Introduction
Thyroid cancer (TC) is one of the most common types
of endocrine tumour with a rapidly increasing morbidity rate in the
last several decades (1). Based on
its histology, TC can be divided into four subtypes, including
papillary, follicular, medullary and anaplastic TCs (2). Papillary TC is the most common
subtype of thyroid cancer and accounts for ~85–90% of all TC cases
(3). Surgical resection together
with radioiodine and thyroid-stimulating hormone inhibition therapy
remains as the main treatment for patients with TC (4). The majority of TC patients exhibit
satisfactory treatment outcomes; however, the prognosis of TC
patients with local recurrence or distant metastasis is markedly
poor (5,6). Unfortunately, the mechanisms
underlying the relapse and metastasis of TC remain largely unknown.
Therefore, understanding of the mechanisms of the initiation and
progression of TC may aid the development of effective therapeutic
strategies to improve the prognosis of TC patients at an advanced
stage.
MicroRNAs (miRNAs/miRs) are an abundant class of
18–25 nucleotide-long, single-stranded RNA molecules that serve a
critical role in tumourigenesis and tumour development (7). These molecules can modulate gene
expression by promoting mRNA cleavage and/or inducing translational
suppression by completely or partially complementary binding to the
3′-untranslated regions (3′-UTRs) of their target genes (8). At present, >2,000 miRNAs have been
identified in the human genome, and these miRNAs have been proposed
to regulate the expression of >30% of all protein-coding genes
(9). An increasing number of
studies have reported that the expression of miRNAs is altered in
almost all types of human malignancy, such as TC (10), hepatocellular carcinoma (11), breast cancer (12) and gastric cancer (13). The dysregulation of miRNAs is
involved in the oncogenesis of TC, and regulates numerous
pathological behaviours, such as cellular growth, cycle, apoptosis,
metastasis and epithelial-mesenchymal transition (14–16).
Considering their important roles in TC, miRNAs may be considered
as potential novel diagnostic and therapeutic biomarkers for
treating patients with this disease.
miR-873-5p (miR-873) is dysregulated and serves
crucial roles in the pathogenesis and progression of various human
cancer types (17–20); however, the expression, roles and
molecular mechanism of miR-873 in TC remain unknown. Therefore, the
present study aimed to determine the expression profile of miR-873
in TC tissues and cell lines, examine the roles of miR-873 in TC
progression and determine the mechanism underlying the action of
miR-873 in TC cells. The present study revealed that miR-873 is
downregulated in TC, and inhibits TC cell proliferation and
invasion by directly targeting zinc finger E-box-binding homeobox 1
(ZEB1), suggesting that this miRNA may be a potential therapeutic
agent for the treatment of patients with this malignancy.
Materials and methods
Patients and tissue samples
In total, 35 pairs of tissue samples, each
comprising TC and adjacent normal tissues, were obtained from
patients (16 males, 19 females; age range, 28–63 years) who
received surgery at the China-Japan Union Hospital of Jilin
University between May 2016 and April 2017. All 35 cases were
pathologically diagnosed as TC. The TC tissue samples were
classified according to the World Health Organization standards
(21). Of note, 24 of the 35 cases
were classified as low-grade (T1 and T2), and 11 cases were
classified as high-grade (T3 and T4); 9 of the 35 TC patients were
diagnosed with lymph node metastasis. After resection, all tissue
specimens were immediately frozen in liquid nitrogen and then
stored at −80°C. None of the TC patients had been treated with any
preoperative treatment, including chemotherapy, radiotherapy or
thyroid-stimulating hormone inhibition therapy before surgery. The
present study was approved by the medical ethics committee of the
China-Japan Union Hospital of Jilin University. In addition,
written consent was obtained from all participants.
Cell lines and culture conditions
One normal human thyroid cell line (HT-ori3) and two
human TC cell lines (TPC-1 and HTH83) were purchased from the
American Type Culture Collection (Manassas, VA, USA). All cell
lines were cultured in Dulbecco's Modified Eagle's medium (DMEM)
containing 10% fetal bovine serum (FBS) and 1%
penicillin/streptomycin mixture (all Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and grown at 37°C in a 5%
CO2 humidified incubator.
Transient transfection
miR-873 mimics and corresponding negative control
mimics (miR-NC) were purchased from Shanghai GenePharma, Co., Ltd.
(Shanghai, China). The miR-873 mimics sequence was
5′-GCAGGAACUUGUGAGUCUCCU-3′ and the miR-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. ZEB1-overexpressing plasmid
pcDNA3.1-ZEB1 and empty plasmid pcDNA3.1 were chemically produced
by Chinese Academy of Sciences (Changchun, China). These miRNA
mimics (100 pmol) and plasmids (4 µg) were transfected into cells
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols.
Cotransfection of miRNA mimics (100 pmol) and plasmid (2 µg) were
performed using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Reverse transcription-quantitative PCR
(RT-qPCR) analysis and MTT assay were performed at 48 h and 24 h
post-transfection. Cell invasion assay was performed 48 h after
transfection. Cells were collected 72 h after transfection to
perform western blot analysis.
RT-qPCR analysis
Total RNA of the tissue samples or cells was
isolated using TRIzol® (Thermo Fisher Scientific, Inc.)
in accordance with the manufacturer's protocol. For the detection
of miR-873 expression, total RNA was reverse transcribed into
complementary DNA (cDNA) using the TaqMan MicroRNA Reverse
Transcription Kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The expression
levels of miR-873 were determined using the TaqMan MicroRNA PCR kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: One cycle at 50°C for 2
min and at 95°C for 10 min, followed by 40 cycles of denaturation
at 95°C for 15 sec, and annealing/extension at 60°C for 60 sec. For
the quantification of ZEB1 mRNA expression levels, cDNA was
synthesized from total RNA using the PrimeScript RT Reagent Kit
(Takara Biotechnology Co., Ltd., Dalian, China). ZEB1 mRNA
expression was assessed by PCR amplification using the SYBR Premix
Ex Taq™ Kit (Takara Biotechnology Co., Ltd.). The thermocycling
conditions were as follows: Initial denaturation at 95°C for 5 min,
followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec. All
reaction was performed using the Applied Biosystems 7500 real-time
PCR system (Thermo Fisher Scientific, Inc.) and repeated three
times. U6 small nuclear RNA and GAPDH were used for the
normalization of miR-873 and ZEB1 mRNA expression, respectively.
Relative gene expression was analysed using the 2−ΔΔCq
method (22). The primers were
designed as follows: miR-873 forward, 5′-GCAGGAACUUGUGAGUCUCCU-3′
and reverse, 5′-AGGAGACUCACAAGUUCCUGC-3′; U6 forward,
5′-TTCACGAATTTGCGTGTCAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; ZEB1 forward,
5′-AAGTGGCGGTAGATGGTA-3′ and reverse, 5′-TTGTAGCGACTGGATTTT-3′; and
GAPDH forward, 5′-CCCCTTCATTGACCTCAACT-3′ and reverse,
5′-ATGAGTCCTTCCACGATACC-3′.
MTT assay
At 24 h following transfection, cells were seeded
into 96-well plates at a concentration of 3×103
cells/well. Analysis of cell proliferation was performed using an
MTT assay at four time points: 0, 24, 48 and 72 h after
inoculation. In total, 20 µl of MTT solution (5 mg/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added into each
well and incubated at 37°C under 5% CO2 for another 4 h.
The culture medium was discarded, and 100 µl of dimethyl sulfoxide
(Sigma-Aldrich; Merck KGaA) was added into each well to dissolve
the formazan precipitates. Finally, the absorbance was detected at
a wavelength of 490 nm using a microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA).
Cell invasion assay
Transwell chambers (8-µm pore size) coated with
Matrigel (BD Biosciences, Franklin Lakes, NY, USA) were used to
investigate cell invasion. Transfected cells were harvested,
suspended in FBS-free DMEM and seeded at a concentration of
1×105 cells/well into the upper chambers. The lower
chambers were coated with 500 µl of DMEM containing 20% FBS (Gibco;
Thermo Fisher Scientific, Inc.) to serve as a chemo-attractant. The
chambers were incubated at 37°C in a 5% CO2 humidified
incubator for 24 h. At the end of the incubation, non-invaded cells
were gently removed using a cotton swab. The invaded cells were
fixed with 100% methanol at room temperature for 20 min and stained
with 0.1% crystal violet at room temperature for 20 min (both from
Beyotime Institute of Biotechnology, Haimen, China). The invasive
ability of cells was determined by counting the number of invaded
cells under an inverted light microscope (IX83; magnification,
×200; Olympus Corporation, Tokyo, Japan) in five independent visual
fields each chamber.
Bioinformatics analysis
TargetScan (version 7.2; release, March 2018;
http://www.targetscan.org/vert_72/)
and microRNA (release, August 2010; http://www.microrna.org/microrna/home.do) were used to
predict the putative targets of miR-873.
Luciferase reporter assay
The wild-type (Wt) and mutant (Mut) 3′-UTRs of ZEB1
were chemically synthesised by Shanghai GenePharma Co., Ltd., and
cloned into the pmirGLO luciferase reporter vector (Promega
Corporation, Madison, WI, USA), to form the pmirGLO-ZEB1-3′-UTR Wt
and pmirGLO-ZEB1-3′-UTR Mut vectors, respectively. Cells were
inoculated into 24-well plates with an initial density of
1×105 cells/well, and the cells were co-transfected with
pmirGLO-ZEB1-3′-UTR Wt or pmirGLO-ZEB1-3′-UTR Mut, and miR-NC or
miR-873 using Lipofectamine 2000, in accordance with the
manufacturer's instructions. After 48 h post-transfection, cells
were assayed using the Dual-Luciferase® Reporter Assay
System (Promega Corporation). Firefly luciferase activity was
normalized to that of Renilla.
Protein extraction and western blot
analysis
Cells or tissue samples were lysed in
radioimmunoprecipitation assay lysis and extraction buffer (Thermo
Fisher Scientific, Inc.) for 30 min on ice to obtain protein. The
concentration of total protein was determined using a BCA Protein
Assay Kit (Beyotime Institute of Biotechnology). Equal amounts of
protein (30 µg) was separated by 10% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA) followed by blocking in 5% nonfat milk diluted at room
temperature for 2 h in Tris-buffered saline containing 0.1%
Tween-20 (TBST). Afterwards, the membranes were incubated overnight
at 4°C with primary antibodies against ZEB1 (ab181451; 1:1,000
dilution, Abcam, Cambridge, UK) or GAPDH (ab110305; 1:1,000
dilution, Abcam). On the following day, the membranes were washed
with TBST and probed with goat anti-mouse
horseradish-peroxidase-conjugated secondary antibody (ab6789;
1:5,000 dilution, Abcam) at room temperature for 2 h. The protein
signals were visualized using Pierce™ ECL Western Blotting
Substrate (Pierce; Thermo Fisher Scientific, Inc.). GAPDH was used
as an internal control. Quantity One software (version 4.62;
Bio-Rad Laboratories, Inc.) was used for densitometry.
Statistical analysis
Each assay was repeated at least three times. All
data were analyzed using SPPS version 19.0 (IBM Corp., Armonk, NY,
USA). The expression of miR-873 and ZEB1 mRNA in TC was presented
as box plots, and a Mann-Whitney U test was used to analyze these
data. The top of the box indicated the upper quartile, while the
bottom presented the lower quartile. The central line in the box
indicated the median. In addition, the whiskers indicated the
minimum and maximum data. All other data were expressed as the mean
± standard deviation, and differences between groups were evaluated
using a two-tailed Student's t-test or one-way ANOVA. A Tukey's
test was applied as the post hoc test following ANOVA. Spearman's
correlation analysis was conducted to determine the association
between miR-873 and ZEB1 mRNA expression in TC tissues. P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-873 is downregulated in human TC
tissues and cell lines
RT-qPCR analysis was performed to detect miR-873
expression in 35 pairs of TC and adjacent normal tissues. The
results of RT-qPCR analysis showed that miR-873 was downregulated
in TC tissues compared with in adjacent normal tissues (P<0.05;
Fig. 1A). To further validate this
observation, the expression of miR-873 was investigated in a normal
human thyroid cell line (HT-ori3) and two human TC cell lines
(TPC-1, and HTH83). Consistently, the expression levels of miR-873
were significantly decreased in the TC cell lines than in HT-ori3
cells (P<0.05; Fig. 1B). These
findings suggested that miR-873 was downregulated in TC tissues and
cell lines.
miR-873 inhibits the proliferation and
invasion of TC cells
To determine the roles of miR-873 in TC, TPC-1 and
HTH83 cells were transfected with miR-873 mimics or miR-NC. miR-873
expression was significantly upregulated in TPC-1 and HTH83 cells
following transfection with miR-873 mimics, as confirmed by RT-qPCR
analysis (P<0.05; Fig. 2A). To
examine the effects of miR-873 on cellular proliferation, MTT
assays were conducted to detect the proliferation of TPC-1 and
HTH83 cells after overexpressing miR-873. Cell proliferation was
significantly reduced in miR-873 mimics-transfected TPC-1 and HTH83
cells at 48 and 72 h relative to that of cells transfected with
miR-NC (P<0.05; Fig. 2B).
Additionally, the invasive abilities of TPC-1 and HTH83 cells
transfected with miR-873 mimics or miR-NC were determined via a
cell invasion assay. The results demonstrated that miR-873
overexpression significantly reduced the invasion of TPC-1 and
HTH83 cells compared with the control group (P<0.05; Fig. 2C). Collectively, the results
indicated that miR-873 may serve inhibitory roles in the
progression of TC.
miR-873 decreases ZEB1 expression in
TC cells by binding to its 3′-UTR
To understand the mechanisms by which miR-873
inhibited TC cell proliferation and invasion in the present study,
bioinformatics analysis was conducted to predict the candidate
targets of miR-873. ZEB1 was predicted as a major putative target
of miR-873 (Fig. 3A) and was
closely associated with the formation and progression of TC
(23). Therefore, ZEB1 was
selected for further experimental confirmation. To clarify whether
miR-873 could bind to the 3′-UTR of ZEB1, luciferase reporter
plasmids were constructed and transfected into TPC-1 and HTH83
cells, together with miR-873 mimics or miR-NC. The results of the
luciferase reporter assay indicated that upregulation of miR-873
significantly decreased the luciferase activities of the plasmid
carrying Wt ZEB1 3′-UTR in TPC-1 and HTH83 cells compared with the
corresponding miR-NC group (P<0.05); however, the luciferase
activity of the plasmid harbouring Mut ZEB1 3′-UTR was markedly
unaffected (Fig. 3B). To further
investigate the regulatory effects of miR-873 on endogenous ZEB1
expression in TC cells, TPC-1 and HTH83 cells were transfected with
miR-873 mimics or miR-NC. The expression of ZEB1 mRNA and protein
levels were then determined. RT-qPCR and western blot analysis
showed that miR-873 upregulation significantly suppressed ZEB1
expression in TPC-1 and HTH83 cells at the mRNA (P<0.05;
Fig. 3C) and protein (P<0.05;
Fig. 3D) levels compared with the
control. Collectively, these results suggested that ZEB1 may be a
direct target gene of miR-873 in TC cells.
ZEB1 exhibits an inverse correlation
with miR-873 expression in TC tissues
To further investigate the correlation between
miR-873 and ZEB1 in TC, we detected ZEB1 expression in 35 pairs of
TC and adjacent normal tissues. Our results showed that the
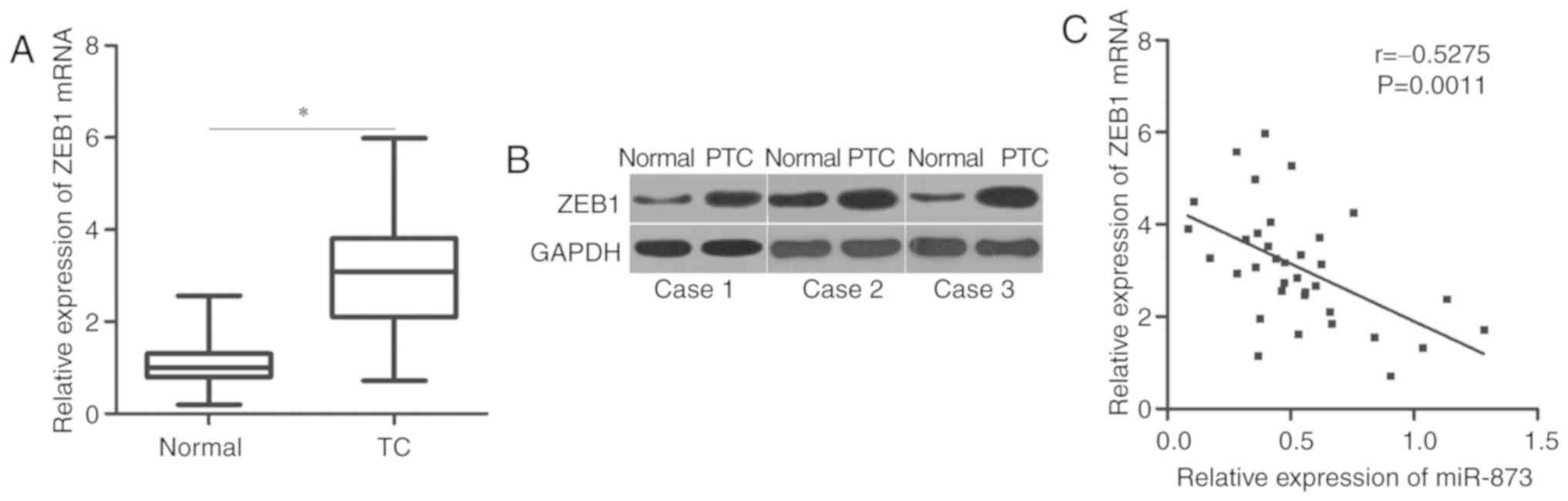
expression of ZEB1 was upregulated in TC tissues compared with in
adjacent normal tissues at the mRNA (P<0.05; Fig. 4A) and protein levels (Fig. 4B). Then, the correlation between
miR-873 and ZEB1 mRNA levels was evaluated using Spearman's
correlation analysis. An inverse correlation between miR-873 and
ZEB1 mRNA levels was observed in TC tissues (r=−0.5275, P=0.0011l;
Fig. 4C).
Restoration of ZEB1 expression rescues
the inhibitory effects of miR-873 on cell proliferation and
invasion in TC
To investigate whether the inhibitory effects of
miR-873 on TC cell proliferation and invasion were mediated by
ZEB1, we restored ZEB1 expression in miR-873-overexpressing TPC-1
and HTH83 cells via co-transfection with ZEB1 overexpression
plasmid pcDNA3.1-ZEB1 (P<0.05; Fig.
5A). Functional assays revealed that restoration of ZEB1
expression could rescue the suppressive effects of miR-873
overexpression on the proliferation (P<0.05; Fig. 5B) and invasion (P<0.05; Fig. 5C) of TPC-1 and HTH83 cells. These
results suggested that miR-873 may exert its tumour-suppressing
roles in TC at least partly by suppressing ZEB1.
Discussion
Previous studies have reported that certain miRNAs
are dysregulated in TC, which is linked with the pathogenesis and
development of TC (24–26). Therefore, improved understanding of
the detailed roles of miRNAs that are aberrantly expressed in TC
may aid the development of valuable therapeutic methods for the
management of patients with this malignancy. The present study
demonstrated that miR-873 expression was reduced in both TC tissues
and cell lines. Ectopic miR-873 expression inhibited cell
proliferation and invasion in TC. In addition, ZEB1 was identified
as a direct target gene of miR-873 in TC. Furthermore, ZEB1 was
upregulated in TC tissues, and negatively correlated with the
expression of miR-873. Additionally, the resumption of ZEB1
expression rescued TC cells from the tumour-suppressive effects of
miR-873 on cell proliferation and invasion. These results suggested
that miR-873 may serve as a potential therapeutic target for
treating patients with TC.
miR-873 was reported to be aberrantly expressed in
several types of human cancer. For example, this miRNA was
downregulated in colorectal cancer tissues and cell lines (17). Decreased miR-873 expression has
been associated with the tumour stage of patients with colorectal
cancer. In addition, patients with this disease and low miR-873
expression levels exhibited reduced overall survival rates than
those with high expression levels (17). miR-873 was determined to be
downregulated in gastric cancer tissues and cell lines (19,20).
Reduced miR-873 expression strongly correlated with the levels of
serum α fetal protein, tumour size and, tumour-node-metastasis
stage of patients with gastric cancer (20). miR-873 downregulation was also
reported in breast cancer (18),
glioblastoma (27) and ovarian
cancer (28); however, miR-873 was
upregulated in lung adenocarcinoma tissues and cell lines (29). These conflicting findings indicate
the potential tissue specificity of miR-873 expression in human
malignancies, and that miR-873 may serve as a biomarker for the
diagnosis and prognosis of these human specific cancer types.
miR-873 exhibits tumour-suppressive effects in
numerous types of human cancer. For instance, miR-873 suppresses
the proliferation of colorectal cancer cells in vitro
(17). Cui et al (18) reported that miR-873 overexpression
inhibits breast cancer cell proliferation in vitro and
tumour growth in vivo. Cao et al (19) and Chen et al (20) revealed that resumption of miR-873
expression suppresses cell viability, promotes cell apoptosis and
induces cell cycle arrest in gastric cancer. Wang et al
(27) revealed that miR-873
overexpression attenuates cell growth and metastasis of
glioblastoma in vitro. Additionally, Wu et al
(28) found that miR-873
overexpression increases the chemosensitivity of ovarian cancer
cells to cisplatin and paclitaxel by enhancing the inhibitory
effects of these drugs on tumour growth. Nevertheless, miR-873
serves oncogenic roles in lung adenocarcinoma by affecting cell
proliferation and migration in vitro (29). These findings demonstrated that
miR-873 exhibits tissue-specific effects on the biological roles
involved in tumourigenesis. These contradictory results may be
explained by the imperfect complementarity of the interactions
between miRNAs and target genes (30). These findings also suggested that
miR-873 may be considered as a potential therapeutic target for the
management of these specific types of tumour.
Direct targets of miR-873 have been previously
validated, including tumour necrosis factor receptor-associated
factor 5 (17) and transforming
growth factor-β activated kinase 1 (17) in colorectal cancer,
cyclin-dependent kinase 3 (18) in
breast cancer, glioma-associated oncogene (19) and C-X-C motif ligand 1 (20) in gastric cancer, insulin-like
growth factor 2 mRNA-binding protein 1 in glioblastoma (27), ATP-binding cassette sub-family B
member 1 (28) in ovarian cancer
and SRC kinase signaling inhibitor 1 (29) in lung cancer. ZEB1, located on the
short arm of human chromosome 10 (31), has been reported as a direct and
functional target of miR-873 in TC. It is a member of the zinc
finger family and is highly expressed in various human cancers,
such as cervical cancer (32),
lung cancer (33), hepatocellular
carcinoma (34), gastric cancer
(35) and glioma (36). ZEB1 is also upregulated in TC and
was strongly associated with tumour-node-metastasis stage, lymph
node metastasis and distant metastasis (23). ZEB1 inhibition suppressed the
proliferation, migration and invasion of TC cells (23). In the present study, we revealed
that miR-873 may inhibit TC progression by downregulating ZEB1
expression. Thus, the inhibition of ZEB1 via miR-873 may be an
effective therapeutic strategy to treat patients with TC.
Overall, miR-873 was downregulated in TC tissues and
cell lines. In addition, overexpression of miR-873 may have
attenuated TC cell proliferation and invasion by directly targeting
ZEB1. Our experimental results suggested that this miRNA may be a
promising therapeutic target for the treatment of TC patients;
however, the association between miR-873, ZEB1 and the clinical
characteristics of patients with TC requires further investigation.
In addition, experiments, including colony formation and wound
healing assays, and in vivo xenograft experiments should be
performed to explore the roles of miR-873 in the progression of TC.
These are limitations of our study, which we aim to resolve in
future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
QF and DJ made substantial contributions to the
design of this research. DJ, FG and QF performed the functional
assays. All authors read and approved the final draft.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of China-Japan Union Hospital of Jilin University, and
was performed in accordance with the Declaration of Helsinki and
the guidelines of the Ethics Committee of China-Japan Union
Hospital of Jilin University. Written informed consent was obtained
from all patients for the use of their clinical tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liebner DA and Shah MH: Thyroid cancer:
Pathogenesis and targeted therapy. Ther Adv Endocrinol Metab.
2:173–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lloyd RV, Buehler D and Khanafshar E:
Papillary thyroid carcinoma variants. Head Neck Pathol. 5:51–56.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Das S, Chaudhary N, Ang LC and Megyesi JS:
Papillary thyroid carcinoma metastasizing to anaplastic meningioma:
An unusual case of tumor-to-tumor metastasis. Brain Tumor Pathol.
34:130–134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nikiforov YE and Nikiforova MN: Molecular
genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol.
7:569–580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu S, Semenciw R, Ugnat AM and Mao Y:
Increasing thyroid cancer incidence in Canada, 1970–1996: Time
trends and age-period-cohort effects. Br J Cancer. 85:1335–1339.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pemayun TG: Current diagnosis and
management of thyroid nodules. Acta Med Indones. 48:247–257.
2016.PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jia L, Zhang D, Qi X, Ma B, Xiang Z and He
N: Identification of the conserved and novel miRNAs in Mulberry by
high-throughput sequencing. PLoS One. 9:e1044092014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xie B, Ding Q, Han H and Wu D: miRCancer:
A microRNA-cancer association database constructed by text mining
on literature. Bioinformatics. 29:638–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mutalib NS, Yusof AM, Mokhtar NM, Harun R,
Muhammad R and Jamal R: MicroRNAs and lymph node metastasis in
papillary thyroid cancers. Asian Pac J Cancer Prev. 17:25–35. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu J, Li J, Zheng TH, Bai L and Liu ZJ:
MicroRNAs in the occurrence and development of primary
hepatocellular carcinoma. Adv Clin Exp Med. 25:971–975. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hamam R, Hamam D, Alsaleh KA, Kassem M,
Zaher W, Alfayez M, Aldahmash A and Alajez NM: Circulating
microRNAs in breast cancer: Novel diagnostic and prognostic
biomarkers. Cell Death Dis. 8:e30452017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Irmak-Yazicioglu MB: Mechanisms of
MicroRNA deregulation and MicroRNA targets in gastric cancer. Oncol
Res Treat. 39:136–139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang M, Wu W, Gao M and Fei Z:
MicroRNA-451 as a prognostic marker for diagnosis and lymph node
metastasis of papillary thyroid carcinoma. Cancer Biomark.
19:437–445. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu J, Li Q, Li R, Ren P and Dong S:
MicroRNA-363-3p inhibits papillary thyroid carcinoma progression by
targeting PIK3CA. Am J Cancer Res. 7:148–158. 2017.PubMed/NCBI
|
|
16
|
Zhao H, Tang H, Huang Q, Qiu B, Liu X, Fan
D, Gong L, Guo H, Chen C, Lei S, et al: MiR-101 targets USP22 to
inhibit the tumorigenesis of papillary thyroid carcinoma. Am J
Cancer Res. 6:2575–2586. 2016.PubMed/NCBI
|
|
17
|
Gong H, Fang L, Li Y, Du J, Zhou B, Wang
X, Zhou H, Gao L, Wang K and Zhang J: miR873 inhibits colorectal
cancer cell proliferation by targeting TRAF5 and TAB1. Oncol Rep.
39:1090–1098. 2018.PubMed/NCBI
|
|
18
|
Cui J, Yang Y, Li H, Leng Y, Qian K, Huang
Q, Zhang C, Lu Z, Chen J, Sun T, et al: MiR-873 regulates ERα
transcriptional activity and tamoxifen resistance via targeting
CDK3 in breast cancer cells. Oncogene. 34:40182015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao D, Yu T and Ou X: MiR-873-5P controls
gastric cancer progression by targeting hedgehog-GLI signaling.
Pharmazie. 71:603–606. 2016.PubMed/NCBI
|
|
20
|
Chen XY, Chen RP, Wu W and Huang ZM:
MicroRNA-873 inhibits proliferation and induces apoptosis by
targeting CXCL1 in gastric cancer. Int J Clin Exp Pathol.
9:10011–10019. 2016.
|
|
21
|
Liu ZY, Zhou GY, Kakudo K and Lam AK:
Update on 2017 Who classification of tumors of thyroid gland.
Zhonghua Bing Li Xue Za Zhi. 47:302–306. 2018.(In Chinese).
PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Liu G, Wu S, Jiang F, Xie J and
Wang Y: Zinc finger E-box-binding homeobox 1: Its clinical
significance and functional role in human thyroid cancer. Onco
Targets Ther. 9:1303–1310. 2016.PubMed/NCBI
|
|
24
|
Wang Z, Zhang H, He L, Dong W, Li J, Shan
Z and Teng W: Association between the expression of four
upregulated miRNAs and extrathyroidal invasion in papillary thyroid
carcinoma. Onco Targets Ther. 6:281–287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Graham ME, Hart RD, Douglas S, Makki FM,
Pinto D, Butler AL, Bullock M, Rigby MH, Trites JR, Taylor SM and
Singh R: Serum microRNA profiling to distinguish papillary thyroid
cancer from benign thyroid masses. J Otolaryngol Head Neck Surg.
44:332015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee JC, Zhao JT, Clifton-Bligh RJ, Gill A,
Gundara JS, Ip JC, Glover A, Sywak MS, Delbridge LW, Robinson BG
and Sidhu SB: MicroRNA-222 and microRNA-146b are tissue and
circulating biomarkers of recurrent papillary thyroid cancer.
Cancer. 119:4358–4365. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang RJ, Li JW, Bao BH, Wu HC, Du ZH, Su
JL, Zhang MH and Liang HQ: MicroRNA-873 (miRNA-873) inhibits
glioblastoma tumorigenesis and metastasis by suppressing the
expression of IGF2BP1. J Biol Chem. 290:8938–8948. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu DD, Li XS, Meng XN, Yan J and Zong ZH:
MicroRNA-873 mediates multidrug resistance in ovarian cancer cells
by targeting ABCB1. Tumour Biol. 37:10499–10506. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao Y, Xue Q, Wang D, Du M, Zhang Y and
Gao S: miR-873 induces lung adenocarcinoma cell proliferation and
migration by targeting SRCIN1. Am J Transl Res. 7:2519–2526.
2015.PubMed/NCBI
|
|
30
|
Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y,
Yu W, Wu X, Ye J, Yang S, et al: Identification of miR-7 as an
oncogene in renal cell carcinoma. J Mol Histol. 44:669–677. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Xu L, Li A and Han X: The roles
of ZEB1 in tumorigenic progression and epigenetic modifications.
Biomed Pharmacother. 110:400–408. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma Y, Zheng X, Zhou J, Zhang Y and Chen K:
ZEB1 promotes the progression and metastasis of cervical squamous
cell carcinoma via the promotion of epithelial-mesenchymal
transition. Int J Clin Exp Pathol. 8:11258–11267. 2015.PubMed/NCBI
|
|
33
|
Larsen JE, Nathan V, Osborne JK, Farrow
RK, Deb D, Sullivan JP, Dospoy PD, Augustyn A, Hight SK, Sato M, et
al: ZEB1 drives epithelial-to-mesenchymal transition in lung
cancer. J Clin Invest. 126:3219–3235. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou YM, Cao L, Li B, Zhang RX, Sui CJ,
Yin ZF and Yang JM: Clinicopathological significance of ZEB1
protein in patients with hepatocellular carcinoma. Ann Surg Oncol.
19:1700–1706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jia B, Liu H, Kong Q and Li B:
Overexpression of ZEB1 associated with metastasis and invasion in
patients with gastric carcinoma. Mol Cell Biochem. 366:223–229.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo E, Wang Z and Wang S: MiR-200c and
miR-141 inhibit ZEB1 synergistically and suppress glioma cell
growth and migration. Eur Rev Med Pharmacol Sci. 20:3385–3391.
2016.PubMed/NCBI
|