dppa4
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | N | Low (n=38) | High (n=42) | P-value |
|---|
| Age (years) |
|
|
| 0.479 |
|
≤60 | 37 | 16 | 21 |
|
|
>60 | 43 | 22 | 21 |
|
| Sex |
|
|
| 0.538 |
|
Male | 49 | 24 | 25 |
|
|
Female | 31 | 13 | 18 |
|
| Smoking
history |
|
|
| 0.685 |
| No | 37 | 24 | 13 |
|
|
Yes | 43 | 26 | 17 |
|
| Histological
type |
|
|
| 0.220 |
|
Adenocarcinoma | 32 | 17 | 15 |
|
|
Squamous cell carcinoma | 32 | 11 | 21 |
|
|
Other | 16 | 9 | 7 |
|
| Grade |
|
|
| 0.04a |
|
Well-differentiated | 51 | 28 | 23 |
|
|
Moderate/Poor | 29 | 9 | 20 |
|
|
differentiation |
|
|
|
|
| T stage |
|
|
| 0.006a |
| T1 | 23 | 11 | 12 |
|
|
T2a | 40 | 24 | 16 |
|
|
T2b | 16 | 2 | 14 |
|
| T3 | 1 | 0 | 1 |
|
| N stage |
|
|
|
<0.001a |
| N0 | 20 | 15 | 5 |
|
| N1 | 33 | 19 | 14 |
|
| N2 | 27 | 3 | 24 |
|
| TNM stage |
|
|
|
<0.001a |
|
I+II | 53 | 34 | 19 |
|
|
IIIA | 27 | 3 | 24 |
|
High Dppa4 expression is associated
with poor clinical outcomes in NSCLC patients
To further explore the association between Dppa4
expression and patient prognosis, a Kaplan-Meier analysis was
conducted. As shown in Fig. 1E,
the analysis revealed that patients with higher Dppa4 expression
exhibited a poorer prognosis. As shown in Table III, higher Dppa4 levels were
significantly associated with lower 1-, 3- and 5-year survival
rates (Table III). Furthermore,
univariate survival analysis indicated that, high Dppa4 expression
predicted poor prognosis in advanced TNM stage of IIIA (P<0.05;
Table IV). Subsequent
multivariate analysis revealed that high Dppa4 expression [hazard
ratio (HR), 1.862; 95% confidence interval (CI), 1.090–3.181;
P=0.023; Table IV] and TNM stage
of IIIA (HR=2.063; 95% CI, 1.148–3.708; P=0.015; Table IV) were correlated with overall
survival. Taken together, these findings demonstrated that Dppa4
expression may be an independent prognostic marker for NSCLC
patients.
 | Table III.Comparison of cumulative survival
rate between the low and high Dppa4 expression groups. |
Table III.
Comparison of cumulative survival
rate between the low and high Dppa4 expression groups.
|
| Cumulative survival
rate, 95% confidence interval |
|---|
|
|
|
|---|
| Dppa4
expression | 1-year | 3-year | 5-year |
|---|
| Low | 86%
(0.742–0.978) | 40%
(0.224–0.576) | 7%
(−0.028–0.168) |
| High | 48%
(0.323–0.637) | 13%
(0.012–0.248) | 2%
(−0.039–0.079) |
 | Table IV.Summary of univariate and
multivariate Cox regression analyses of overall survival duration
in all non-small cell lung carcinoma patients. |
Table IV.
Summary of univariate and
multivariate Cox regression analyses of overall survival duration
in all non-small cell lung carcinoma patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Dppa4
expression |
|
|
|
|
|
|
|
Low | 1 |
|
| 1 |
|
|
|
High | 2.271 | 1.384–3.727 |
<0.001a | 1.862 | 1.090–3.181 | 0.023a |
| Age(years) |
|
|
|
|
|
|
|
≤60 | 1 |
|
|
|
|
|
|
>60 | 0.777 | 0.479–1.260 | 0.306 |
|
|
|
| Sex |
|
|
|
|
|
|
|
Male | 1 |
|
|
|
|
|
|
Female | 1.086 | 0.658–1.793 | 0.747 |
|
|
|
| Smoking
history |
|
|
|
|
|
|
|
Never | 1 |
|
|
|
|
|
|
Ever | 0.795 | 0.483–1.308 | 0.366 |
|
|
|
| Histological
type |
|
|
|
|
|
|
|
Adenocarcinoma | 1 |
|
|
|
|
|
|
Squamous cell carcinoma | 0.951 | 0.552–1.638 | 0.857 |
|
|
|
| Others | 1.009 | 0.516–1.972 | 0.980 |
|
|
|
| Grade |
|
|
|
|
|
|
|
Well-differentiated | 1 |
|
|
|
|
|
|
Moderate/Poor
differentiation | 1.363 | 0.835–2.224 | 0.216 |
|
|
|
| T stage |
|
|
|
|
|
|
| T1 | 1 |
|
|
|
|
|
|
T2a | 0.731 | 0.416–1.284 | 0.275 |
|
|
|
|
T2b | 2.004 | 1.013–3.964 | 0.046a |
|
|
|
| T3 | 7.885 | 0.973–63.877 | 0.053 |
|
|
|
| N stage |
|
|
|
|
|
|
| N0 | 1 |
|
|
|
|
|
| N1 | 2.225 | 1.233–4.015 | 0.008a |
|
|
|
| N2 | 0.691 | 0.380–1.257 | 0.226 |
|
|
|
| TNM stage |
|
|
|
|
|
|
|
I+II | 1 |
|
| 1 |
|
|
|
IIIA | 2.616 | 1.520–4.502 |
<0.001a | 2.063 | 1.148–3.708 | 0.015a |
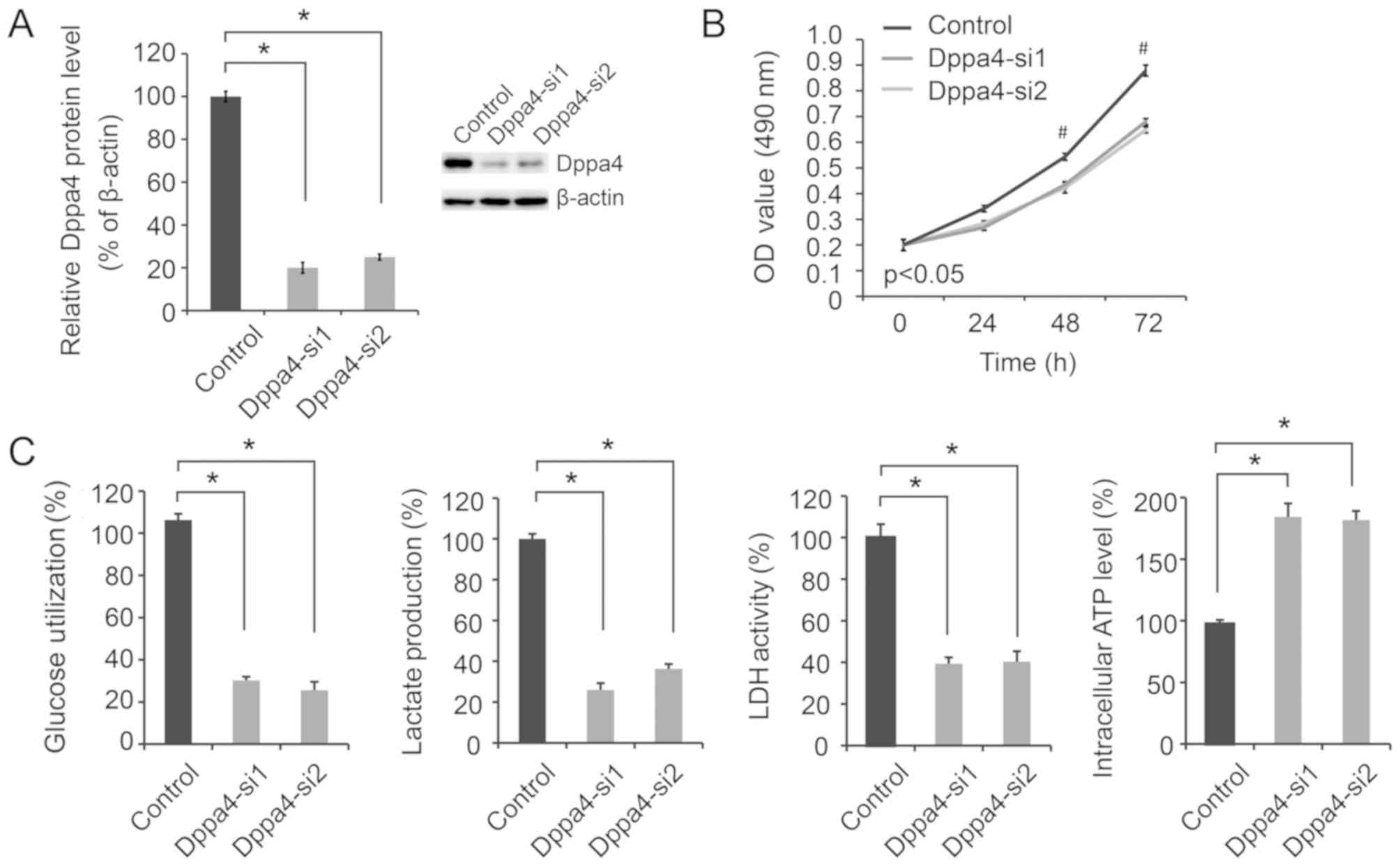
Dppa4 knockdown decreases NSCLC cell
viability and glycolysis
To determine the effect of altered Dppa4 expression
on NSCLC biology, siRNA transfection was performed in A549 cells.
As shown in Fig. 2A, Dppa4
expression was significantly decreased in A549 cells transfected
with Dppa4-si1 and Dppa4-si2 compared with the control group,
verifying the efficacy of Dppa4 knockdown. Next, the biological
effects of Dppa4 knockdown on NSCLC were evaluated. It was observed
that Dppa4 downregulation significantly inhibited cell viability
(Fig. 2B; P<0.05).
As altered aerobic glycolysis is a characteristic of
cancer metabolism, the present study then focused on the effect of
Dppa4 regulation of glycolysis on NSCLC metabolism. Following Dppa4
knockdown, significant decreases in glucose utilization, lactate
production and LDH activity were observed, along with a marked
increase in the intracellular ATP level (P<0.05; Fig. 2C), suggesting a decrease in
glycolysis.
Dppa4 overexpression increases cell
proliferation and glycolysis
Overexpression of Dppa4 was subsequently induced in
H1299 cells. As shown in Fig. 3A,
Dppa4 expression was increased in H1299 cells transfected with the
Dppa4-over plasmid, therefore verifying that overexpression was
successfully induced. It was then observed that Dppa4
overexpression significantly promoted cell growth (P<0.05;
Fig. 3B). Furthermore, the Dppa4
upregulation was associated with significant increases in glucose
utilization, lactate production and LDH activity, along with a
decrease in the intracellular ATP level (P<0.05; Fig. 3C).
Dppa4 regulates the levels of
glycolytic enzymes in NSCLC cells
The effect of Dppa4 on the expression levels of
glycolytic enzymes in NSCLC cells was then screened. It was
observed that knockdown of Dppa4 expression had no evident effect
on the majority of enzymes examined; however, significant
upregulation of glucose transporter type 4 (GLUT-4) and pyruvate
kinase isozyme M2 (PKM2), and marked downregulation of hexokinase
II (HK-II) and LDHB (P<0.05; Fig.
4A) were observed in the Dppa4 knockdown groups. In addition,
Dppa4 overexpression had no marked effect on the majority of the
enzymes, with the exception of GLUT-4 and LDHA downregulation, and
the upregulation of HK-II, enolase and LDHB (P<0.05; Fig. 4B). Among the abovementioned
enzymes, LDHB exhibited the most prominent changes.
 | Figure 4.Expression of glycolysis-associated
enzymes in non-small-cell lung cancer cells. (A) Knockdown of Dppa4
expression did not alter the expression of the majority of
glycolysis-associated enzymes, with the exception of GLUT-4 and
PKM2 upregulation, and HK-II and LDHB downregulation. (B) Dppa4
overexpression did not affect the expression of the majority of
these enzymes, apart from the downregulation of GLUT-4 and LDHA,
and the upregulation of HK-II, enolase and LDHB. *P<0.05 vs.
corresponding control group. Dppa4, developmental
pluripotency-associated 4; GLUT-4, glucose transporter type 4;
PKM2, pyruvate kinase isozyme M2; HK-II, hexokinase II; LDHB,
lactate dehydrogenase B. |
LDHB reverses the promoting effects of
Dppa4 in NSCLC cells
To elucidate the importance of LDHB expression in
NSCLC progression, Dppa4 overexpression/control plasmid and
LDHB/control siRNA were co-transfected in the cells (Fig. 5A). The results demonstrated that
LDHB siRNA reversed the promoting effect of Dppa4 overexpression on
cell proliferation (Fig. 5B) and
glycolysis (Fig. 5C). These
findings indicated that Dppa4 promotes NSCLC progression and this
effect is partly mediated via LDHB.
Discussion
Dppa4, a protein that is located in the embryonic
stem cell nucleus and is associated with chromatin, has been
reported to regulate the differentiation of ES cells into a
primitive ectodermal lineage (11). Although the expression of Dppa4 in
ES cells has been extensively investigated, little is known
regarding its in vivo expression in cancer. Previous
research has indicated that human cancer may be considered as a
stem-cell disease (12).
Cancer-initiating cells, also referred to as cancer stem cells,
serve an important role in hematopoietic cancer, as well as in
several solid tumors, such as lung cancer (13).
The aim of the present study was to investigate the
role of Dppa4 in NSCLC tumorigenesis. Initially, Dppa4 was found to
be highly expressed in NSCLC tissue samples and cell lines, both at
the mRNA and protein levels. Next, the correlation between Dppa4
expression and clinicopathological characteristics in NSCLC
patients was assessed. It was observed that Dppa4 was positively
correlated with tumor differentiation grade, T stage, N stage and
clinical stage. Subsequent Kaplan-Meier analysis revealed that
patients with higher Dppa4 expression exhibited poorer prognosis.
Univariate and multivariate survival analyses also revealed that,
in addition to tumor stage, upregulated Dppa4 expression was a
predictor of poor prognosis. However, the underlying mechanism
remained unclear.
The key role of Dppa4 in NSCLC development and
progression was demonstrated in the present study; however, the
currently available literature contains little evidence regarding
the role of Dppa4 in cancer metabolism. Abnormal metabolism is
regarded as one of the characteristic features of cancer. Since the
energy produced by glycolysis may contribute to the abnormal
proliferation of cancer cells, the current study focused on whether
glycolysis affected cell proliferation. It is widely accepted that
cancer growth depends on metabolism, and its metabolic pattern
shifts from aerobic to anaerobic respiration. Under the condition
of sufficient oxygen, the glycolysis of malignant tumor cells is
also active. The metabolic characteristic of this aerobic
glycolysis is known as the Warburg effect, which displays high
glucose uptake rate, active glycolysis and high lactic acid content
of metabolites. Furthermore, Jones et al (14) reported that cancer stem cells have
a third metabolic mode. On the basis of maintaining the aerobic
mode, glucose metabolism is converted into protein catabolism,
which provides energy for tumor cells (14). Malignant tumor cells perform
glycolysis at a rate that is 10 times faster than that observed in
their non-cancerous counterparts (15). To generate energy, cancer cells
convert glucose into lactate, supplying a primary route for the
carbon source that is required for macromolecular biosynthesis.
This process enables cancer cells to meet the increasing energy
demands of rapid tumor growth (16,17).
In the present study, Dppa4 knockdown had no evident
effect on the expression of the majority of the
glycolysis-associated enzymes, with the exception of upregulation
of GLUT-4 and PKM2, and downregulation of HK-II and LDHB.
Similarly, Dppa4 overexpression exerted no marked effect on the
levels of the majority of these enzymes, apart from downregulation
of GLUT-4 and LDHA, and upregulation of HK-II, enolase and LDHB.
The most notable changes as a result of Dppa4 knockdown or
overexpression were reported in LDHB. Among the abovementioned
enzymes, PKM1 and PKM2 are alternative splicing products of the PKM
gene, which have distinct functions and expression characteristics
(18). Only PKM2 is expressed in
embryonic tissues, whereas the PKM1 type is expressed during
adulthood, however, PKM2 expression is observed in tumor tissues,
rather than PKM1 (18). During
tumor formation, PKM1 or PKML/R gradually disappear, while PKM2 is
markedly upregulated, ultimately becoming a tumor-specific pyruvate
kinase (18). In addition, HK-II
has been reported to accelerate glucose metabolism and promote
cancer progression (19).
Furthermore, insulin resistance is known to serve an important role
in tumor cachexia. A study by Yoshikawa et al (20) demonstrated that insulin resistance
in tumor-tolerant mice was due to a decrease in the expression of
GLUT-4.
LDH, comprising LDHA and LDHB, is a terminal enzyme
catalyzing the interconversion of pyruvate and lactate in the
anaerobic glycolytic pathway, and is considered as the key
glycolytic enzyme (21,22). With regard to LDHB expression, it
differs markedly among different malignant tumors. Certain studies
have reported that the high expression of LDHB in lung and breast
cancer enhances the proliferation of tumor cells, and is a
significant predictor of poor prognosis (23–25).
Other studies have demonstrated that LDHB expression is suppressed
in hepatocellular carcinoma (HCC) (26), and prostate (21), pancreatic (27) and gastric cancer (28). Furthermore, LDHB may act as a
suppressor of glycolysis, thereby suppressing pancreatic cancer and
HCC progression. The mechanism may involve promoter
hypermethylation and decreased expression of LDHB, leading to
glycolytic transition by converting lactate to pyruvate, and a
shift from LDH1 to LDH5 (29). As
a therapeutic target, LDHB should be approached differently in
different types of cancer. Investigating the importance of LDHB in
NSCLC progression revealed that transfection with LDHB siRNA
reversed the promoting effect of Dppa4 overexpression on cancer
cell proliferation and glycolysis. These findings indicate that
Dppa4 promotes NSCLC cell proliferation and glycolysis in part via
LDHB.
In conclusion, the present study, to the best of our
knowledge, is the first to provide critical insight into the role
of Dppa4 in NSCLC cell proliferation and glycolysis, and to
indicate a role for Dppa4-LDHB signaling in NSCLC development and
progression. The findings not only indicated a newly identified
molecular mechanism involved in NSCLC glycolysis and progression,
but also highlighted the Dppa4-LDHB axis as a novel promising
molecular target for the design of new therapeutic strategies for
NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL and YFW performed the cellular and histological
studies, and statistical analysis, and drafted the manuscript. QW
and FS collected tumor tissues and follow up information on the
patients. JQ, XW, JX and YJW assisted in performing the cellular
and histological studies. YZ participated in the study design. All
authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Research
Ethics Committee of Xuzhou Cancer Hospital (Xuzhou, China).
Informed consent was obtained from all the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ezeh UI, Turek PJ, Reijo RA and Clark AT:
Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are
expressed in both seminoma and breast carcinoma. Cancer.
104:2255–2265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pardal R, Clarke MF and Morrison SJ:
Applying the principles of stem-cell biology to cancer. Nat Rev
Cancer. 3:895–902. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amini S, Fathi F, Mobalegi J,
Sofimajidpour H and Ghadimi T: The expressions of stem cell
markers: Oct4, Nanog, Sox2, nucleostemin, Bmi, Zfx, Tcl1, Tbx3,
Dppa4, and Esrrb in bladder, colon, and prostate cancer, and
certain cancer cell lines. Anat Cell Biol. 47:1–11. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Du J, Chen T, Zou X, Xiong B and Lu G:
Dppa2 knockdown-induced differentiation and repressed proliferation
of mouse embryonic stem cells. J Biochem. 147:265–271. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang M, Cui F, Lu S, Lu H, Xue Y, Wang J,
Chen J, Zhao S, Ma S, Zhang Y, et al: Developmental
pluripotency-associated 4: A novel predictor for prognosis and a
potential therapeutic target for colon cancer. J Exp Clin Cancer
Res. 34:602015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tung PY, Varlakhanova NV and Knoepfler PS:
Identification of DPPA4 and DPPA2 as a novel family of
pluripotency-related oncogenes. Stem Cells. 31:2330–2342. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang Y, Xiao J, Zhao S, Feng Y and Zhang
H: A study on construction of plasmid pcDNA3.1 (−) CMV.CD and
transfection into laryngeal cancer cell Hep-2. Lin Chuang Er Bi Yan
Hou Ke Za Zhi. 19:988–991. 2005.(In Chinese). PubMed/NCBI
|
|
9
|
Peirson SN, Butler JN and Foster RG:
Experimental validation of novel and conventional approaches to
quantitative real-time PCR data analysis. Nucleic Acids Res.
31:e732003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chakravarthy H, Boer B, Desler M, Mallanna
SK, McKeithan TW and Rizzino A: Identification of DPPA4 and other
genes as putative Sox2:Oct-3/4 target genes using a combination of
in silico analysis and transcription-based assays. J Cell Physiol.
216:651–662. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abdul Khalek FJ, Gallicano GI and Mishra
L: Colon cancer stem cells. Gastrointest Cancer Res. (Suppl
1):S16–S23. 2010.PubMed/NCBI
|
|
13
|
Eramo A, Lotti F, Sette G, Pilozzi E,
Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C and De
Maria R: Identification and expansion of the tumorigenic lung
cancer stem cell population. Cell Death Differ. 15:5042008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jones CL, Stevens BM, D'Alessandro A,
Reisz JA, Culp-Hill R, Nemkov T, Pei S, Khan N, Adane B, Ye H, et
al: Inhibition of amino acid metabolism selectively targets human
leukemia stem cells. Cancer Cell. 34:724–740.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu J, Tan M and Cai Q: The Warburg effect
in tumor progression: Mitochondrial oxidative metabolism as an
anti-metastasis mechanism. Cancer Lett. 356:156–164. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin L, Huang H, Liao W, Ma H, Liu J, Wang
L, Huang N and Liao Y: MACC1 supports human gastric cancer growth
under metabolic stress by enhancing the Warburg effect. Oncogene.
34:2700–2710. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Christofk HR, Vander Heiden MG, Wu N,
Asara JM and Cantley LC: Pyruvate kinase M2 is a
phosphotyrosine-binding protein. Nature. 452:181–186. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mathupala SP, Heese C and Pedersen PL:
Glucose catabolism in cancer cells. The type II hexokinase promoter
contains functionally active response elements for the tumor
suppressor p53. J Biol Chem. 272:22776–22780. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yoshikawa T, Noguchi Y and Satoh S:
Inhibition of IRS-1 phosphorylation and the alterations of GLUT4 in
isolated adipocytes from cachectic tumor-bearing rats. Biochem
Biophys Res Commun. 256:676–681. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Leiblich A, Cross SS, Catto JW, Phillips
JT, Leung HY, Hamdy FC and Rehman I: Lactate dehydrogenase-B is
silenced by promoter hypermethylation in human prostate cancer.
Oncogene. 25:2953–2960. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song T, Gan W, Chen J, Huang L, Yin H, He
T, Huang H and Hu X: Antibodies against Clonorchis sinensis LDH
could cross-react with LDHB localizing on the plasma membrane of
human hepatocarcinoma cell SMMC-7721 and induce apoptosis.
Parasitol Res. 115:1595–1603. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McCleland ML, Adler AS, Deming L, Cosino
E, Lee L, Blackwood EM, Solon M, Tao J, Li L, Shames D, et al:
Lactate dehydrogenase B is required for the growth of
KRAS-dependent lung adenocarcinomas. Clin Cancer Res. 19:773–784.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McCleland ML, Adler AS, Shang Y, Hunsaker
T, Truong T, Peterson D, Torres E, Li L, Haley B, Stephan JP, et
al: An integrated genomic screen identifies LDHB as an essential
gene for triple-negative breast cancer. Cancer Res. 72:5812–5823.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dennison JB, Molina JR, Mitra S,
González-Angulo AM, Balko JM, Kuba MG, Sanders ME, Pinto JA, Gómez
HL, Arteaga CL, et al: Lactate dehydrogenase B: A metabolic marker
of response to neoadjuvant chemotherapy in breast cancer. Clin
Cancer Res. 19:3703–3713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen R, Zhou X, Yu Z, Liu J and Huang G:
Low expression of LDHB correlates with unfavorable survival in
hepatocellular carcinoma: Strobe-compliant article. Medicine
(Baltimore). 94:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cui J, Quan M, Jiang W, Hu H, Jiao F, Li
N, Jin Z, Wang L and Wang Y: Suppressed expression of LDHB promotes
pancreatic cancer progression via inducing glycolytic phenotype.
Med Oncol. 32:1432015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Maekawa M, Taniguchi T, Ishikawa J,
Sugimura H, Sugano K and Kanno T: Promoter hypermethylation in
cancer silences LDHB, eliminating lactate dehydrogenase isoenzymes
1–4. Clin Chem. 49:1518–1520. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Revill K, Wang T, Lachenmayer A, Kojima K,
Harrington A, Li J, Hoshida Y, Llovet JM and Powers S: Genome-wide
methylation analysis and epigenetic unmasking identify tumor
suppressor genes in hepatocellular carcinoma. Gastroenterology.
145:1424–135.e1-e25. 2013. View Article : Google Scholar : PubMed/NCBI
|