Introduction
Retinoblastoma (RB) originates from primitive
retinal cells and is the most prevalent primary intraocular
malignant tumor in children (1).
The incidence rate of RB is one case per 15,000–20,000 live births,
or ~9,000 novel cases globally every year (2). The symptoms of RB include leukocoria,
strabismus, nystagmus, inflamed eyes and loss of sight, and the
clinical presentation depends almost entirely on the location of
the tumor (3). Marked improvements
in surgical resection and chemotherapy have been made, and remain
as the standard treatment options; however, the clinical outcome
for patients with RB remains unsatisfactory due to the spread of
tumors to the brain via the optic nerve (4,5). The
RB1 gene has been identified as defective in the pathogenesis of RB
(6); however, the complex
mechanisms underlying the formation and progression of RB remain
unclear. Therefore, improved understanding of the mechanisms
underlying the pathogenesis and development of RB is urgently
required for the identification of novel therapeutic strategies for
the treatment of patients with this disease.
MicroRNAs (miRNAs/miRs) are a series of
single-stranded noncoding RNA molecules comprising 19–25
nucleotides (7). A total of 1,881
precursor and 2,588 mature miRNAs have been identified in the human
genome, according to miRBase (http://www.mirbase.org/index.shtml) (8). Mature miRNAs act as important
modulators of gene expression by directly interacting with
complementary sequences in the 3′-untranslated regions (3′-UTRs) of
their target genes, suppressing translation and inducing the
degradation of mRNAs (9). It has
been reported that miRNAs are aberrantly expressed in almost all
types of human cancer, and their dysregulation contributes to
tumorigenesis and development (10–12).
In particular, a large number of miRNAs have been identified as
dysregulated in RB; these miRNAs are involved in the regulation of
a wide range of biological behaviors, including the proliferation,
cell cycle, apoptosis, invasion, metastasis and chemoresistance of
tumor cells (13–15). Therefore, investigation into the
expression profiles and roles of miRNAs in RB may aid the
identification of novel targets in the diagnosis or treatment of
RB.
Numerous studies have reported that the dysregulated
expression of miR-330 is involved in the onset and progression of
various human cancers, including breast (16,17),
esophageal (18) and lung cancer
(19,20), and glioblastoma (21); however, the expression profile,
roles and underlying mechanisms of miR-330 in RB remain unclear.
Therefore, the present study aimed to investigate the expression of
miR-330 in RB tissues and cell lines, determine the roles served by
miR-330 in RB cells and reveal the underlying mechanisms.
Materials and methods
Clinical specimens
The present study was approved by the Ethics
Committee of the China-Japan Union Hospital of Jilin University
(Changchun, China). Written informed consent was obtained from all
patients that participated in the study. RB tissues were collected
from 24 patients with RB (16 males and 8 females; aged 14–35 years)
that had not been treated with chemotherapy and radiotherapy prior
to surgical treatment at the China-Japan Union Hospital of Jilin
University. Normal retinal tissues were obtained from 7 patients (5
males and 2 females; aged 26–57 years) who were diagnosed with
globe rupture and treated via enucleation. Tissues were collected
between May 2015 and October 2017. All tissues were flash frozen in
liquid nitrogen and stored at −80°C until further use.
Cell lines
The human RB cell lines (Y79, SO-RB50 and Weri-RB1)
and normal retinal pigmented epithelium cell line ARPE-19 were all
obtained from the American Type Culture Collection (Manassas, VA,
USA). The cells were cultured in Dulbecco's Modified Eagle's medium
(DMEM) containing 10% v/v heat-inactivated fetal bovine serum (FBS)
and 1% v/v penicillin-streptomycin mixture (all from Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The cell lines were
cultured at 37°C in a humidified incubator supplemented with 5%
CO2.
Cell transfection
The miR-330 mimics and miRNA mimics negative control
(miR-NC) used in the study were purchased from Shanghai GenePharma
Co., Ltd. (Shanghai, China). The miR-330 mimics sequence was
5′-GCAAAGCACACGGCCUGCAGAGA-3′, and the miR-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. The ρ-associated coiled-coil
containing protein kinase 1 (ROCK1) overexpression plasmid
pCMV-ROCK1 and blank plasmid pCMV were designed and chemically
synthesized by Obio Technology Corp., Ltd. (Shanghai, China). Y79
and Weri-RB1 cells (5×105 cells/well) were seeded into
6-well plates and incubated overnight at 37°C under 5%
CO2. Y79 and Weri-RB1 cells were transfected with
miR-330 mimics (100 pmol), miR-NC (100 pmol), pCMV-ROCK1 (4 µg) or
pCMV (4 µg) using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocols. At 6 h following transfection, cells were washed with
PBS (Gibco; Thermo Fisher Scientific, Inc.), and DMEM with 10% FBS
was added to each well. Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis and MTT assays were
conducted following incubation at 37°C for 24 h.
Matrigel®-based invasion assays and western blot
analysis were conducted at 48 and 72 h post-transfection,
respectively.
RT-qPCR analysis
Total RNA was isolated from tissue samples and
cultured cells using TRIzol® reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols. To
determine the expression levels of miRNA, cDNA was produced from
total RNA using a miScript Reverse Transcription kit (Qiagen GmbH,
Hilden, Germany), according to the manufacturer's protocols.
Subsequently, qPCR was performed to determine miR-330 expression
using a miScript SYBR® Green PCR kit (Qiagen GmbH). The
thermocycling conditions for qPCR were as follows: 95°C for 2 min,
95°C for 10 sec, 55°C for 30 sec and 72°C for 30 sec, for 40
cycles. For the determination of ROCK1 mRNA expression, total RNA
was reverse-transcribed into cDNA using an M-MLV Reverse
Transcription system (Promega Corporation, Madison, WI, USA)
according to the manufacturer's protocols, which was then subjected
to qPCR using an SYBR Green I mix (Takara Biotechnology Co., Ltd.,
Dalian, China). The thermocycling conditions for qPCR were as
follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec
and 60°C for 1 min. The levels of miR-330 and ROCK1 mRNA were
analyzed using the 2−ΔΔCq method (22), and normalized to the expression
levels of U6 small nuclear RNA and GAPDH, respectively. The primers
were designed as follows: miR-330, forward
5′-GGGCTCGAGCCACTCACCCACACTGAAGA-3′, reverse,
5′-GGGGCGGCCGCGTTTCTCCCTCTGCTTGACG-3′; U6, forward
5′-GCTTCGGCAGCACATATACTAAAAT-3′, reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; ROCK1, forward
5′-AGGAAGGCGGACATATTGATCCCT-3′, reverse,
5′-AGACGATAGTTGGGTCCCGGC-3′; and GAPDH, forward
5′-CAGCCTCAAGATCATCAGCA-3′ and reverse, 5′-GTCTTCTGGGTGGCAGTGAT-3′.
RT-qPCR experiments were performed in triplicate and repeated at
least three times.
MTT assay
Transfected Y79 and Weri-RB1 cells were collected
following 24 h of incubation and plated in a 96-well plate at a
density of 3×103 cells/well. Y79 and Weri-RB1 cells were
then incubated at 37°C with 5% CO2. MTT assays were
performed to determine the viability of cells at 0, 24, 48 and 72 h
following inoculation at 37°C. In detail, 20 µl of MTT solution (5
mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to
each well, prior to incubation at 37°C for 4 h. The culture medium
was then discarded, and 150 µl dimethyl sulfoxide (Beyotime
Institute of Biotechnology, Shanghai, China) was added to dissolve
the formazan crystals. A microplate reader (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) was used to detect the absorbance at 490
nm. MTT assays were performed in triplicate and repeated at least
three times.
Matrigel-based invasion assay
The invasive ability of cells was determined using
24-well Transwell inserts (8-µm pores) coated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA). Transfected Y79 and Weri-RB1
cells were harvested following 48 h of incubation and re-suspended
in FBS-free DMEM. A total of 1×105 cells were plated in
the upper chamber of Transwell inserts, whereas DMEM with 20% FBS
(500 µl; Gibco; Thermo Fisher Scientific, Inc.) was added to the
lower chambers. Non-invasive cells were gently removed 24 h later
with a cotton swab. Invasive cells were fixed with 4%
paraformaldehyde at room temperature for 20 min, and stained with
0.05% crystal violet (Beyotime Institute of Biotechnology) at room
temperature for 20 min. The average number of invasive cells in
five randomly selected fields was calculated for each insert under
a light microscope (magnification, ×200; Olympus Corporation,
Tokyo, Japan). Each experiment was repeated at least three
times.
Bioinformatics analysis and luciferase
reporter assay
miRanda (version 3.3a; http://www.microrna.org/microrna/home.do) and
TargetScan (Release 6.0; www.targetscan.org) were employed to identify putative
targets of miR-330. It was revealed that miR-330 may directly bind
to the sequence at 141–147 bp in the 3′-UTR of ROCK1. Wild-type and
mutant 3′-UTR fragments of ROCK1 were generated by Shanghai
GenePharma Co., Ltd. and subcloned into a pmirGLO dual-luciferase
reporter vector (Promega Corporation). Y79 and Weri-RB1 cells were
inoculated in 24-well plates 1 night prior to transfection.
Luciferase plasmids containing the wild-type or mutant 3′-UTR of
ROCK1 were cotransfected with miR-330 mimics or miR-NC into cells
using Lipofectamine 2000, according to the manufacturer's
protocols. Following culture for 48 h at 37°C, the Dual-Luciferase
Reporter Assay System (Promega Corporation) was used to evaluate
luciferase activity. The relative luciferase activity was
normalized to Renilla luciferase activity. Each experiment
was repeated at least three times.
Western blot analysis
Total protein was extracted from tissues or cells
using radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology). The concentration of total protein was
determined using a Bicinchoninic Acid Protein Quantification kit
(Vazyme, Piscataway, NJ, USA). Proteins (20 µg/lane) were separated
via 10% SDS-PAGE and transferred to polyvinylidene difluoride
membranes (Beyotime Institute of Biotechnology). Then, the
membranes were blocked at room temperature for 2 h in Tris-buffered
saline containing 0.1% Tween-20 (TBST) with 5% dried skimmed milk,
followed by incubation with primary antibodies overnight at 4°C.
The following primary antibodies from Abcam (Cambridge, UK) were
used: Rabbit anti-human ROCK1 (1:1,000; cat. no. ab45171) and
rabbit anti-human GAPDH (1:1,000; cat. no. ab181603). Following
three washes with TBST, the membranes were further incubated with a
goat anti-rabbit horseradish peroxidase-conjugated secondary
antibody (1:5,000; ab6721, Abcam) at room temperature for 2 h.
Protein bands were visualized using a Pierce™ Fast Western Blot kit
(Pierce; Thermo Fisher Scientific, Inc.). GAPDH was used as a
loading control. All experiments were repeated at least three
times. Quantity One version 4.62 software (Bio-Rad Laboratories,
Inc.) was used to quantify protein expression.
Statistical analysis
Statistical analysis was conducted using SPSS v.16.0
(SPSS, Inc., Chicago, IL, USA). All experimental data were
presented as the mean ± standard deviation from at least three
independent experiments. Differences between groups were analyzed
using Student's t-tests or one-way analyses of variance followed by
a Tukey's post hoc test. Pearson correlation analysis was performed
to investigate the association between miR-330 and ROCK1 expression
in RB tissues. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-330 is downregulated in RB tissues
and cell lines
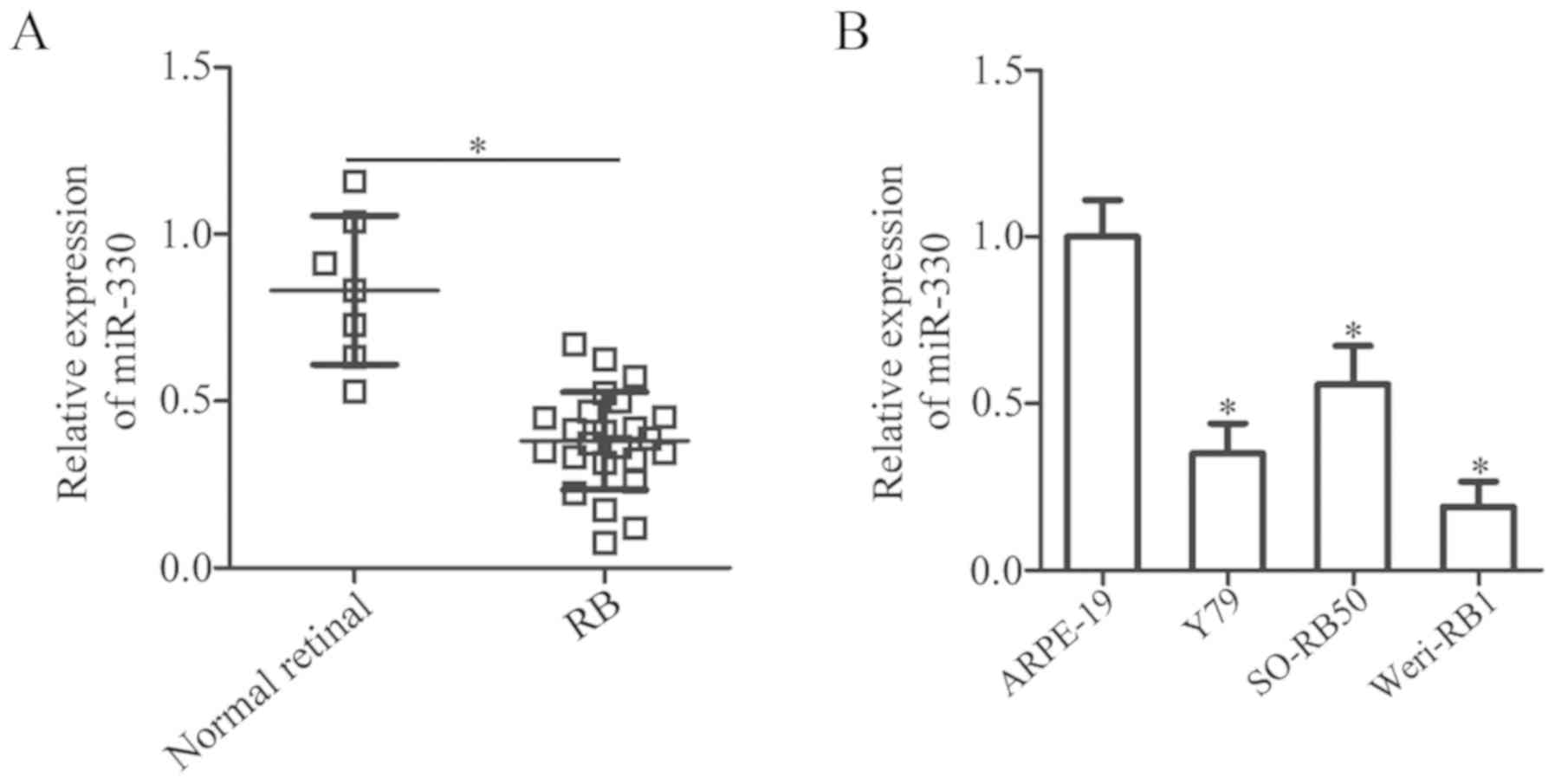
The expression of miR-330 in 24 RB and 7 normal
retinal tissues was determined. RT-qPCR analysis revealed that the
levels of miR-330 expression was significantly downregulated in RB
tissues compared with in normal retinal tissues (P<0.05;
Fig. 1A). Additionally, the
expression levels of miR-330 in three human RB cell lines (Y79,
SO-RB50 and Weri-RB1) were significantly decreased compared with in
ARPE-19 cells (P<0.05; Fig.
1B). The results suggested that the downregulation of miR-330
is an important event in RB.
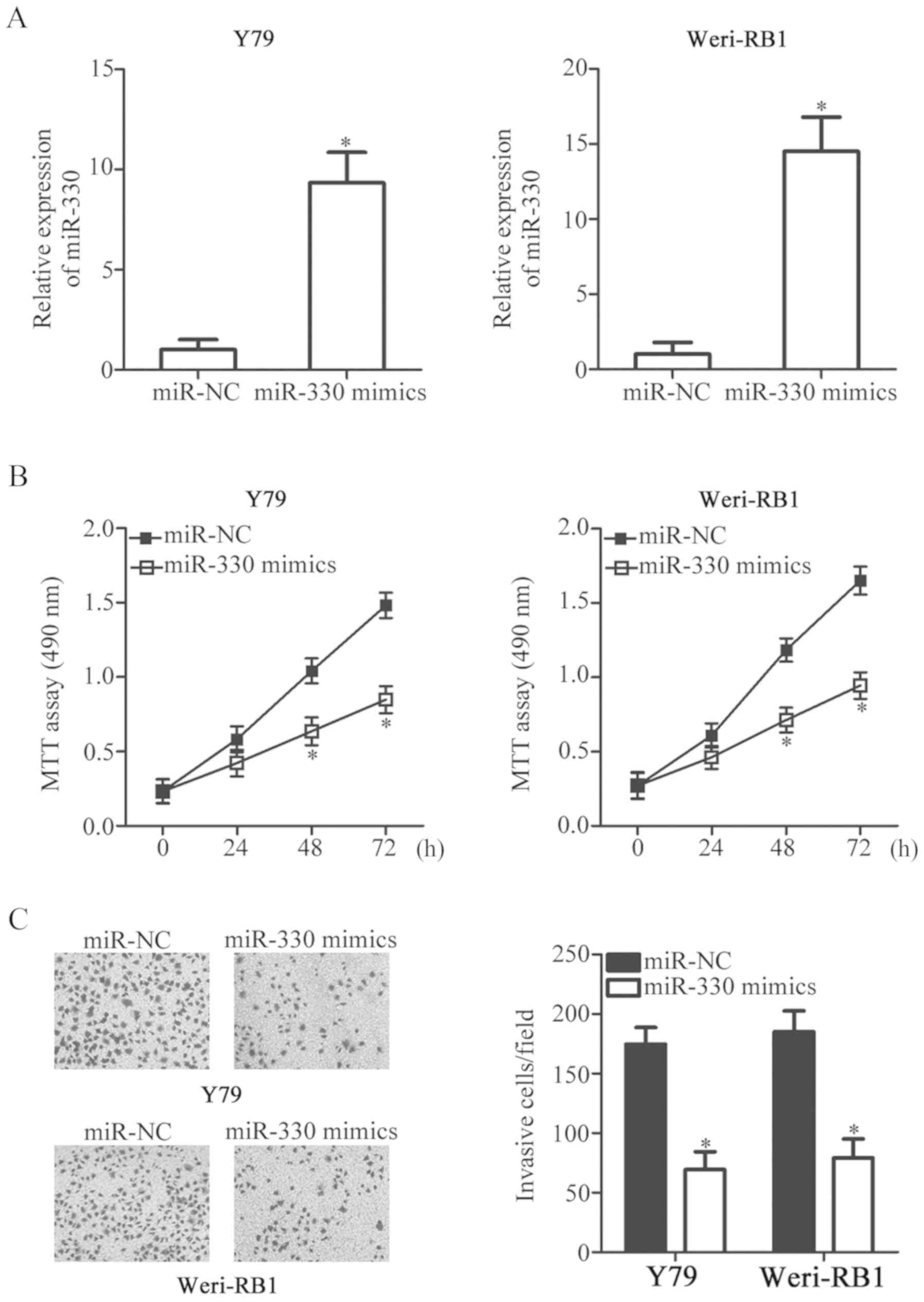
miR-330 overexpression inhibits RB
cell viability and invasion in vitro
To investigate the potential roles of miR-330 in RB,
Y79 and Weri-RB1 cells, due to their relatively reduced miR-330
expression compared with SO-RB50 cells, were selected for
subsequent experiments and transfected with miR-330 mimics or
miR-NC. miR-330 was effectively overexpressed in miR-330
mimics-transfected Y79 and Weri-RB1 cells (P<0.05; Fig. 2A). MTT assays were subsequently
performed to determine the effects of upregulation of miR-330 on
the viability of RB cells. It was revealed that transfection with
miR-330 mimics significantly reduced the viability of Y79 and
Weri-RB1 cells compared with miR-NC (P<0.05; Fig. 2B). Furthermore, the invasive
abilities of Y79 and Weri-RB1 cells transfected with miR-330 mimics
or miR-NC were further determined using Matrigel invasion assays.
Overexpression of miR-330 significantly suppressed the invasion of
Y79 and Weri-RB1 cells compared with cells transfected with miR-NC
(P<0.05; Fig. 2C). The results
indicated that miR-330 serves a tumor suppressor role in the
progression of RB.
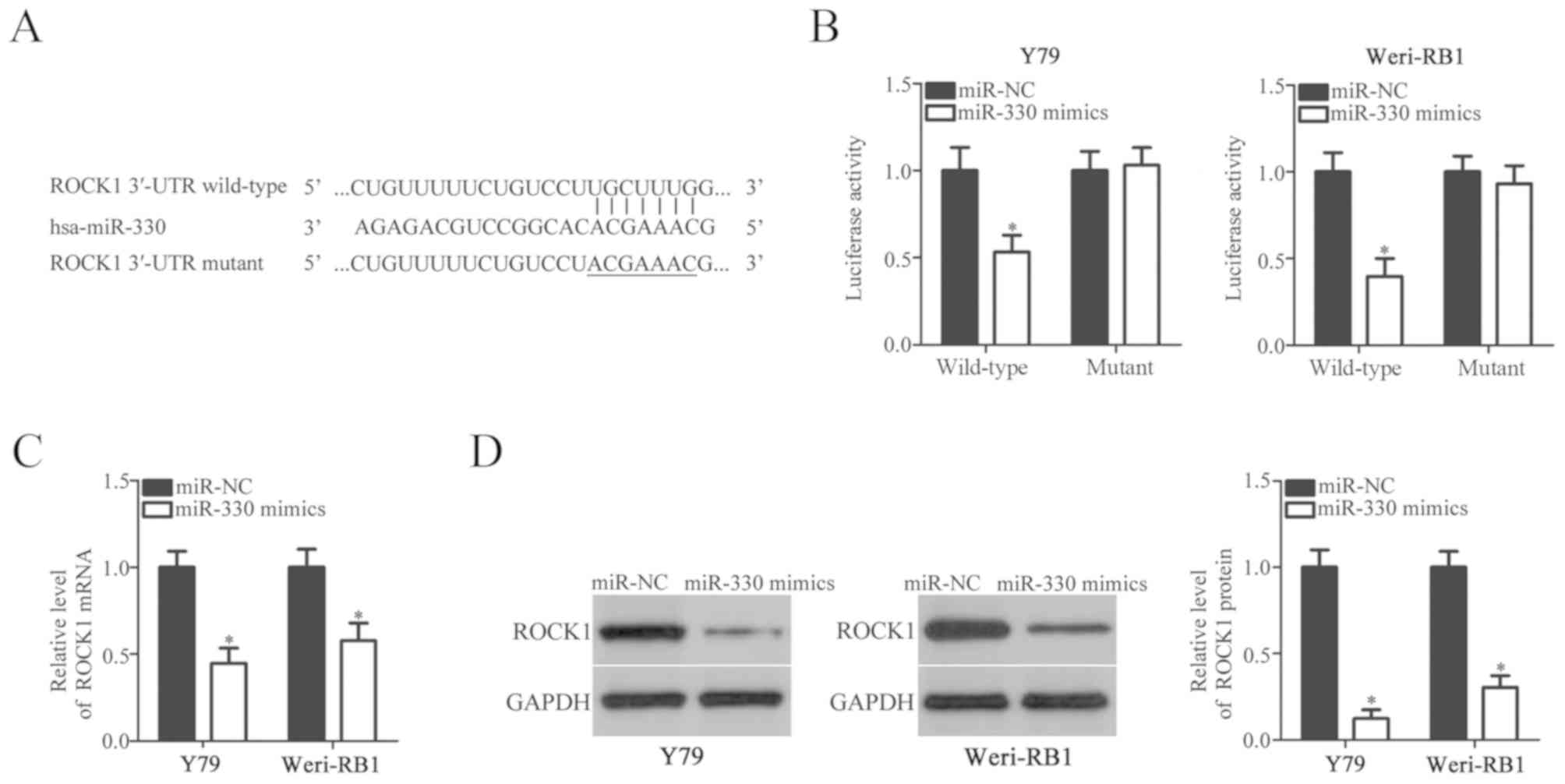
miR-330 directly targets ROCK1 and
decreases its expression in RB cells
Bioinformatics analysis was employed to identify the
putative targets of miR-330. Analysis indicated that there was a
binding site for miR-330 within the 3′-UTR region of ROCK1
(Fig. 3A). ROCK1 was selected for
further experimentation as previous studies have reported ROCK1 to
be associated with the genesis and development of RB (23,24).
A luciferase reporter assay was conducted to verify whether miR-330
was able to bind to the 3′-UTR region of ROCK1. The results
revealed that overexpression of miR-330 significantly decreased the
luciferase activity of plasmids containing the wild-type, but not
mutant 3′-UTR of ROCK1 in Y79 and Weri-RB1 cells compared with
cells transfected with miR-NC (P<0.05; Fig. 3B). To investigate whether miR-330
regulates the expression of ROCK1 in RB cells, RT-qPCR and western
blot analysis were performed in Y79 and Weri-RB1 cells following
transfection with miR-330 mimics or miR-NC. It was revealed that
miR-330 mimics-transfected Y79 and Weri-RB1 cells exhibited
significantly decreased ROCK1 mRNA (P<0.05; Fig. 3C) and protein (P<0.05; Fig. 3D) expression compared with cells
transfected with miR-NC. Collectively, these results indicated that
ROCK1 is a target gene of miR-330 in RB cells.
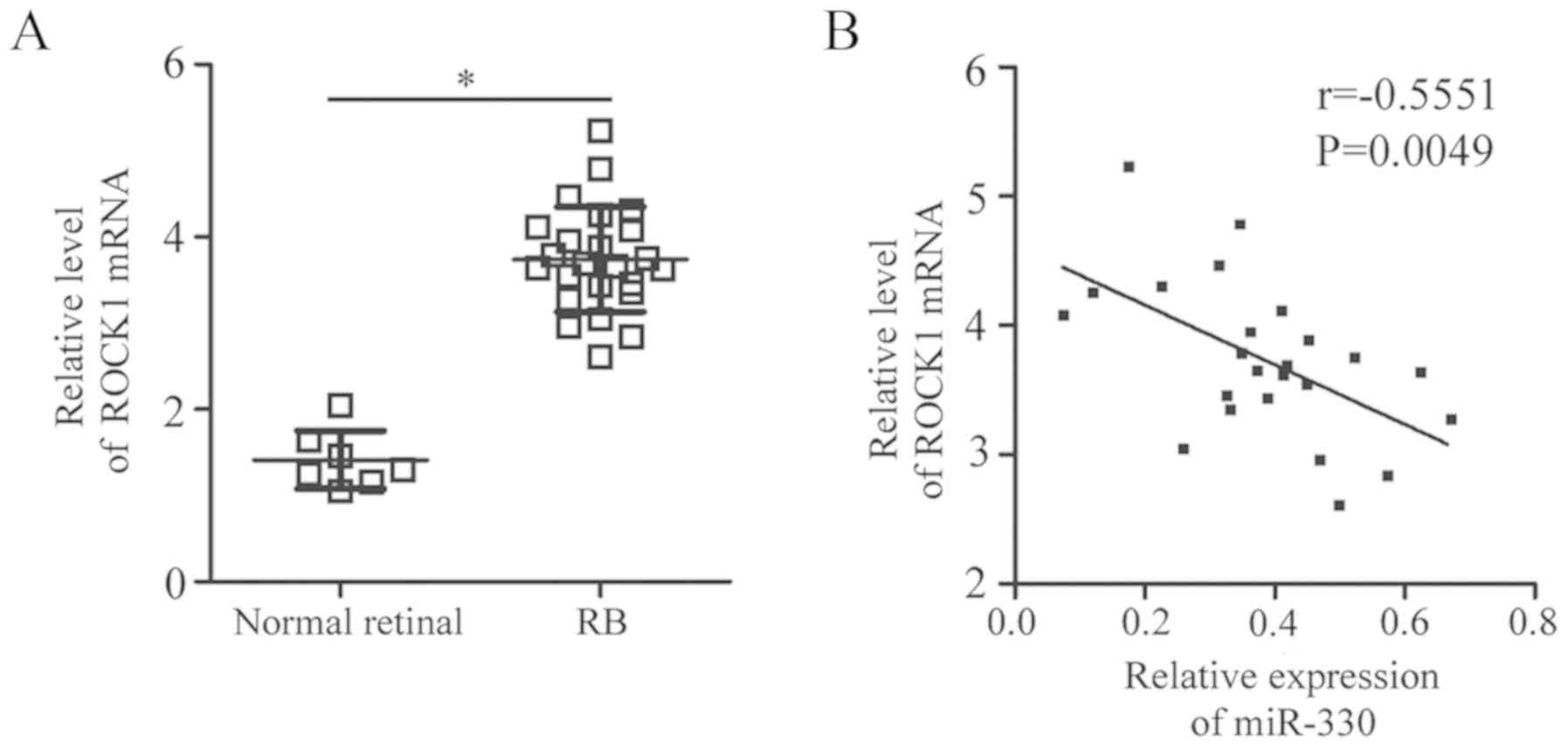
Expression of miR-330 is inversely
correlated with ROCK1 expression in RB tissues
To further investigate the association between
miR-330 and ROCK1 in RB, RT-qPCR analysis was performed using RB
and normal retinal tissues. ROCK1 mRNA expression levels were
upregulated in RB tissues compared with in normal retinal tissues
(P<0.05; Fig. 4A).
Additionally, Pearson correlation analysis revealed an inverse
correlation between the levels of miR-330 and ROCK1 mRNA expression
in RB tissues (r=−0.5551, P=0.0049; Fig. 4B). The results indicated that the
upregulation of ROCK1 in RB tissues is associated with the
downregulation of miR-330.
Rescue of ROCK1 expression reverses
the suppressive effects of miR-330 overexpression in RB cells
Rescue experiments were conducted to determine
whether ROCK1 is associated with the tumor-suppressive roles of
miR-330 in RB cells. Y79 and Weri-RB1 cells were transfected with
miR-330 mimics in combination with pCMV or pCMV-ROCK1. The
downregulation of ROCK1 expression in Y79 and Weri-RB1 cells
induced by miR-330 overexpression was rescued by co-transfection
with pCMV-ROCK1 (P<0.05; Fig.
5A). Additionally, MTT and Matrigel invasion assays revealed
that rescue of ROCK1 expression eliminated the inhibitory effects
of miR-330 overexpression on the viability (P<0.05; Fig. 5B) and invasion (P<0.05; Fig. 5C) of Y79 and Weri-RB1 cells. The
results indicated that miR-330 serves tumor-suppressor roles in RB
cells by downregulating ROCK1 expression.
Discussion
The abnormal expression of miRNAs has been reported
in almost all types of human cancer, including RB (25–27).
Dysregulated miRNAs have been reported to be important epigenetic
regulators of numerous biological events associated with RB, acting
as oncogenes or tumor suppressors (13). Therefore, determining the detailed
roles of miRNAs in RB could aid the identification of potential
therapeutic approaches in the treatment of patients with RB.
miR-330 was reported to be upregulated in breast cancer, and
increased miR-330 expression was significantly associated with
tumor, node and metastasis stage, and lymph node metastasis
(16). Patients with breast cancer
and high levels of miR-330 expression demonstrated reduced 5-year
overall survival than patients with low miR-330 expression levels
(16). Additionally, univariate
and multivariate regression analyses have identified the expression
of miR-330 as an independent prognostic factor for patients with
breast cancer (16,17). miR-330 has also been reported to be
upregulated in esophageal cancer (18), non-small cell lung cancer (NSCLC)
(19,20) and glioblastoma (21). Conversely, miR-330 expression is
downregulated in prostate cancer (28), osteosarcoma (29) and colorectal cancer (30); however, its expression status in RB
remains unclear. These contradictory observations indicate that
miR-330 exhibits tissue-specific expression profiles in human
malignant tumors. In the present study, it was revealed that
miR-330 expression was downregulated in RB tissues and cell lines,
suggesting that miR-330 may be a novel biomarker for the diagnosis
of this specific type of cancer.
miR-330 has been reported to serve oncogenic roles
in the process of carcinogenesis and cancer progression. For
example, miR-330 overexpression promoted the metastasis of breast
cancer in vitro (17). In
esophageal cancer, upregulation of miR-330 increased the
proliferation, survival and metastasis of cells in vitro,
and promoted tumor growth in vivo (18). Additionally, in NSCLC,
overexpression of miR-330 enhanced tumor growth and metastasis
in vitro and in vivo (19,20),
whereas in glioblastoma, miR-330 overexpression promoted the
proliferation and motility of cells, induced cell cycle arrest and
suppressed the apoptosis of cells in vitro (21). Conversely, miR-330 served as a
tumor suppressor in osteosarcoma (29), and prostate (28) and colorectal cancer (30), demonstrating the tissue-specific
activity of miR-330. In the present study, MTT and Matrigel
invasion assays were employed to determine the regulatory roles of
miR-330 in RB cells; it was revealed that miR-330 acts as a tumor
suppressor in RB cells by inhibiting the viability and invasion of
cells in vitro. These findings suggested that miR-330 may be
a therapeutic target in the treatment of patients with RB.
Numerous targets of miR-330 have been identified,
including collagen and calcium-binding epidermal growth factor
domain-containing protein 1 (17)
and programmed cell death protein 4 (18) in breast cancer, early growth
response protein 2 (19) and
glutamate ionotropic receptor AMPA type subunit 3 (20) in lung cancer, SH3 domain containing
GRB2 like 2, endophilin-A1 (21)
in glioblastoma, specificity protein 1 (28) in prostate cancer, B lymphoma Mo-MLV
insertion region homolog 1 (29)
in osteosarcoma and thymidylate synthetase (30) in colorectal cancer. In the present
study, ROCK1 was validated as a direct target gene of miR-330 in RB
cells. TargetScan and miRanda were employed to identify putative
targets of miR-330. It was considered that miR-330 may directly
bind to the 141–147 bp sequence in the 3′-UTR of ROCK1. Then,
luciferase reporter assays, RT-qPCR and western blotting
demonstrated that miR-330 could directly target the 3′-UTR of ROCK1
and decrease its expression in RB cells. Furthermore, it was
revealed that ROCK1 was upregulated in RB tissues and its
expression was inversely correlated with miR-330 expression.
Finally, restoration of ROCK1 expression eliminated the suppressive
effects of miR-330 overexpression on the malignant phenotypes of RB
cells.
ROCK1, located on 18q11.1, is an essential
downstream effector of the Rho small guanosine 5′-triphosphatase
and serves as a molecular switch, binding GTP and guanosine
5′-diphosphate to regulate various biological behaviors (31–33).
It is frequently overexpressed in a variety of human cancers,
including pancreatic cancer (34),
nasopharyngeal carcinoma (35),
bladder cancer (36) and
osteosarcoma (37). Additionally,
the present study reported that ROCK1 was demonstrated to be
upregulated in RB. The dysregulation of ROCK1 is involved in the
initiation and progression of RB, and regulates a number of
physiological and pathological processes, including the
proliferation, invasion, adhesion and apoptosis of cells (23,24).
In the present study, it was demonstrated that miR-330 targets
ROCK1 to inhibit the progression of RB cells; thus, the
miR-330/ROCK1 axis may be a potential therapeutic target in the
treatment of patients with RB.
In conclusion, the findings from the present study
indicated that miR-330 may function as a tumor suppressor in RB by
directly targeting ROCK1 to inhibit the viability and invasion of
cells. Further investigation into the mechanisms underlying the
precise functions of miR-330 in RB may aid the identification of
therapeutic approaches for patients with this malignancy. A single
miRNA can regulate numerous targets, and a single gene can be
negatively modulated by various miRNAs (9); however, in the present study, only
ROCK1 was identified as a direct target gene of miR-330 in RB
cells. Further studies are required to address this issue.
Furthermore, the patient sample size was small. Therefore, analysis
of miR-330/ROCK expression in additional tissues from patients with
RB is required to further validate these findings.
Acknowledgements
Not applicable.
Funding
Not funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
LingW and XL designed the present study. LingW and
LinaW conducted RT-qPCR and western blot analyses, and the
luciferase reporter assays. LL and HZ performed the MTT and
Matrigel-based invasion assays. All authors have read and approved
the final draft.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of China-Japan Union Hospital of Jilin University
(Changchun, China), and was performed in accordance with the
Declaration of Helsinki and the guidelines of the Ethics Committee
of China-Japan Union Hospital of Jilin University. Written informed
consent was obtained from all patients for the use of their
clinical tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jabbour P, Chalouhi N, Tjoumakaris S,
Gonzalez LF, Dumont AS, Chitale R, Rosenwasser R, Bianciotto CG and
Shields C: Pearls and pitfalls of intraarterial chemotherapy for
retinoblastoma. J Neurosurg Pediatr. 10:175–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kivelä T: The epidemiological challenge of
the most frequent eye cancer: Retinoblastoma, an issue of birth and
death. Br J Ophthalmol. 93:1129–1131. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abramson DH, Beaverson K, Sangani P, Vora
RA, Lee TC, Hochberg HM, Kirszrot J and Ranjithan M: Screening for
retinoblastoma: Presenting signs as prognosticators of patient and
ocular survival. Pediatrics. 112:1248–1255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meel R, Radhakrishnan V and Bakhshi S:
Current therapy and recent advances in the management of
retinoblastoma. Indian J Med Paediatr Oncol. 33:80–88. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abramson DH, Marr BP, Brodie S, Dunkel IJ
and Gobin PY: Intraarterial chemotherapy for kissing macula tumors
in retinoblastoma. Retin Cases Brief Rep. 6:209–211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Benavente CA and Dyer MA: Genetics and
epigenetics of human retinoblastoma. Annu Rev Pathol. 10:547–562.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Munker R and Calin GA: MicroRNA profiling
in cancer. Clin Sci (Lond). 121:141–158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng Y, Lu X, Xu L, Chen Z, Li Q and Yuan
J: MicroRNA-675 promotes glioma cell proliferation and motility by
negatively regulating retinoblastoma 1. Hum Pathol. 69:63–71. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bagnyukova TV, Pogribny IP and Chekhun VF:
MicroRNAs in normal and cancer cells: A new class of gene
expression regulators. Exp Oncol. 28:263–269. 2006.PubMed/NCBI
|
|
10
|
Laengsri V, Kerdpin U, Plabplueng C,
Treeratanapiboon L and Nuchnoi P: Cervical cancer markers:
Epigenetics and microRNAs. Lab Med. 49:97–111. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lou W, Liu J, Gao Y, Zhong G, Chen D, Shen
J, Bao C, Xu L, Pan J, Cheng J, et al: MicroRNAs in cancer
metastasis and angiogenesis. Oncotarget. 8:115787–115802. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ramassone A, Pagotto S, Veronese A and
Visone R: Epigenetics and MicroRNAs in cancer. Int J Mol Sci.
19(pii): E4592018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Golabchi K, Soleimani-Jelodar R, Aghadoost
N, Momeni F, Moridikia A, Nahand JS, Masoudifar A, Razmjoo H and
Mirzaei H: MicroRNAs in retinoblastoma: Potential diagnostic and
therapeutic biomarkers. J Cell Physiol. 233:3016–3023. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singh U, Malik MA, Goswami S, Shukla S and
Kaur J: Epigenetic regulation of human retinoblastoma. Tumour Biol.
37:14427–14441. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang J, Yang Y, Fang F and Liu K: MALAT1
modulates the autophagy of retinoblastoma cell through
miR-124-mediated stx17 regulation. J Cell Biochem. 119:3853–3863.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang H, Chen SH, Kong P, Zhang LY, Zhang
LL, Zhang NQ and Gu H: Increased expression of miR-330-3p: A novel
independent indicator of poor prognosis in human breast cancer. Eur
Rev Med Pharmacol Sci. 22:1726–1730. 2018.PubMed/NCBI
|
|
17
|
Mesci A, Huang X, Taeb S, Jahangiri S, Kim
Y, Fokas E, Bruce J, Leong HS and Liu SK: Targeting of CCBE1 by
miR-330-3p in human breast cancer promotes metastasis. Br J Cancer.
116:1350–1357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meng H, Wang K, Chen X, Guan X, Hu L,
Xiong G, Li J and Bai Y: MicroRNA-330-3p functions as an oncogene
in human esophageal cancer by targeting programmed cell death 4. Am
J Cancer Res. 5:1062–1075. 2015.PubMed/NCBI
|
|
19
|
Liu X, Shi H, Liu B, Li J, Liu Y and Yu B:
miR-330-3p controls cell proliferation by targeting early growth
response 2 in non-small-cell lung cancer. Acta Biochim Biophys Sin
(Shanghai). 47:431–440. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei CH, Wu G, Cai Q, Gao XC, Tong F, Zhou
R, Zhang RG, Dong JH, Hu Y and Dong XR: MicroRNA-330-3p promotes
cell invasion and metastasis in non-small cell lung cancer through
GRIA3 by activating MAPK/ERK signaling pathway. J Hematol Oncol.
10:1252017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qu S, Yao Y, Shang C, Xue Y, Ma J, Li Z
and Liu Y: MicroRNA-330 is an oncogenic factor in glioblastoma
cells by regulating SH3GL2 gene. PLoS One. 7:e460102012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Liu XH, Yang ZJ, Xie B and Zhong
YS: The effect of ROCK-1 activity change on the adhesive and
invasive ability of Y79 retinoblastoma cells. BMC Cancer.
14:892014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu S, Ai N, Liu Q and Zhang J:
MicroRNA-448 inhibits the progression of retinoblastoma by directly
targeting ROCK1 and regulating PI3K/AKT signalling pathway. Oncol
Rep. 39:2402–2412. 2018.PubMed/NCBI
|
|
25
|
Wang Z, Yao YJ, Zheng F, Guan Z, Zhang L,
Dong N and Qin WJ: Mir-138-5p acts as a tumor suppressor by
targeting pyruvate dehydrogenase kinase 1 in human retinoblastoma.
Eur Rev Med Pharmacol Sci. 21:5624–5629. 2017.PubMed/NCBI
|
|
26
|
Yang L, Wei N, Wang L, Wang X and Liu QH:
miR-498 promotes cell proliferation and inhibits cell apoptosis in
retinoblastoma by directly targeting CCPG1. Childs Nerv Syst.
34:417–422. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao Y, Zhang S and Zhang Y: MicroRNA-320
inhibits cell proliferation, migration and invasion in
retinoblastoma by targeting specificity protein 1. Mol Med Rep.
16:2191–2198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mao Y, Chen H, Lin Y, Xu X, Hu Z, Zhu Y,
Wu J, Xu X, Zheng X and Xie L: microRNA-330 inhibits cell motility
by downregulating Sp1 in prostate cancer cells. Oncol Rep.
30:327–333. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng Z, Bao F, Chen X, Huang H and Zhang
X: MicroRNA-330-3p expression indicates good prognosis and
suppresses cell proliferation by targeting Bmi-1 in osteosarcoma.
Cell Physiol Biochem. 46:442–450. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu W, Jiang H, Zhang F, Gao J and Hou J:
MicroRNA-330 inhibited cell proliferation and enhanced
chemosensitivity to 5-fluorouracil in colorectal cancer by directly
targeting thymidylate synthase. Oncol Lett. 13:3387–3394. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang C, Zhang S, Zhang Z, He J, Xu Y and
Liu S: ROCK has a crucial role in regulating prostate tumor growth
through interaction with c-Myc. Oncogene. 33:5582–5591. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rossman KL, Der CJ and Sondek J: GEF means
go: Turning on RHO GTPases with guanine nucleotide-exchange
factors. Nat Rev Mol Cell Biol. 6:167–180. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Patel RA, Forinash KD, Pireddu R, Sun Y,
Sun N, Martin MP, Schönbrunn E, Lawrence NJ and Sebti SM: RKI-1447
is a potent inhibitor of the Rho-associated ROCK kinases with
anti-invasive and antitumor activities in breast cancer. Cancer
Res. 72:5025–5034. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Whatcott CJ, Ng S, Barrett MT, Hostetter
G, Von Hoff DD and Han H: Inhibition of ROCK1 kinase modulates both
tumor cells and stromal fibroblasts in pancreatic cancer. PLoS One.
12:e01838712017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X, Huang Y, Guo R and Liu Y, Qian Y,
Liu D, Dai X, Wei Z, Jin F and Liu Y: Clinicopathological
significance of ROCK1 and PIK3CA expression in nasopharyngeal
carcinoma. Exp Ther Med. 13:1064–1068. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu D, Niu X, Pan H, Zhou Y, Qu P and Zhou
J: MicroRNA-335 is downregulated in bladder cancer and inhibits
cell growth, migration and invasion via targeting ROCK1. Mol Med
Rep. 13:4379–4385. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang S, Zhao Y and Wang L: MicroRNA-198
inhibited tumorous behaviors of human osteosarcoma through directly
targeting ROCK1. Biochem Biophys Res Commun. 472:557–565. 2016.
View Article : Google Scholar : PubMed/NCBI
|