Introduction
Lung cancer is the leading cause of
cancer-associated mortality worldwide (1). The prevention and treatment of lung
cancer have drawn wide attention and have been a focus in clinical
practice. Non-small cell lung cancer (NSCLC), which initiates from
non-small cells of the lung, accounts for approximately 85% of all
lung cancer cases with a high mortality rate (2). In the past decade, surgical resection
combined with chemotherapy has greatly improved the prognosis of
lung cancer patients; however, due to the fact that most patients
are diagnosed with middle or late stage of the disease, the 5-year
survival rate of NSCLC patients still remains poor. Therefore,
exploration of the underlying molecular mechanisms which regulate
tumor growth is important in searching for effective targets that
may benefit the treatment of NSCLC.
MicroRNAs (miRNAs) are a class of non-coding
regulatory RNAs with a length of 20–24 nucleotides (3–6).
miRNAs regulate gene expression via binding the 3′-untranslated
region (3′-UTR) of the targeted mRNAs to suppress the translation
or induce the degradation of messenger RNAs (mRNAs) (5). It has been well-documented that
miRNAs play important roles in a variety of physiological
conditions, including cell growth, differentiation and apoptosis
(7). The oncogenic or
tumor-suppressive function of miRNAs is involved in the initiation
and progression of human cancers (8,9).
Notably, increasing evidence has demonstrated that miRNAs play
important roles in regulating the proliferation, invasion and
metastasis of NSCLC (10,11). For example, highly expressed miR-10
was found to promote the growth of NSCLC cells via targeting
phosphatase and tensin homolog (PTEN) (12). miR-17 was found to regulate
cisplatin-resistance and metastasis of NSCLC (13). A recent study showed that
miR-187-3p mitigated the proliferation of NSCLC cells by modulating
the expression of BCL6 (14).
Collectively, these studies indicate that miRNAs are important
regulators which modulate the growth of NSCLC cells.
miR-221-3p is a newly identified oncogenic miRNA
involved in the development of tumors (15,16).
A recent study showed that miR-221-3p is upregulated in pancreatic
cancer and serves as a potential biomarker for the diagnosis of
cancer patients (16). miR-221-3p
was also found to promote the metastasis of cervical cancer via
targeting the Twist homolog 2 (THBS2) (15). Additionally, the oncogenic function
of miR-221-3p has also been identified in gastric cancer, and
enhanced the growth of gastric cancer cells by inhibiting the
expression of PTEN (17).
Interestingly, downregulation of miR-221-3p has been suggested as a
prognostic biomarker in triple-negative breast cancer (TNBC) and
was found to be associated with the poor prognosis of TNBC patients
(18). The oncogenic or
tumor-suppressive function of miR-221-3p may depend on the tumor
system. Recent research has proposed miR-221-3p as a possible
therapeutic tool (19). These
findings indicate the critical involvement of miR-221-3p in
cancers; however, the involvement of miR-221-3p in NSCLC has not
been well investigated.
The present study aimed to evaluate the expression
and function of miR-221-3p in NSCLC. The results uncovered that
miR-221-3p was highly expressed in NSCLC tissues and cell lines.
Overexpression of miR-221-3p promoted the growth of NSCLC cells via
targeting the cell cycle regulator p27 and enhanced cell cycle
progression.
Materials and methods
Cell lines and tissue samples
The NSCLC cell lines A549, H1299, H23 and SK-MES-1,
and the normal bronchial epithelium BEAS-2B were purchased from the
American Type Culture Collection (ATCC; Manassas, VA, USA). The
cells were cultured in Dulbecco's modified Eagle's medium (Gibco™
DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc.), 100 U/ml of penicillin and 100 µg/ml of
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C
with 5% CO2. For cell transfection, 20 nM control miRNA
(5′-CGGUACGAUCGCGGCGGGAUAUC-3′) or 20 nM miR-221-3p
(5′-AGCUACAUUGUCUGCUGGGUUUC-3′) was transfected into
1×104 NSCLC cells using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The cells were harvested for the
following experiments after transfection for 48 h.
The 50-paired NSCLC tissues and matched adjacent
normal lung tissues were collected from patients (age range, 35–75
years; female to male ratio, 1:0.78) who were diagnosed at Longnan
Hospital (Daqing, Heilongjiang, China) between April 2011 and
August 2013. None of these patients received radiotherapy or
chemotherapy prior to surgical resection. All samples were
confirmed by histopathological examination and stored in liquid
nitrogen before use. Written informed consent was obtained from all
the participants. The sample collection was approved by the
Institutional Review Board of Longnan Hospital (Daqing,
Heilongjiang, China). The experiments using these samples were
performed according to the Declaration of Helsinki.
RNA extraction and quantitative
PCR
Total miRNA was extracted from the tissues using the
miRcute miRNA isolation kit (Tiangen, Beijing, China) according to
the manufacturer's instructions. Reverse transcription (RT) PCR was
performed using the One Step Prime Script miRNA cDNA Synthesis kit
(Takara, Dalian, China). The quantitative PCR (qPCR) was conducted
with TaqMan™ Multiplex Master Mix (Invitrogen; Thermo Fisher
Scientific, Inc.) using the Applied Biosystems 7500 Real-Time
RT-PCR system (Thermo Fisher Scientific, Inc.). The expression of
U6 RNA was detected as the normalization control. The primers for
miR-221-3p and U6 were designed as follows: miR-221-3p forward,
5′-ACACTCCAGCTGGGAGCTACA and reverse, 5′-CTCAACTGGTGTCGTGGA; U6
forward, 5′-ATTGGAACGATACAGAG and reverse, 5′-GGAACGCTTCACGAATTT.
The thermocycling conditions were as follows: Initial denaturation
at 95°C for 5 min followed by 40 cycles at 95°C for 10 sec and 60°C
for 1 min. The relative expression of miR-221-3p was calculated
with the 2−∆∆Cq method (20). The experiments were performed in
triplicate.
Cell proliferation
Both A549 and H1299 cells were seeded into a 96-well
plate at a density of 2×103 cells per well. The cell
proliferation was quantified by adding 10 µl of Cell Counting Kit-8
reagent (CCK-8, Dojindo, Kumamoto, Japan) and incubated at 37°C for
2 h. The absorbance of each well at 450 nm was measured with a
microplate reader.
Cell cycle analysis
Cells transfected with the corresponding miRNAs were
cultured in 60-mm dishes. When the cell confluence reached
approximately 70%, the cells were harvested and fixed with pre-cold
70% ethanol overnight at 4°C. The cells were washed twice with PBS
and incubated with 1 mg/ml RNase A at 37°C for 30 min. Afterwards,
cells were stained with 50 µg/ml propidium iodide (PI) for 40 min
at 37°C. Cells were then passed through a 300-mesh nylon net and
the cell cycle distribution was determined by flow cytometry
(Becton Dickinson FACS Calibur; BD Biosciences, Franklin Lakes, NJ,
USA).
Wound-healing assay
NSCLC cells were cultured in 6-well plates and
transfected with the indicated miRNA. After transfection, the cells
were incubated for 36 h, and the wound was generated using a 200 µl
pipette tip. Cells were washed twice with PBS buffer. The wound
region was detected after 24 h by light microscopy (magnification,
×40).
Cell invasion
The cell invasion assay was performed using 24-well
polycarbonate Transwell chambers with 8-µm pores (Corning, Inc.).
In total, 1×103 NSCLC cells/well transfected with the
control miRNA or miR-221-3p were plated in serum-free DMEM in the
upper chamber. Inserts were precoated with Matrigel. DMEM with 10%
FBS was added to the lower chambers. After incubation for 36 h at
37°C, invaded cells were stained with 0.1% crystal violet for 10
min at room temperature and quantified. Cells were visualized and
counted using a light microscope (magnification, ×40).
Dual-luciferase reporter assay
The 3′-UTR of p27 containing the targeting sites of
miR-221-3p was amplified and cloned into the p-MIR-reporter plasmid
(Ambion; Thermo Fisher Scientific, Inc.). NSCLC cells cotransfected
with miR-221-3p and the luciferase reporter vector of p27 were
plated (1×103 cells/well) in 96-well plates. The
Renilla luciferase vector was also transfected with
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) as
control of the transfection efficiency. After transfection for 48
h, the luciferase activity was measured with the Dual-Luciferase
Reporter Assay System (Promega Corporation) according to the
manufacturer's protocol. The p-MIR-firefly (Ambion; Thermo Fisher
Scientific, Inc.) luciferase activity was normalized to
p-MIR-Renilla (Ambion; Thermo Fisher Scientific, Inc.)
activity.
Bioinformatics prediction
The databases of TargetScan (http://www.Targetscan.org) and miRBase (http://www.mirbase.org) were used to predict the
potential targets of miR-221-3p by inputting the name of miRNA in
the query.
Western blot analysis
After transfection for 48 h, cells were harvested
and lysed with the NP-40 buffer [150 mM NaCl, 1% NP-40, 50 mM
Tris-HCl (pH 8.0), 1 mM EDTA] containing 0.15 U/ml aprotinin, 20 mM
leupeptin and 1 mM phenylmethylsulfonyl fluoride. Proteins were
loaded onto the 15% SDS-PAGE and transferred onto nitrocellulose
filter membranes (Pall Life Sciences, Port Washington, NY, USA).
The membrane were initially blocked with 5% non-fat milk for 1 h at
room temperature (RT) and then incubated with the primary antibody
overnight at 4°C. The membranes were then incubated with the
secondary antibody for 1 h at RT. The western blot bands were
visualized with the Amersham™ ECL Plus Western Blotting Detection
System (GE Healthcare, UK). The antibodies used in this study
included anti-p27 (cat. no. sc-1641, Santa Cruz Biotechnology,
Inc., Dallas, TX, USA; dilution ratio: 1:2,000), anti-GAPDH (cat.
no. 3H12, MBL, Japan; dilution ratio: 1:3,000) and anti-Flag (cat.
no. ab1257; Abcam, Cambridge, MA, USA; dilution ratio: 1:2,000)
which were purchased from the mentioned companies. The intensities
of the protein bands were analyzed using the Image J software
(version D1.47; National Institutes of Health).
Cell apoptosis analysis
The percentage of cell apoptosis was assessed using
PI/Annexin V-based flow cytometry with the Annexin V-FITC Apoptosis
Detection kit (Thermo Fisher Scientific, USA) according to the
manufacturer's instructions. Briefly, cells were harvested and
washed with pre-cold PBS. Cells were re-centrifuged and resuspended
to a final density of ~1×106 cells/ml with the
Annexin-binding buffer. 5 µl of FITC/Annexin V and 1 µl of 100
µg/ml PI working solution was added to each 100 µl of cell
suspension. After incubation for 15 min at RT, 400 µl of 1X
Annexin-binding buffer was added into the cells and mixed gently.
The cell apoptosis was analyzed by flow cytometry as soon as
possible.
Statistical analysis
Data are presented as mean ± standard deviation
(SD). Statistical analysis was examined with SPSS 19.0 software
version (IBM Corp., Armonk, NY, USA). Student's t-test was used to
analyze the difference between two groups. One-way analysis of
variance followed by Dunnett's test was adopted when comparing more
than two groups. P<0.05 was considered to be statistically
significant.
Results
miR-221-3p is overexpressed in NSCLC
tissues and cell lines
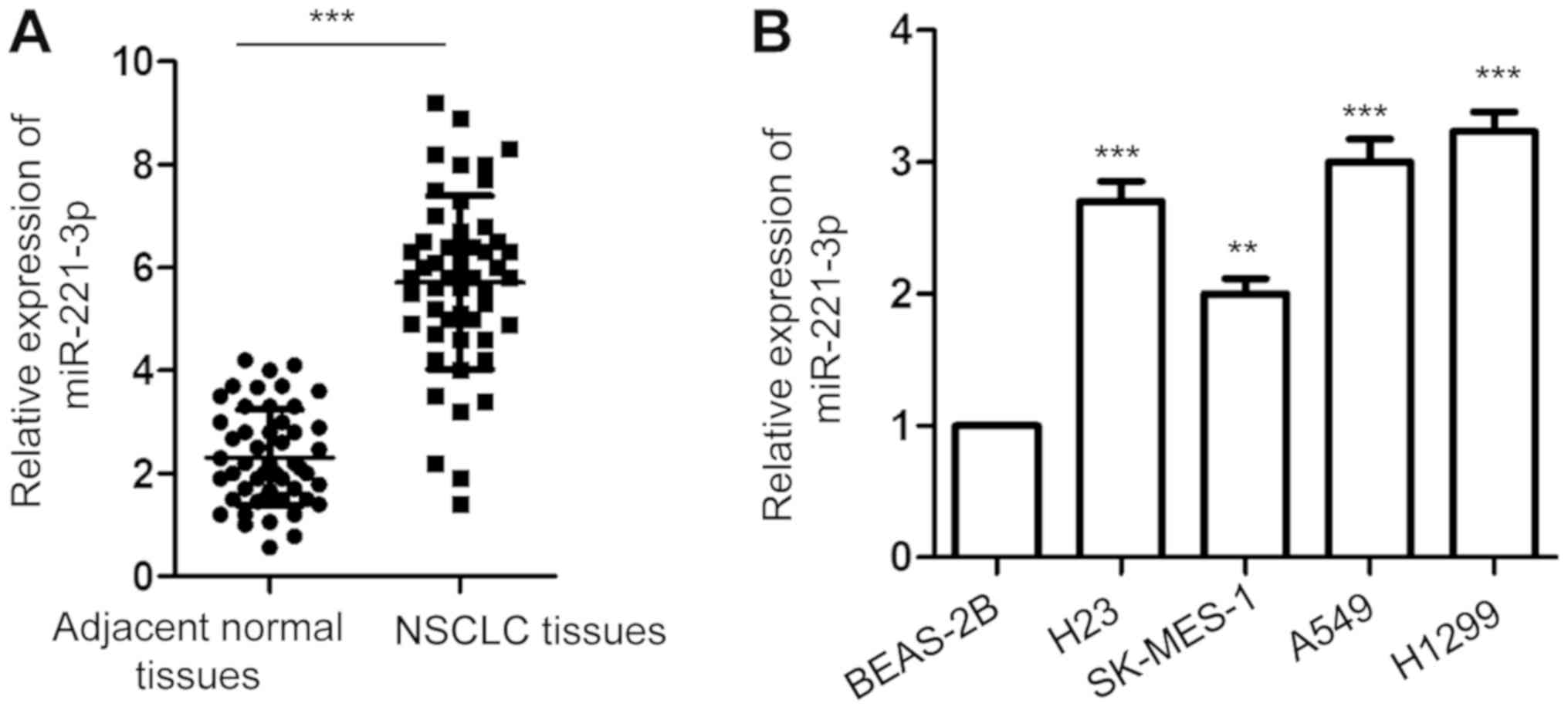
To investigate the involvement of miR-221-3p in
NSCLC, the expression of miR-221-3p in 50-paired NSCLC tissues and
matched corresponding normal lung tissues was detected with
RT-qPCR. The data showed that the expression of miR-221-3p was
significantly increased in NSCLC tissues compared with that in the
adjacent normal tissues (Fig. 1A).
Additionally, the abundance of miR-221-3p in NSCLC cell lines
including A549, H1299, H23 and SK-MES-1 and normal bronchial
epithelium BEAS-2B cells were also investigated. As presented in
Fig. 1B, a significantly higher
level of miR-221-3p was obtained in the NSCLC cell lines than that
noted in the normal cells. These results indicated the
overexpression of miR-221-3p in NSCLC.
Downregulation of miR-221-3p
suppresses the growth of NSCLC cells
As the expression of miR-221-3p was overexpressed in
NSCLC, we investigated the influence of miR-221-3p on the growth of
NSCLC cells. Thus, miR-221-3p was downregulated by transfecting
miR-221-3p antagomir into A549 and H1299 cells. The downregulation
efficiency of miR-221-3p was validated by RT-qPCR analysis
(Fig. 2A). The effect of
miR-221-3p on the proliferation of NSCLC cells was detected with
the CCK-8 assay. As presented in Fig.
2B and C, suppression of miR-221-3p significantly inhibited the
proliferation of both A5459 and H1299 cell lines in comparison with
the cells transfecting with the negative control miRNA. To further
confirm these results, an in vitro wound-healing assay was
performed with A549 and H1299 cells expressing downregulated
miR-221-3p. Decreased expression of miR-221-3p markedly reduced the
migration of both A549 and H1299 cell lines (Fig. 2D). Furthermore, the effect of
miR-221-3p downregulation by antagomir transfection on the
apoptosis of NSCLC cells was also investigated. As shown in
Fig. 2E, depletion of miR-221-3p
significantly increased the relative cell apoptosis percentage of
both A549 and H1299 cell lines. The effect of miR-221-3p on the
invasion of NSCLC cells was also detected by transfecting negative
control miRNA or miR-221-3p antagomir into A549 and H1299 cells.
The result showed that downregulation of miR-221-3p significantly
inhibited the invasion of NSCLC cells (Fig. 2F). Collectively, these data
demonstrated that depletion of miR-221-3p suppressed the growth of
NSCLC cells.
p27 is a target of miR-221-3p in NSCLC
cells
The prediction of RNA-associated interactions with
computational prediction algorithms plays important roles in
characterizing the function of miRNAs (21). To further understand the molecular
mechanisms by which miR-221-3p regulates the growth of NSCLC cells,
the potential targets of miR-221-3p were predicted with the
bioinformatics resource TargetScan (http://www.Targetscan.org) and miRBase (http://www.mirbase.org) (Tables SI and SII). The data showed that p27, the
inhibitor of cell cycle progression, which plays important roles in
the initiation and progression of cancers (22,23),
ranked highly among all the possible targets. The putative binding
sites of miR-221-3p at the 3′-UTR of p27 are presented as Fig. 3A. To detect the binding between
miR-221-3p with the 3′-UTR of p27, luciferase assay was performed
by transfecting luciferase reporter plasmids containing wild-type
(WT) or mutant 3′-UTR of p27 in the presence of miR-221-3p mimics
in A549 and H1299 cells. The results showed that the luciferase
activity of WT but not the mutant 3′-UTR of p27 was significantly
decreased with the co-transfection of miR-221-3p mimics (Fig. 3B and C). This result suggested the
binding between miR-221-3p and the 3′-UTR of p27. To detect the
consequence of the binding, the mRNA and protein levels of p27 in
the NSCLC cell lines with overexpression of miR-221-3p were
examined, respectively. As presented in Fig. 3D, high expression of miR-221-3p
significantly decreased the mRNA level of p27. Consistently, the
protein level of p27 was also inhibited upon the overexpression of
miR-221-3p in the A549 and H1299 cell lines (Fig. 3E). These results indicated that
miR-221-3p was able to bind the 3′-UTR of p27 and suppress the
expression of p27 in NSCLC cells.
Overexpression of miR-221-3p promotes
the cell cycle progression of NSCLC cells
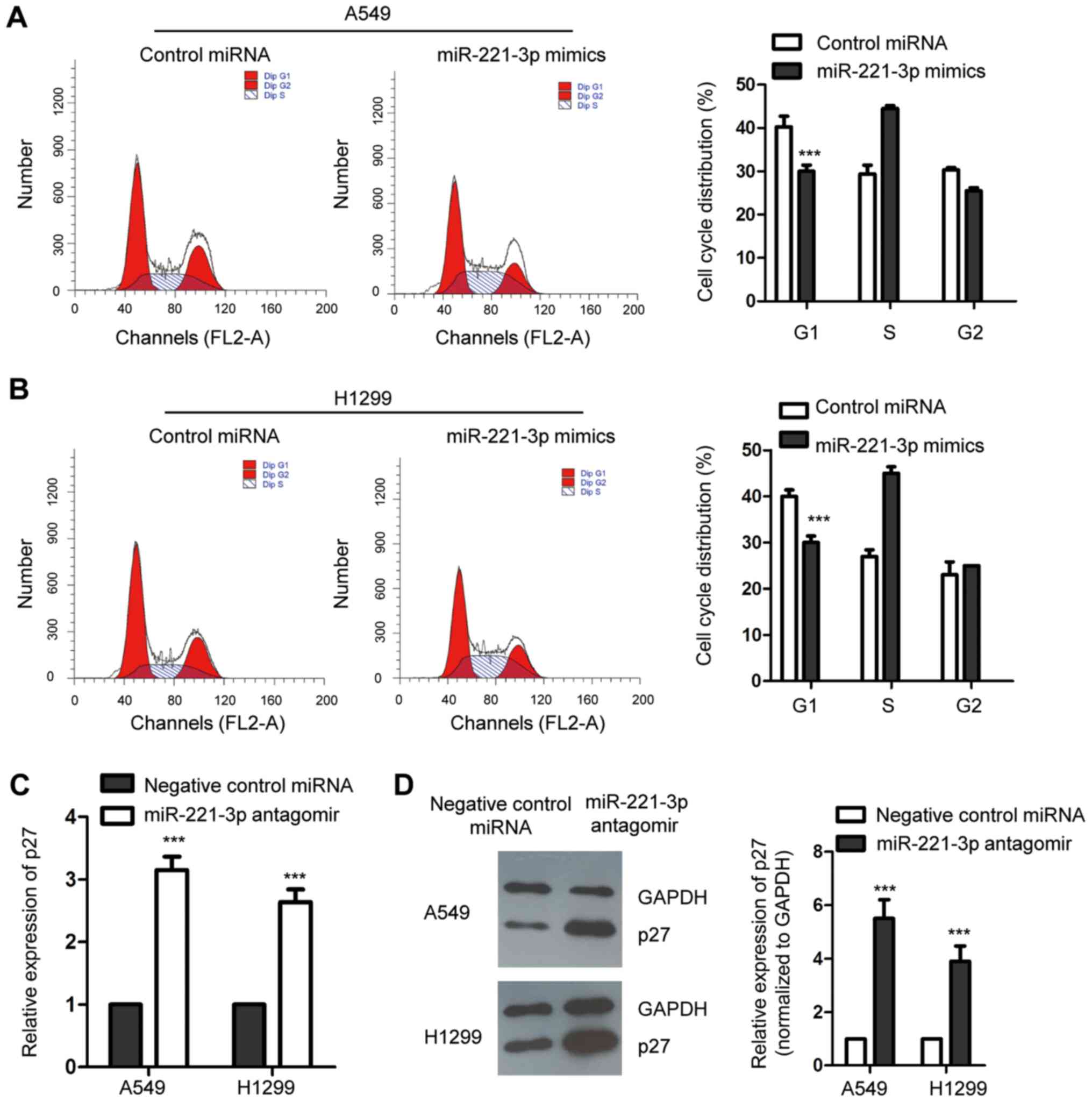
An increased level of p27 has been reported to
control the cell cycle progression at G1 phase (24). As miR-221-3p was found to
negatively regulate the expression of p27, to ascertain the
influence of miR-221-3p on the cell cycle progression of NSCLC
cells, both A549 and H1299 cell lines were transfected with
miR-221-3p mimics or control miRNA, and the cell cycle progression
was assessed by flow cytometry. As presented in Fig. 4A and B, compared with the control
cells, overexpression of miR-221-3p significantly promoted the cell
cycle progression from G1 to S phase in the A549 and H1299 cell
lines. This result was consistent with the decreased expression of
p27 with the ectopic expression of miR-221-3p in NSCLC cells. To
further confirm this observation, endogenous miR-221-3p was
downregulated by inducing miR-221-3p antagomir into the NSCLC
cells. RT-qPCR and western blot analysis were performed to detect
the expression level of p27. The data showed that depletion of
miR-221-3p increased the abundance of p27 in the A549 and H1299
cell lines (Fig. 4C and D). The
cell cycle progression of NSCLC cells expressing negative control
miRNA or miR-221-3p antagomir was examined by flow cytometry. As
shown in Fig. 4E and F,
downregulation of miR-221-3p significantly induced cell cycle
arrest at the G1 phase. These results collectively indicated that
miR-221-3p is a novel regulator of the cell cycle progression of
NSCLC cells. To detect whether the modulation of miR-221-3p on the
proliferation of NSCLC cells was by regulating p27, both A549 and
H1299 cell lines were transfected with Flag empty vector or
Flag-p27 (Fig. 4G). The CCK-8
assay indicated that overexpression of miR-221-3p promoted the
proliferation of the NSCLC cell lines, and transfection of p27
significantly decreased the growth of NSCLC cells expressing
miR-221-3p (Fig. 4H and I). These
results suggest that p27 mediates the role of miR-221-3p in
NSCLC.
Discussion
Non-small cell lung cancer (NSCLC) is a heavy burden
for human health worldwide. Exploring novel factors involved in the
progression of NSCLC will benefit the prognosis and treatment of
NSCLC patients. In the present study, we evaluated the expression
of miR-221-3p in NSCLC tissues and cell lines. High expression of
miR-221-3p was observed in NSCLC tissues compared with the normal
control tissues. Further mechanistic study found that
overexpression of miR-221-3p suppressed the growth of NSCLC cells
via targeting p27.
Consistent with the high expression of miR-221-3p in
NSCLC tissues, a previous study showed that overexpression of
miR-221-3p maintained tumor necrosis factor (TNF)-related
apoptosis-inducing ligand (TRAIL) in NSCLC, which indicated that
miR-221-3p is a possible target for the diagnosis of TRAIL
resistance in NSCLC (25). A
recent study demonstrated that miR-221-3p is overexpressed in
pancreatic cancer and serves as a potential biomarker for the
diagnosis of cancer patients (16). In addition, the oncogenic function
of miR-221-3p was also identified in gastric cancer, which targeted
PTEN and enhanced the growth of gastric cancer cells (17). It was also reported that cervical
squamous cell carcinoma-secreted exosomal miR-221-3p promoted the
lymphangiogenesis and lymphatic metastasis (26). miR-221-3p was upregulated by the
transcription factor TWIST2 and targeted THBS2 in cervical cancer,
which promoted lymph node metastasis (15). Different from the potential
oncogenic role of miR-221-3p in the above reports, a recent study
by Wu et al (27) found
that a higher level of miR-221-3p in epithelial ovarian cancer
(EOC) inhibited the growth of cells and was associated with the
better overall survival of EOC patients, which indicated the
tumor-suppressive role of miR-221-3p in EOC. Consistently, a high
level of miR-221-3p was associated with better 5-year disease-free
survival of the triple-negative breast cancer patients (18). Combined with these findings, the
regulation of miR-221-3p on cell growth might be associated with
cancer types. In the present study, downregulation of miR-221-3p
increased the apoptosis of NSCLC cells. It may be interesting to
further analyze this miR-221-3p-associated cell death with the
bioinformatics resource such as ncRDeathDB (28).
The cyclin-dependent kinase inhibitor p27 is a cell
cycle regulator belonging to the Cip/Kip family that inhibits the
cell cycle progression at the G1/S boundary (29,30).
A low level of p27 was found to be correlated with rapid
proliferation in a variety of cancer types (31). The abundance of p27 expression is
regulated by diverse mechanisms including transcription,
translation and protein degradation (32). Additionally, multiple
oncogene-activated pathways target p27 to trigger its cytoplasmic
translocation, which consequently attenuates the nuclear cell cycle
inhibitory functions and allows activation of CDK1 and CDK2
(33). Emerging evidence suggests
that miRNAs control the cell cycle progression of cancer cells by
modulating the expression of p27 (34). For example, miR-199a-5p was found
to target p27 in osteosarcoma and to contribute to cancer cell
growth (35). Recent results by
Wang et al (36)
demonstrated that miR-200c directly regulates the expression of p27
and enhances the proliferation of gastric cancer cells. p27 has
also been identified as the target of miR-222 in hepatocellular
carcinoma (HCC) to regulate the proliferation of HCC cells
(37). Downregulation of
miR-221-3p inhibited the proliferation of pancreatic cancer cells
via upregulating the expression of p27 (38). In the present study, it was found
that miR-221-3p is an upstream regulator of p27 in NSCLC cells.
miR-221-3p was able to bind the 3′-UTR of p27 and inhibit the
expression of p27 in NSCLC cells. Consistent with the decreased
level of p27 with miR-221-3p, overexpression of miR-221-3p
significantly promoted the cell cycle progression of NSCLC cells.
Due to the inhibition of p27 expression upon the overexpression of
miR-221-3p, the correlation between the expression of miR-221-3p
with the clinical parameters of NSCLC is a significant issue which
warrants further study. Additionally, the subcellular location of
miR-221-3p deserves further investigation with the available
software as the location might affect the function and structure of
miRNAs (39). The involvement of
miR-221-3p in other types of cancer or diseases can be predicted
with the bioinformatic databases (40,41).
In summary, our findings found that miR-221-3p was
highly expressed in NSCLC tissues and cell lines. Molecular
mechanistic studies revealed that miR-221-3p targets p27 and
promotes the cell cycle progression of NSCLC cells. Our results
uncovered the possible mechanisms of miR-221-3p in NSCLC, which
indicate that miR-221-3p might be considered as a promising
anticancer target in NSCLC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GY and JL designed the study, performed the
experiments and analyzed the data. BZ collected the tissue samples
and performed some experiments. JL wrote the manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all the
participants. The sample collection was approved by the
Institutional Review Board of Longnan Hospital (Daqing,
Heilongjiang, China). The experiments using these samples were
performed according to the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vestergaard HH, Christensen MR and Lassen
UN: A systematic review of targeted agents for non-small cell lung
cancer. Acta Oncol. 57:176–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xiong Y, Wang T, Wang M, Zhao J, Li X,
Zhang Z, Zhou Y, Liu J, Jia L and Han Y: Long non-coding RNAs
function as novel predictors and targets of non-small cell lung
cancer: A systematic review and meta-analysis. Oncotarget.
9:11377–11386. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mohr AM and Mott JL: Overview of microRNA
biology. Semin Liver Dis. 35:3–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kwak PB, Iwasaki S and Tomari Y: The
microRNA pathway and cancer. Cancer Sci. 101:2309–2315. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gentilin E, Degli Uberti E and Zatelli MC:
Strategies to use microRNAs as therapeutic targets. Best Pract Res
Clin Endocrinol Metab. 30:629–639. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma R, Wang C, Wang J, Wang D and Xu J:
miRNA-mRNA interaction network in non-small-cell lung cancer.
Interdiscip Sci. 8:209–219. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang W, Zhang Q, Zhang M, Zhang Y, Li F
and Lei P: Analysis for the mechanism between the small cell lung
cancer and non-small cell lung cancer combing the miRNA and mRNA
expression profiles. Thorac Cancer. 6:70–79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu T, Liu L, Li J, Yan M, Lin H, Liu Y,
Chu D, Tu H, Gu A and Yao M: MiRNA-10a is upregulated in NSCLC and
may promote cancer by targeting PTEN. Oncotarget. 6:30239–30250.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang Z, Yin J, Fu W, Mo Y, Pan Y, Dai L,
Huang H, Li S and Zhao J: MiRNA 17 family regulates
cisplatin-resistant and metastasis by targeting TGFbetaR2 in NSCLC.
PLoS One. 9:e946392014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun C, Li S, Yang C, Xi Y, Wang L, Zhang F
and Li D: MicroRNA-187-3p mitigates non-small cell lung cancer
(NSCLC) development through down-regulation of BCL6. Biochem
Biophys Res Commun. 471:82–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei WF, Zhou CF, Wu XG, He LN, Wu LF, Chen
XJ, Yan RM, Zhong M, Yu YH, Liang L and Wang W: MicroRNA-221-3p, a
TWIST2 target, promotes cervical cancer metastasis by directly
targeting THBS2. Cell Death Dis. 8:32202017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li F, Xu JW, Wang L, Liu H, Yan Y and Hu
SY: MicroRNA-221-3p is up-regulated and serves as a potential
biomarker in pancreatic cancer. Artif Cells Nanomed Biotechnol.
46:482–487. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi J, Zhang Y, Jin N, Li Y, Wu S and Xu
L: MicroRNA-221-3p plays an oncogenic role in gastric carcinoma by
inhibiting PTEN expression. Oncol Res. 25:523–536. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deng L, Lei Q, Wang Y, Wang Z, Xie G,
Zhong X, Wang Y, Chen N, Qiu Y, Pu T, et al: Downregulation of
miR-221-3p and upregulation of its target gene PARP1 are prognostic
biomarkers for triple negative breast cancer patients and
associated with poor prognosis. Oncotarget. 8:108712–108725. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Garofalo M, Quintavalle C, Romano G, Croce
CM and Condorelli G: miR221/222 in cancer: Their role in tumor
progression and response to therapy. Curr Mol Med. 12:27–33. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yi Y, Zhao Y, Li C, Zhang L, Huang H, Li
Y, Liu L, Hou P, Cui T, Tan P, et al: RAID v2.0: An updated
resource of RNA-associated interactions across organisms. Nucleic
Acids Res. 45:D115–D118. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Raghu D, Paul PJ, Gulati T, Deb S, Khoo C,
Russo A, Gallo E, Blandino G, Chan AL, Takano E, et al: E6AP
promotes prostate cancer by reducing p27 expression. Oncotarget.
8:42939–42948. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vasile Bochis O, Achimas-Cadariu P, Vlad
C, Fetica B, Corneliu Leucuta D, Ioan Busuioc C and Irimie A: The
prognostic role of Skp2 and the tumor suppressor protein p27 in
colorectal cancer. J BUON. 22:1122–1130. 2017.PubMed/NCBI
|
|
24
|
Ventura C, Nunez M, Gaido V, Pontillo C,
Miret N, Randi A and Cocca C: Hexachlorobenzene alters cell cycle
by regulating p27-cyclin E-CDK2 and c-Src-p27 protein complexes.
Toxicol Lett. 270:72–79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Garofalo M, Quintavalle C, Di Leva G,
Zanca C, Romano G, Taccioli C, Liu CG, Croce CM and Condorelli G:
MicroRNA signatures of TRAIL resistance in human non-small cell
lung cancer. Oncogene. 27:3845–3855. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou CF, Ma J, Huang L, Yi HY, Zhang YM,
Wu XG, Yan RM, Liang L, Zhong M, Yu YH, et al: Cervical squamous
cell carcinoma-secreted exosomal miR-221-3p promotes
lymphangiogenesis and lymphatic metastasis by targeting VASH1.
Oncogene. 38:1256–1268. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu Q, Ren X, Zhang Y, Fu X, Li Y, Peng Y,
Xiao Q, Li T, Ouyang C, Hu Y, et al: MiR-221-3p targets ARF4 and
inhibits the proliferation and migration of epithelial ovarian
cancer cells. Biochem Biophys Res Commun. 497:1162–1170. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu D, Huang Y, Kang J, Li K, Bi X, Zhang
T, Jin N, Hu Y, Tan P, Zhang L, et al: ncRDeathDB: A comprehensive
bioinformatics resource for deciphering network organization of the
ncRNA-mediated cell death system. Autophagy. 11:1917–1926. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sgambato A, Cittadini A, Faraglia B and
Weinstein IB: Multiple functions of p27(Kip1) and its alterations
in tumor cells: A review. J Cell Physiol. 183:18–27. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guan X, Wang Y, Xie R, Chen L, Bai J, Lu J
and Kuo MT: p27(Kip1) as a prognostic factor in breast cancer: A
systematic review and meta-analysis. J Cell Mol Med. 14:944–953.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Borriello A, Bencivenga D, Criscuolo M,
Caldarelli I, Cucciolla V, Tramontano A, Borgia A, Spina A, Oliva
A, Naviglio S and Della Ragione F: Targeting p27Kip1 protein: Its
relevance in the therapy of human cancer. Expert Opin Ther Targets.
15:677–693. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wander SA, Zhao D and Slingerland JM: p27:
A barometer of signaling deregulation and potential predictor of
response to targeted therapies. Clin Cancer Res. 17:12–18. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sharma SS and Pledger WJ: The
non-canonical functions of p27(Kip1) in normal and tumor biology.
Cell Cycle. 15:1189–1201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fernandez S, Risolino M and Verde P: A
novel miRNA-mediated STOP sign in lung cancer: miR-340 inhibits the
proliferation of lung cancer cells through p27(KIP1). Mol Cell
Oncol. 2:e9771472015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang C, Ba X, Guo Y, Sun D, Jiang H, Li W,
Huang Z, Zhou G, Wu S, Zhang J and Chen J: MicroRNA-199a-5p
promotes tumour growth by dual-targeting PIAS3 and p27 in human
osteosarcoma. Sci Rep. 7:414562017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Zeng J, Pan J, Geng X, Liu Y, Wu
J, Song P, Wang Y, Jia J and Wang L: MicroRNA-200c is involved in
proliferation of gastric cancer by directly repressing p27(Kip1).
Biochem Biophys Rep. 8:227–233. 2016.PubMed/NCBI
|
|
37
|
Yang YF, Wang F, Xiao JJ, Song Y, Zhao YY,
Cao Y, Bei YH and Yang CQ: MiR-222 overexpression promotes
proliferation of human hepatocellular carcinoma HepG2 cells by
downregulating p27. Int J Clin Exp Med. 7:893–902. 2014.PubMed/NCBI
|
|
38
|
Sarkar S, Dubaybo H, Ali S, Goncalves P,
Kollepara SL, Sethi S, Philip PA and Li Y: Down-regulation of
miR-221 inhibits proliferation of pancreatic cancer cells through
up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer
Res. 3:465–477. 2013.PubMed/NCBI
|
|
39
|
Zhang T, Tan P, Wang L, Jin N, Li Y, Zhang
L, Yang H, Hu Z, Zhang L, Hu C, et al: RNALocate: A resource for
RNA subcellular localizations. Nucleic Acids Res. 45:D135–D138.
2017.PubMed/NCBI
|
|
40
|
Cui T, Zhang L, Huang Y, Yi Y, Tan P, Zhao
Y, Hu Y, Xu L, Li E and Wang D: MNDR v2.0: An updated resource of
ncRNA-disease associations in mammals. Nucleic Acids Res.
46:D371–D374. 2018.PubMed/NCBI
|
|
41
|
Li Y, Wang C, Miao Z, Bi X, Wu D, Jin N,
Wang L, Wu H, Qian K, Li C, et al: ViRBase: A resource for
virus-host ncRNA-associated interactions. Nucleic Acids Res.
43:D578–D582. 2015. View Article : Google Scholar : PubMed/NCBI
|