Introduction
Glucocorticoids (GCs) are the most common risk
factors in glucocorticoid-induced osteoporosis (GIOP) (1). In total, 30–50% of patients treated
with GCs suffer osteoporotic fractures, and GIOP primarily affects
adults between the ages of 20 and 45 years (2). GCs are commonly used for the
treatment of rheumatoid arthritis in the UK (3). Several studies reported the
relationship between GCs and GIOP; however, the molecular mechanism
underlying this association remains unclear (1,4,5). GCs
may have an important role in bone metabolism (6). However, bone metabolism involves a
number of endogenous and exogenous factors. In total, three cell
types, osteoblasts, osteoclasts and osteocytes, regulate the bone
remodeling process (3). GCs,
including dexamethasone and methylprednisolone, regulate bone
remodeling by affecting the osteogenic process or by inhibiting the
proliferation of these three cell types (3). Furthermore, GCs can promote
osteoclast survival, decreasing bone strength (7). Jia et al (4) reported that the number of osteoblasts
decreased significantly following treatment with GCs in cell
cultures, and osteogenic differentiation was inhibited by GCs.
Moreover, osteogenic markers, including bone morphogenetic protein
2 (BMP2), runt related transcription factor 2 (RUNX2) and
osteoprotegerin (OPG) were also downregulated compared with the
control group (4). O'Brien et
al (5) demonstrated that GCs
directly affected the survival of osteocytes, promoting apoptosis
of these cells.
Circular RNAs (circRNAs) are a novel type of
non-coding RNAs transcribed from exons that exhibit cell- or
tissue-specific expression (8,9). As
previously reported, due to their resistance to RNase, the
expression levels of circRNAs in tissues are highly stable
(8). Previous studies have
demonstrated the importance of circRNAs in the control of cellular
functions by sponging microRNAs (miRNAs), thus regulating gene
expression, or by interacting with RNA-binding proteins (8,9). Li
and Li (10) demonstrated
that hsa_circ_0007534 regulates cell growth and apoptosis by
affecting the AKT/glycogen synthase kinase 3β signaling pathway in
osteosarcoma. Song and Xiao (11) showed that the downregulation of
hsa_circ_0007534 suppresses breast cancer cell proliferation and
invasion by targeting the miRNA (miR)-593/mucin 19, oligomeric
signaling pathway. However, to the best of our knowledge, no
previous studies have explored the role of circRNAs in GIOP, and
the role of circRNAs in the regulation of bone metabolism remains
unclear.
hsa_circ_0006393 is a circRNA that was identified in
a previous RNA-seq study (12). To
the best of our knowledge, the present study is the first to
investigate the function of hsa_circ_0006393. In the present study,
miR-145-5p was identified to be a target of hsa_circ_0006393, and
miR-145-5p inhibited the expression level of forkhead box O1
(FOXO1), thus decreasing bone mass and osteogenesis.
Materials and methods
Animals
The animal experiments were approved by The Ethics
Committee of Gansu Provincial Hospital (Lanzhou, China). All animal
procedures were performed according to the guidelines of The Animal
Care Committee of The Gansu Provincial Hospital. Animals were
maintained in an environment at 26±2°C, 40–60% relative humidity,
with a 12 h at light/dark cycle and are free access to food and
water. In total, 18 female C57BL/6J mice supplied by animal center
of the Gansu Provincial Hospital (age, 8 weeks; weight, 20 ± 3g)
were randomly divided into three groups: i) Control group; ii)
dexamethasone (Dex; 5 mg/kg/day intramuscular injection) treated
group; and iii) Dex + lentiviral vector-circRNA (Guangzhou RiboBio
Co., Ltd.) group. The vector-circRNA was diluted in PBS at a
concentration of 5 nM, and 5 µl of vector-circRNA was injected into
the medullary cavity of the bilateral distal femur every week.
Tissue isolation and cell culture
From June 2017 to January 2018, six bone tissue
samples from male patients with GIOP (aged 62–71) and six bone
tissue samples from male patients (aged 43–55 years) with traumatic
fractures without GIOP were collected in the Orthopedics Department
of Gansu Provincial Hospital. Only male patients were included in
the present study. Patients with complications that affect bone
metabolism, including nephropathy, liver disease and hematopathy,
were excluded. All experimental and sampling procedures were
approved by The Human and Animal Research Ethics Committee of Gansu
Provincial Hospital. Informed consent was obtained from all
patients. Bone mesenchymal stem cells (BMSCs) were isolated from
human bone marrow as previously described (13). BMSCs were incubated in α-minimal
essential media (α-MEM; HyClone; GE Healthcare Life Sciences)
supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher
Scientific, Inc.) and 100 µg/ml penicillin/streptomycin for 3 days
at 37°C with 5% CO2. The culture medium was changed
every other day.
RNA extraction, reverse
transcription-quantitative (RT-q) PCR and RNase R assay
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from 1 g
bone tissues or from a total of 106 cells. For RNase R
treatment, 2 µg total RNA extracted from BMSCs were incubated for
15 min at room temperature with or without 3 U/mg RNase R. The
PrimeScript RT reagent kit (Takara Bio, Inc.) was used to
synthesize the cDNA. The conditions for the reverse transcription
reaction were as follows: 37°C for 15 min, 85°C for 5 sec, and
storage at 4°C. qPCR analysis was performed using the SYBR Premix
Ex Taq II kit (Takara Bio, Inc.) and detected using the Roche
LightCycler 480 (Roche Diagnostics) sequence detection system. The
thermocycling conditions were as follows: 50°C for 2 min and 95°C
for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for
10 min. GAPDH is the reference gene for mRNA and circRNA and U6 is
the reference gene for miRNA. Relative gene expression was
calculated using the 2−ΔΔCq method (14). The primer sequences are listed in
Table I.
 | Table I.Reverse transcription-quantitative PCR
primer sequences. |
Table I.
Reverse transcription-quantitative PCR
primer sequences.
| Gene symbol | Sequence (5′→3′) |
|---|
| BMP2 | F:
TGTATCGCAGGCACTCAGGTCA |
|
| R:
CCACTCGTTTCTGGTAGTTCTTC |
| RUNX2 | F:
CCCAGTATGAGAGTAGGTGTCC |
|
| R:
GGGTAAGACTGGTCATAGGACC |
| OPG | F:
AGGTGGTTCACACGACAGCAGA |
|
| R:
GTGAATCACTGTTCCTTGGTGTG |
| Sp7 | F:
TTCTGCGGCAAGAGGTTCACTC |
|
| R:
GTGTTTGCTCAGGTGGTCGCTT |
| FOXO1 | F:
CTACGAGTGGATGGTCAAGAGC |
|
| R:
CCAGTTCCTTCATTCTGCACACG |
| HIF1a | F:
TATGAGCCAGAAGAACTTTTAGGC |
|
| R:
CACCTCTTTTGGCAAGCATCCTG |
|
hsa_circ_0006393 | F:
TCCATGTGACCATGAGGAAA |
|
| R:
GAGATCTGGCTGCATCTCG |
| miR-145-5p | F:
GGGGTCCAGTTTTCCCAGGA |
|
| R:
CAGTGCGTGTCGTGGAGT |
| U6 | F:
AGAGAAGATTAGCATGGCCCCTG |
|
| R:
ATCCAGTGCAGGGTCCGAGG |
| GAPDH | F:
GTCTCCTCTGACTTCAACAGCG |
|
| R:
ACCACCCTGTTGCTGTAGCCAA |
Vector construction and stable
transfection
The structure of the circRNA was analyzed using
circPrimer (version 1.2) (15).
The CircInteractome database (https://circinteractome.nia.nih.gov/) was used to
predict the binding site between circRNA and miRNA.
Lentivirus-small interfering (si)-circRNA0006393 and
lentivirus-overexpression-circRNA0006393 were purchased from
Shanghai GenePharma Co., Ltd. A total of 50 nM (0.33 µg/µl) siRNA
were transfected into BMSCs at 105 cells/ml, using
Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.).
The expression levels of the genes of interest were measured 48 h
after transfection. The sequence of si-circRNA0006393 was
5′-ACACACAGAAATGGATAAGTT-3′. The sequence of scramble siRNA was
5′-TACATCGGATTTGTAACGTAA-3′.
Western blot assay
Total protein from bone tissues and cells were
extracted using RIPA buffer as previously described (16). The protein concentration was
determined using bicinchoninic acid kit (Invitrogen; Thermo Fisher
Scientific). In total, 20 µl protein were loaded in each lane.
Protein bands were separated using 10% SDS-PAGE electrophoretically
transferred to PVDF membranes for 60 min at 200 mA. The PVDF
membranes were blocked in 5% skimmed milk (BDBiosciences) dissolved
in TBST at room temperature for 3 h. Subsequently, the PVDF
membranes were incubated at 37°C for 2 h with primary antibodies
anti-FOXO1 (1:1,000; Abcam; cat. no. ab207204), anti-RUNX2
(1:1,000; Abcam; cat. no. ab192256), BMP2 (1:1,000; Abcam; cat. no.
ab214821) and GAPDH (1:1,000; Abcam; cat. no. ab181602). The PVDF
membranes were washed three times for 30 min with TBST.
Subsequently, the membranes were incubated with a horseradish
peroxidase-labeled secondary antibody (1:5,000; ProteinTech Group,
Inc.; cat. no. SA00001-2) at room temperature for 1 h. The Amersham
Imager 600 (GE Healthcare) was used to observe the bands on the
PVDF membrane.
Dual luciferase reporter assay
The hsa_circ_0006393 was obtained from BMSCs, and
its PCR product, containing 300 bp of the 5′ sequence flanking
hsa_circ_0006393 promoter, was amplified using Premix Taq™ (Ex Taq™
Version 2.0, Takara Bio, Inc.) using the following thermocycling
conditions: A total of 30 cycles at 98°C for 10 sec, 55°C for 30
sec and 72°C for 1 min. The PCR product was inserted into the pGL3
vector (pGL3-circRNA Promoter, Shanghai GenePharma Co., Ltd.) for
Luciferase binding assays. Then, 200 ng pGL3-circRNA promoter with
40 ng pRL-TK Vector (cat. no. E2241; Promega Corporation) were
co-transfected into BMSCs at a density of 104 cells/ml
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.). The luciferase assays were performed 36 h after transfection
using a dual-luciferase reporter assay system (cat. no. E1910;
Promega Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity, and luminescence levels were
measures using the Sirius L LB 9525 Tube Luminometer (Berthold
Detection Systems GmbH).
To assess the binding between circRNA0006393 and
miR-145-5p, 50 nM miR-145-5p mimics or control (Guangzhou RiboBio
Co., Ltd.) were co-transfected with the luciferase reported
plasmids using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The sequence of miR-145-5p mimics is
5′-GUCCAGUUUUCCCAGGAAUCCCU-3′, while that of the miR-145-5p control
is 5′-AAUUGAAGUUCCCAGGAAUCCCU-3′. Subsequently, three databases,
including PicTar (https://pictar.mdc-berlin.de/), miRanda (http://miranda.org.uk/) and TargetScan (http://www.targetscan.org/vert_72/) were used to
predict the mRNA binding targets of miR-145-5p.
Fluorescence in situ hybridization
(FISH)
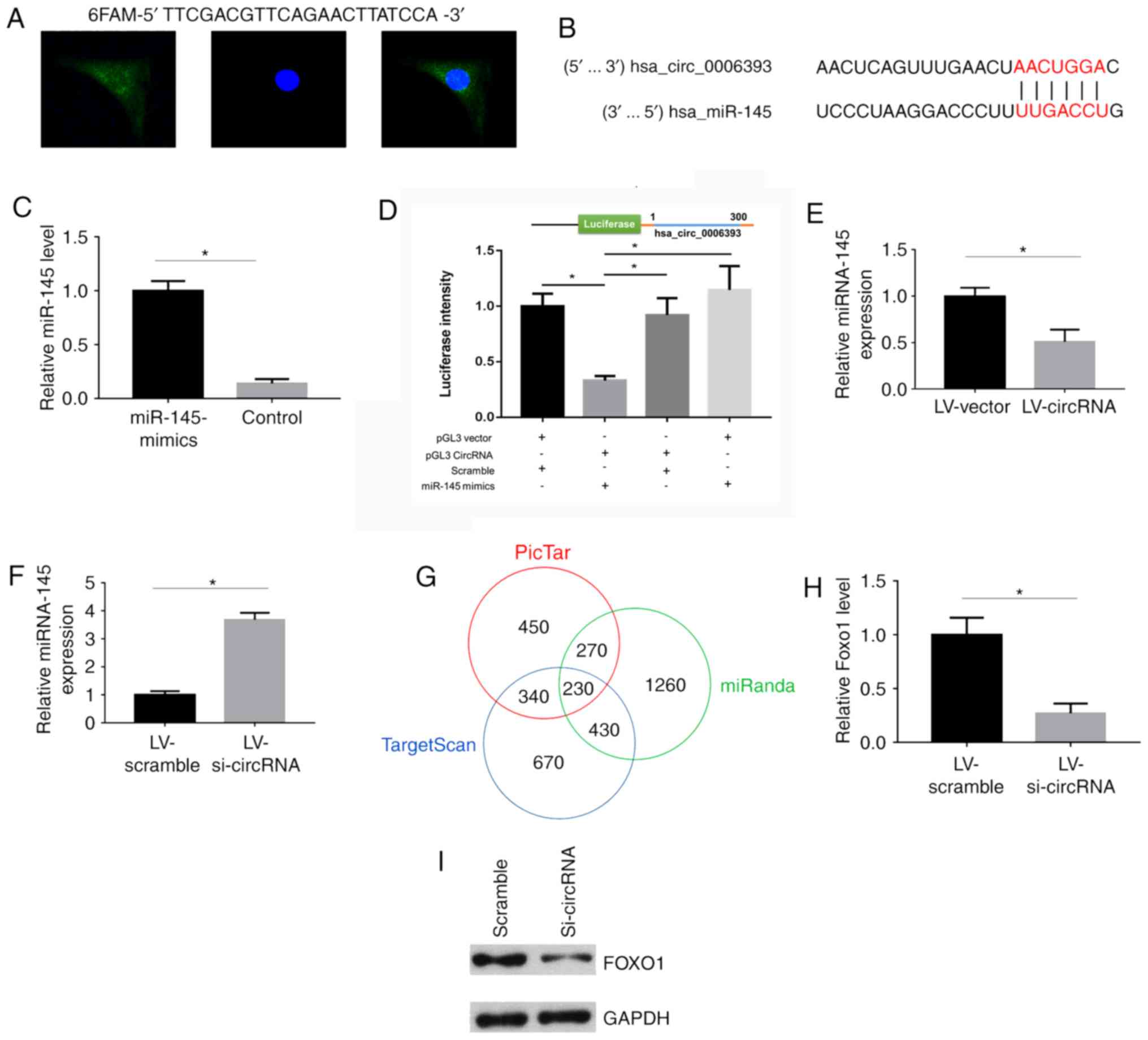
The probe 5′-TTCGACGTTCAGAACTTATCCA-3′ (Qiagen GmbH)
was labeled with 6-carboxyfluorescein and was designed using the
sequence of hsa_circ_0006393. Subsequently, in situ
hybridization was performed using the FISH detection kit (Qiagen
GmbH) according to the manufacturer's protocol. Cells were
visualized under a confocal fluorescence microscope.
Dual-energy X-ray absorptiometry (DXA)
analysis
The animals were analyzed with a Faxitron DXA
Imaging system (Faxitron Bioptics, LLC). Whole-body scans were
performed in animals using DXA. The structures were reconstructed
using the built-in analysis software of the Faxitron DXA imaging
system. The bone, lean and fat maps, and the DXA status were
calculated using the built-in software of the instrument.
Statistical analysis
All experiments were repeated three times. The data
are presented as the mean ± SD. Comparisons between two groups were
performed using Student's t-test. Multiple comparisons were
performed using one-way ANOVA followed by Fisher's least
significant difference post hoc test. Statistical analyses were
performed using SPSS 20.0 (IBM Corp.). P<0.05 was considered to
indicate a statistically significant difference.
Results
hsa_circ_0006393 characterization and
expression in patients with GIOP
The structure of hsa_circ_0006393 was analyzed in
order to examine the exons present in this circRNA. The present
results suggested that hsa_circ_0006393 was 422 nucleotides in
length and spanned three exons, including exon 2, 3 and 4, of the
hypoxia inducible factor 1α gene, on chromosome 14 (Fig. 1A). Next, the expression levels of
hsa_circ_0006393 in patients with GIOP and patients presenting
traumatic fractures were investigated. In addition, RT-qPCR was
performed to examine the relative expression levels of
hsa_circ_0006393, and the present results suggested that
hsa_circ_0006393 was downregulated in patients with GIOP (Fig. 1B). In addition, RNase R assay was
performed to examine the expression level of hsa_circ_0006393, and
the results suggested that hsa_circ_0006393 was resistant to
digestion with RNase R exonuclease (Fig. 1C).
hsa_circ_0006393 overexpression
increases the expression levels of osteogenic genes involved in
bone remodeling
The efficiency of hsa_circ_0006393 overexpression
was examined by RT-qPCR (Fig. 2A and
B). Next, the role of hsa_circ_0006393 in bone remodeling was
investigated. RT-qPCR was used to determine the expression levels
of RUNX2, OPG, BMP2 and Sp7 transcription factor in BMSCs
overexpressing hsa_circ_0006393. The present results suggested that
hsa_circ_0006393 overexpression increased the expression levels of
these four markers, suggesting that hsa_circ_0006393 may have an
important role in osteogenesis (Fig.
2C-F).
hsa_circ_0006393 acts as a sponge of
miR-145-5p, which targets FOXO1
FISH was performed to investigate the subcellular
location of hsa_circ_0006393. The present results suggested that
hsa_circ_0006393 was localized in the cytoplasm and nucleus of
BMSCs (Fig. 3A). Next, the
sponging ability of hsa_circ_0006393 was investigated, and
bioinformatics analysis was performed to investigate the miRNAs
that may be sponged by hsa_circ_0006393. According to the
CircInteractome database (https://circinteractome.nia.nih.gov/), miR-145-5p
showed one of the highest ‘context+ score percentile’ values. A
literature search also revealed that miR-145-5p was conserved
between species expressed in several tissues. Therefore, the role
of miR-145-5p was further studied in the present study. The
predicted binding sites between miR-145-5p and hsa_circ_0006393
were analyzed using CircInteractome (Fig. 3B). Furthermore, the transfection
efficiency of miR-145 mimics was examined by RT-qPCR (Fig. 3C). A dual luciferase reporter assay
was performed to examine the binding site between hsa_circ_0006393
and miR-145-5p. A vector containing 300 bp of the 5′ sequence
flanking hsa_circ_0006393 containing a luciferase gene was
transfected into BMSCs. The luciferase reporter assay confirmed the
interaction between hsa_circ_0006393 and miR-145-5p (Fig. 3D). The results also demonstrated
that miR-145-5p mimics decreased the luciferase intensity when
co-cultured with pGL3 circRNA vector. However, the luciferase
intensity of pGL3 vector+miR-145-5p group was higher compared with
the control transfections, but this was not statistically
significant. In addition, the RT-qPCR results suggested that
miR-145-5p expression was downregulated after transfection with
hsa_circ_0006393 overexpression vector (Fig. 3E) and that miR-145-5p expression
level was upregulated after the knockdown of has-circ_0006393
(Fig. 3F). Moreover, the
miR-145-5p targets were predicted using several miRNA databases.
Three databases, including PicTar, miRanda and TargetScan, were
used to predict the miR-145-5p target genes. In total, 230 genes
were predicted by all databases to exhibit potential miR-145-5p
binding sites (Fig. 3G). FOXO1 is
an important gene that regulates bone mass in humans (17) and was found to be downregulated
following knockdown of hsa_circ_0006393 in the present study
(Fig. 3H). In addition, western
blotting was performed to investigate the association between FOXO1
and hsa_circ_0006393, and the present results suggested that the
knockdown of hsa_circ_0006393 reduced the protein expression level
of FOXO1 (Fig. 3I).
Overexpression of hsa_circ_0006393
increases bone mass in an animal model of GIOP
A plasmid overexpressing hsa_circ_0006393 was
constructed and injected intramedullary into the femur of six mice.
After 4 weeks, bone mass was evaluated by DXA, and the results
suggested that hsa_circ_0006393 overexpression increased the bone
mineral density (BMD) compared with the Dex-treated group (Fig. 4A and B). The RNA was extracted from
femoral tissues and the effect of hsa_circ_0006393 overexpression
was investigated using RT-qPCR (Fig.
4C). The results indicated that Dex treatment decreased the
expression of hsa_circ_0006393, which was consistent with the
results of the clinical data. Moreover, hsa_circ_0006393
overexpression promoted the expression of this circRNA despite Dex
treatment. In addition, western blotting was performed to analyze
the protein expression levels of two osteogenic markers, and the
results suggested that overexpression of hsa_circ_0006393 increased
the protein expression levels of RUNX2 and BMP2 (Fig. 4D).
Discussion
GIOP is a type of secondary osteoporosis caused by
long-term or acute administration of glucocorticoids (5,18).
Bone fractures are the most common adverse event caused by GIOP
(19). An increasing number of
previous studies have demonstrated that miRNAs play an important
role in the regulation of GIOP (13,20).
However, the role of long non-coding RNAs (lncRNAs) and circRNAs in
GIOP pathogenesis remains poorly understood. A previous study
demonstrated the therapeutic potential of circRNAs in
glucocorticoid induced bone necrosis (12). Moreover, to the best of our
knowledge, no previous studies have demonstrated the association
between circRNAs and GIOP. Therefore, the present study is the
first to investigate the function of hsa_circ_0006393 in GIOP.
A previous study has reported that non-coding RNAs,
including miRNAs, lncRNAs and circRNAs, exhibit therapeutic effects
in human diseases (21). Numerous
recent studies investigated the role of circRNAs in various
cellular processes, such as cell apoptosis, stem cell
differentiation and cell migration (9,22,23).
Zhang et al (8)
investigated the functional role of circular La ribonucleoprotein
domain family member 4 (circLARP4) in gastric cancer, with
circLARP4 identified as a sponge for miR-424-5p, inhibiting gastric
cancer invasion and proliferation, and the downregulation of
miR-424-5p decreased the expression level of large tumor suppressor
kinase 1. In our previous study (12), circular ubiquitin specific
peptidase 45 (circUSP45) was identified as an important biomarker
in glucocorticoid-induced osteonecrosis of the femoral head, and
circUSP45 was found to be upregulated in glucocorticoid-induced
osteonecrotic femurs. In addition, circUSP45 is able to inhibit the
proliferation of BMSCs by sponging miR-127-5p (12). Song et al (24) demonstrated that hsa_circ_0000337
was upregulated in esophageal squamous cell carcinoma and regulated
cell proliferation, migration and invasion. These previous findings
demonstrated that circRNAs are important regulators of cellular
functions and can be used as biomarkers in human diseases.
However, to the best of our knowledge, no studies
have investigated the functional role of circRNAs in GIOP, and no
previous studies have examined the mechanism of hsa_circ_0006393 in
GIOP. In the present study, the expression of circRNA in patients
with GIOP was investigated using RT-qPCR. The present results
showed that hsa_circ_0006393 was decreased in patients with GIOP
compared with patients with traumatic fractures. Cellular
localization is considered an important indicator of the function
of circRNAs (23,25). The cellular localization of
hsa_circ_0006393 was examined using FISH, and the results suggested
that hsa_circ_0006393 is mainly localized in the cytoplasm and
nucleus of cells. Moreover, in the present study, hsa_circ_0006393
was found to promote osteogenesis in BMSCs. Collectively, the
present results identified that hsa_circ_0006393 may be used as a
biomarker in GIOP.
In addition, the mechanism underlying hsa_circ_
0006393-mediated regulation of bone metabolism was investigated in
the present study. Following FISH, the miRNA-sponging ability of
hsa_circ_0006393 was investigated. The present results suggested
that the hsa_circ_0006393/miR-145-5p/FOXO1 pathway might have an
important role in regulating bone metabolism in GIOP. A previous
study demonstrated that miR-145-5p inhibited cell proliferation and
promoted cell apoptosis (26). The
present results suggested that hsa_circ_0006393 acted as a sponge
of miR-145-5p and exhibited osteogenic effects in GIOP by targeting
FOXO1. The knockout of FOXO1 in osteoblasts decreases bone mass and
osteogenesis (17). This previous
finding is consistent with the present results. Overexpression of
hsa_circ_0006393 upregulated the expression of FOXO1, and bone mass
was increased in a mouse model of GIOP. The function of
hsa_circ_0006393 was investigated in vivo, and a mouse model
of GIOP was established to investigate the role of
hsa_circ_0006393.
The present results suggested that the
overexpression of hsa_circ_0006393 improved BMD in GIOP mice, and
western blot assay identified the osteogenic potential of
hsa_circ_0006393. Collectively, the in vivo and in
vitro results suggested that hsa_circ_0006393 exhibited
osteogenic effects by sponging miR-145-5p and upregulating the
expression level of FOXO1.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SBS and DW conceived the study and provided
experimental materials. XBW performed the experiments and wrote the
paper. PBL, SFG, QSY and ZXC analyzed the data. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
All experimental procedures were approved by the
Animal Care and Use Committee of Gansu Provincial Hospital. All
patients enrolled in the present study provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Weinstein RS: Glucocorticoid-induced
osteoporosis and osteonecrosis. Endocrinol Metab Clin North Am.
41:595–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Krasselt M and Baerwald C: An update on
glucocorticoid-induced osteoporosis. Dtsch Med Wochenschr.
141:352–357. 2016.(In German). PubMed/NCBI
|
|
3
|
Güler-Yüksel M, Hoes JN, Bultink IEM and
Lems WF: Glucocorticoids, inflammation and bone. Calcif Tissue Int.
102:592–606. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jia D, O'Brien CA, Stewart SA, Manolagas
SC and Weinstein RS: Glucocorticoids act directly on osteoclasts to
increase their life span and reduce bone density. Endocrinology.
147:5592–5599. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
O'Brien CA, Jia D, Plotkin LI, Bellido T,
Powers CC, Stewart SA, Manolagas SC and Weinstein RS:
Glucocorticoids act directly on osteoblasts and osteocytes to
induce their apoptosis and reduce bone formation and strength.
Endocrinology. 145:1835–1841. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weinstein RS: Clinical practice.
Glucocorticoid-induced bone disease. N Engl J Med. 365:62–70. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Whittier X and Saag KG:
Glucocorticoid-induced osteoporosis. Rheum Dis Clin North Am.
42:177–189. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang R, Zhang S, Chen X, Li N, Li J, Jia
R, Pan Y and Liang H: circNT5E acts as a sponge of microRNA-422a to
promote glioblastoma tumorigenesis. Cancer Res. 78:4812–4825. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li B and Li X: Overexpression of
hsa_circ_0007534 predicts unfavorable prognosis for osteosarcoma
and regulates cell growth and apoptosis by affecting AKT/GSK-3β
signaling pathway. Biomed Pharmacother. 107:860–866. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song L and Xiao Y: Downregulation of
hsa_circ_0007534 suppresses breast cancer cell proliferation and
invasion by targeting miR-593/MUC19 signal pathway. Biochem Biophys
Res Commun. 503:2603–2610. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuang MJ, Xing F, Wang D, Sun L, Ma JX and
Ma XL: circUSP45 inhibited osteogenesis in glucocorticoid-induced
osteonecrosis of femoral head by sponging miR-127-5p through
PTEN/AKT signal pathway: Experimental studies. Biochem Biophys Res
Commun. 509:255–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi C, Qi J, Huang P, Jiang M, Zhou Q,
Zhou H, Kang H, Qian N, Yang Q, Guo L and Deng L: MicroRNA-17/20a
inhibits glucocorticoid-induced osteoclast differentiation and
function through targeting RANKL expression in osteoblast cells.
Bone. 68:67–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhong S, Wang J, Zhang Q, Xu H and Feng J:
CircPrimer: A software for annotating circRNAs and determining the
specificity of circRNA primers. BMC Bioinformatics. 19:2922018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tao SC, Yuan T, Rui BY, Zhu ZZ, Guo SC and
Zhang CQ: Exosomes derived from human platelet-rich plasma prevent
apoptosis induced by glucocorticoid-associated endoplasmic
reticulum stress in rat osteonecrosis of the femoral head via the
Akt/Bad/Bcl-2 signal pathway. Theranostics. 7:733–750. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liao L, Su X, Yang X, Hu C, Li B, Lv Y,
Shuai Y, Jing H, Deng Z and Jin Y: TNF-α inhibits FoxO1 by
upregulating miR-705 to aggravate oxidative damage in bone
marrow-derived mesenchymal stem cells during osteoporosis. Stem
cells. 34:1054–1067. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu J, Gong H, Lu S, Deasey MJ and Cui Q:
Animal models of steroid-induced osteonecrosis of the femoral
head-a comprehensive research review up to 2018. Int Orthop.
42:1729–1737. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Buckley L, Guyatt G, Fink HA, Cannon M,
Grossman J, Hansen KE, Humphrey MB, Lane NE, Magrey M, Miller M, et
al: 2017 American college of rheumatology guideline for the
prevention and treatment of glucocorticoid-induced osteoporosis.
Arthritis Rheumatol. 69:1521–1537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu D, Gao Y, Hu N, Wu L and Chen Q:
miR-365 ameliorates dexamethasone-induced suppression of
osteogenesis in MC3T3-E1 cells by targeting HDAC4. Int J Mol Sci.
18(pii): E9772017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Matsumoto A, Pasut A, Matsumoto M,
Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI,
Clohessy JG and Pandolfi PP: mTORC1 and muscle regeneration are
regulated by the LINC00961-encoded SPAR polypeptide. Nature.
541:228–232. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng X, Zhang L, Zhang K, Zhang G, Hu Y,
Sun X, Zhao C, Li H, Li YM and Zhao J: Circular RNA VMA21 protects
against intervertebral disc degeneration through targeting miR-200c
and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis.
77:770–779. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Greco S, Cardinali B, Falcone G and
Martelli F: Circular RNAs in muscle function and disease. Int J Mol
Sc. 19(pii): E34542018. View Article : Google Scholar
|
|
24
|
Song H, Xu D, Shi P, He B, Li Z, Ji Y,
Agbeko CK and Wang J: Upregulated circ RNA hsa_circ_0000337
promotes cell proliferation, migration, and invasion of esophageal
squamous cell carcinoma. Cancer Manag Res. 11:1997–2006. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xie F, Li Y, Wang M, Huang C, Tao D, Zheng
F, Zhang H, Zeng F, Xiao X and Jiang G: Circular RNA BCRC-3
suppresses bladder cancer proliferation through miR-182-5p/p27
axis. Mol Cancer. 17:1442018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu Z, Huang W, Wang X, Wang T, Chen Y,
Chen B, Liu R, Bai P and Xing J: Circular RNA CEP128 acts as a
sponge of miR-145-5p in promoting the bladder cancer progression
via regulating SOX11. Mol Med. 24:402018. View Article : Google Scholar : PubMed/NCBI
|