Introduction
Calcium-activated potassium channels can be
categorized as large-conductance calcium-activated potassium
channels, intermediate-conductance calcium-activated potassium
channels (IK or SK4, encoded by KCNN4), or small-conductance
calcium-activated potassium channels (SK1-3) according to their
level of conductance (1).
SK4 is a non-voltage-dependent channel, and
intracellular free calcium concentrations are sufficiently high to
activate these channels. The channel is widely distributed in red
blood cells, lymphocytes, monocytes, macrophages, epithelial cells
and vascular smooth muscle cells (2). In addition, SK4 transcripts are found
in embryonic stem cell-derived cardiomyocytes (ESC-CM), the mouse
sinoatrial node, the adult human right atrium and in ventricular
biopsies (3–6). Previous studies have shown that the
overexpression or activation of SK4 in ESC-CM can significantly
increase the duration of action potentials, elevate the trigger
frequency and increase the beating area of cardiac embryoid bodies;
these effects disappear after SK4 channel blockage, indicating that
SK4 serves an important role in regulation of the cardiac rhythm
(3,4). A recent study has shown that SK4 is
closely related to the pace function of the sinoatrial node in
vivo (7). It was shown that
the trigger frequency of the sinoatrial node decreased
significantly following application of a specific inhibitor of SK4,
and a mathematical model predicted that the trigger frequency of
the sinoatrial node cells could be increased by the increasing SK4
current (6). A previous study
postulated that the SK4 outward potassium current, which is
responsible for the notch of the maximum diastolic potential (MDP),
provides a driving force that is sufficiently strong to activate
funny current [I(f) current] at the early phase of the diastolic
depolarization (DD) slope (2).
Hyperpolarization-activated cyclic nucleotide-gated
channel (HCN)2 is a member of the HCN channel family and its
activity is affected by cAMP. Compared with HCN1, HCN2 and HCN4
both respond strongly to camp, but HCN2 has faster kinetics than
HCN4, thus HCN2 was chosen for the present study. Previous studies
have shown that the overexpression of HCN2 alone (8,9) or
in combination with other genes (10–13)
can increase the beating frequency of cardiomyocytes, and promote
in vivo pacemaker activity in a canine model of complete
heart block. The aim of the present study was to explore whether
the SK4 overexpression alone or combined with HCN2 could generate
biological pacemaker activity. This was investigated in a rat model
of complete heart block.
Materials and methods
Animals
The present study was approved by the Animal Studies
Subcommittee of Wuhan University School of Medicine and was
conducted in accordance with the Guide for the Care and Use of
Laboratory Animals of the National Institutes of Health (14). All of the surgeries were performed
under sodium pentobarbital anesthesia (40 mg/kg). After a 30-min
procedure, the animals were routinely treated with penicillin for 3
days to prevent infection, and then housed in a barrier environment
until the end of the experiment. Prior to the heart extraction, the
rats were anesthetized intraperitoneally with pentobarbital sodium
(40 mg/kg). When deep anesthesia was established [respiratory
depression, severe muscle relaxation, bradycardia, no reflexes
(palpebral, corneal) and pupil dilation], the heart was removed for
further experiments.
Adenovirus construction and
purification
pHBAd-MCMV (Hanbio Biotechnology Co., Ltd.) was
double digested with BamHI/AgeI. The ORF of the mouse
KCNN4 gene (GenScript) was amplified via PCR using Taq
polymerase (SinoBio Biotech). The sequences were: Kcnn4, forward
5′-GTTCTGCACGCTGAGATGTTG-3′ and reverse 5′CTTGGCATGGAAGACCACAAT-3′.
The thermocycling conditions were pre-denaturation at 98°C for 5
min, denaturation at 98°C for 10 sec, annealing at 55°C for 10 sec,
and a final extension at 72°C for 90 sec, a total of 30 cycles.
After enzyme digestion, gel extraction was performed. The digested
fragment and vector were ligated to form pHBAd-MCMV-KCNN4, which
was then transformed into competent DH5α cells (Tiangen Biotech
Co., Ltd.). Positive clones were identified by liquid sequencing.
Large-scale preparation of recombinant plasmid was conducted using
the Plasmid Midi Preparation kit (Beijing CW Biotech Co., Ltd.).
293 cells (from our laboratory; 60% confluence) were transfected
with pHBAd-MCMV-KCNN4 (1 µg/µl) and the backbone vector pHBAd-BHG
(Hanbio Biotechnology Co., Ltd.) using Lipofectamine
2000® (Thermo Fisher Scientific, Inc.). The supernatant
was harvested after virus amplification (10 days). pHBAd-MCMV
vector was also used for mouse HCN2 (M-HCN2, forward
5′-GTTCTGCACGCTGAGATGTTG-3′ and reverse
5′-CTTGGCATGGAAGACCACAAT-3′) and GFP construction. HCN2 vectors
were prepared as described previously (15). The titers of Ad-GFP, Ad-HCN2 and
Ad-KCNN4 were measured as 1×1010 PFU/ml and preserved at
−80°C.
Experimental protocol
A total of 40 male 8-week Sprague Dawley rats each
weighing 220–250 g were purchased from Hunan Slac Jingda Laboratory
Animal Co., Ltd. and raised in an SPF laboratory animal room at
20–26°C with humidity of 40–70% and fed with irradiated feed and
sterile water. The rats were randomly divided into 4 groups: A GFP
group (n=10), a SK4 group (n=10), a HCN2 group (n=10) and a
SK4/HCN2 group (n=10). All of the surgeries were performed under
sodium pentobarbital anesthesia (40 mg/kg). Once anesthetized, the
rats were intubated, and a thoracotomy was performed on the left
side to expose the heart. The free left ventricle wall was used as
the transgene injection site. The injection site was marked by a
line on the surface of the left ventricle, and the virus was
injected with a microsyringe (1×109 PFU/ml, 100 µl).
Ex vivo electrocardiogram
recording
At 5–7 days following cardiac injection (the levels
of gene expression were highest during this time window; data not
shown), when complete anesthesia was established in rats, a
thoracotomy was conducted, and the hearts were quickly isolated,
inserted into a Langendorff perfusion system (ADInstruments Ltd.)
and secured for retrograde perfusion at 37°C with oxygenated
Tyrode's solution at a rate of 6–8 ml/min. The perfused heart was
placed on a Sylgard-coated plate filled with warm Tyrode's solution
(mM:NaCl 130, KCL 5.4, CaCl2 1.8, MgCl2 1,
Na2HPO4 0.3, HEPES 10 and Glucose 10; pH=7.4,
adjusted with NaOH). ECG leads I and II were placed at appropriate
sites. After a 20-min equilibration period, the atrioventricular
node region was ablated with 75% alcohol using a microsyringe.
After a complete heart block was stably established, the heart rate
was recorded for a period of 20 min and electrode pacing was
performed at the site of the transgene injection at 200-msec
intervals.
Action potential duration (APD)
alternans
Paired platinum-stimulating electrodes were
positioned on the basal surface of the right ventricle. The S1-S1
pacing protocol was performed with a series of pulse trains at a
regular pacing cycle length (PCL). Starting at 150 msec, the PCL
was shortened to 70 msec in 10-msec intervals. The regular pacing
at each PCL lasted for 15 sec to ensure a steady rhythm, and each
pacing was separated by at least 30 sec to minimize the pacing
memory. The APD at a PCL of 300 msec was determined at 90%
repolarization (APD90) at each site. The APD alternans was assessed
at each site by subtracting the APD90 for 2 consecutive beats when
the alternate APD90 differed by 5% over 10 beats. The threshold was
defined as the maximal PCL (PCLmax) that induced APD alternans.
Ventricular arrhythmia (VA)
inducibility
Burst pacing protocols were performed to determine
the VA susceptibility. Burst pacing (2-msec pulses at 50 Hz and two
spacing durations) was performed at the left anterior free-wall,
and repeated 3 times after a 2-sec rest period. VA was defined as a
run of >2 sec of consecutive premature ventricular contractions
(wide QRS complexes). VA was considered non-sustained when the
contractions lasted 2–30 sec and sustained when they lasted >30
sec, according to established criteria (16). The VA vulnerability was evaluated
based on the incidence of VA and the ratio of sustained VA.
Western blot analysis
Tissue specimens were obtained from the myocardium
from the injection site and temporarily stored at −80°C until
assay. The tissue protein was extracted by RIPA Lysis Buffer (Aspen
Technology, Inc.). Determination of protein concentration was the
BCA method. Then, 5% of concentration gel and 10% of separation gel
were chosen and the protein samples (40 µg protein/lane) were mixed
with 5X SDS-PAGE buffer (Aspen Technology, Inc.) in a water bath at
95–100°C for 5 min, prior to being transferred to a polyvinylidene
difluoride membrane. The expression of SK4 and HCN2 was measured
using western blotting (n=10/group). Membranes were incubated with
the primary antibodies against GAPDH (loading control; 1:500;
ab181602, Abcam), SK4 (1:500; ab215990, Abcam) and HCN2 (1:1,000;
ab19346, Abcam). The membranes were blocked with 5% non-fat dry
milk in tris-buffered saline with 0.05% Tween 20 (TBST) for 1 h at
room temperature and incubated with the primary antibodies
overnight at 4°C. The membranes were then washed in TBST three
times, incubated with horseradish peroxidase-conjugated anti-rat
(1:10,000; 14-16-06) and anti-rabbit (1:10,000; 074-1506) secondary
antibodies (SeraCare Life Sciences, Inc.) for 1 h at 37°C, and
imaged using Immun-Star (Bio-Rad Laboratories, Inc.) horseradish
peroxidase substrate. The relative expression of the protein levels
were determined using image analyzer software (AlphaEaseFC V; Alpha
Innotech; ProteinSimple).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the myocardium (100 mg)
at the end of the study using TRIzol reagent (1 ml) (Invitrogen;
Thermo Fisher Scientific, Inc.). RT-qPCR was performed to evaluate
the mRNA expression of KCNN4 and HCN2 (n=10/group). Isolated RNA (2
µg) was converted into cDNA using a First Strand cDNA Synthesis kit
(Toyobo Life Science) in a 15 µl mixture as follows: 42°C for 2
min, 37°C for 15 min, 85°C for 5 min and 4°C for 10 min. The
primers (Table I) used for qPCR
amplification were synthesized by Invitrogen (Thermo Fisher
Scientific, Inc.). qPCR was performed using a StepOne Real-Time PCR
system (Life Technologies, Thermo Fisher Scientific, Inc.) and
SYBR® Premix Ex Taq™ II (Takara Bio, Inc.) as follows:
Pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30
sec, annealing at 58°C for 20 sec, and a final extension at 72°C
for 45 sec, a total of 40 cycles. The dissolution curve was from
60–95°C and the temperature was raised by 1°C per 20 sec. Semilog
amplification curves were analyzed using the 2-∆∆Cq comparative
quantification method, and the expression of each gene was
normalized to GAPDH (17).
 | Table I.Polymerase chain reaction primers used
in this study. |
Table I.
Polymerase chain reaction primers used
in this study.
| Gene | Primer sequences | Product size
(bp) |
|---|
| R-GAPDH |
|
Forward |
5′-CGCTAACATCAAATGGGGTG-3′ | 201 |
|
Reverse |
5′-TTGCTGACAATCTTGAGGGAG-3′ |
|
| M-HCN2 |
|
Forward |
5′-TCCTCATAGTGGAGAAGGGAATC-3′ | 191 |
|
Reverse |
5′-ACAGATGCGCATCACTGCAC-3′ |
|
| M-SK4 |
|
Forward |
5′-GTTCTGCACGCTGAGATGTTG-3′ | 126 |
|
Reverse |
5′-CTTGGCATGGAAGACCACAAT-3′ |
|
Immunohistochemistry
SK4 and HCN2 overexpression were validated by
immunohistochemistry. All the samples for histology were fixed in
4% paraformaldehyde for 15 min at room temperature and embedded in
paraffin. Sections (4 µm) were cut from paraffin blocks of
myocardium. The sections were stained with a rabbit anti-HCN2
antibody (1:200; ab84817, Abcam) and a mouse anti-SK4 antibody
(1:200; ab219108, Abcam) overnight at 4°C. The sections were then
incubated with Cy3-conjugated goat anti-mouse IgG (1:50, AS-1112,
Aspen Technology, Inc.; red fluorescence for SK4) and Alexa
Fluor® 488-conjugated goat anti-rabbit IgG (1:50,
AS-1109, Aspen Technology, Inc.; green fluorescence for HCN2)
secondary antibodies for 50 min at 37°C. DAPI was used to visualize
the nuclei at room temperature for 3 min. Images were obtained from
three random visual fields in three different samples. Each slide
was examined under a fluorescence microscope (magnification,
×200).
Statistical analysis
The data are expressed as the mean ± standard
deviation. The statistical significance of the differences between
two groups was determined using Student's t-test. Comparisons among
multiple groups were made using one-way analysis of variance with
Turkey tests. P<0.05 was considered to indicate a statistically
significant difference. All of the statistical analysis was
performed using SPSS 20.0 software (IBM Corp.).
Results
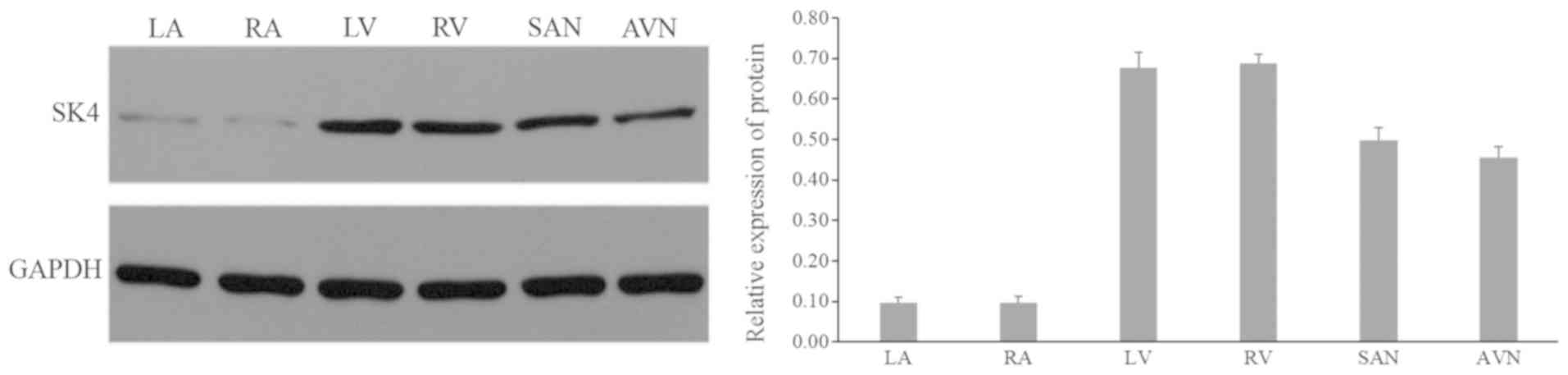
Expression of SK4 in the rat
heart
The expression of SK4 was detected using western
blotting in different portions of the Ad-GFP rat heart (n=10); the
expression level of SK4 in the ventricle area was the highest,
followed by the sinoatrial node and the atrioventricular node area.
The expression was lowest in the atrial area (Fig. 1).
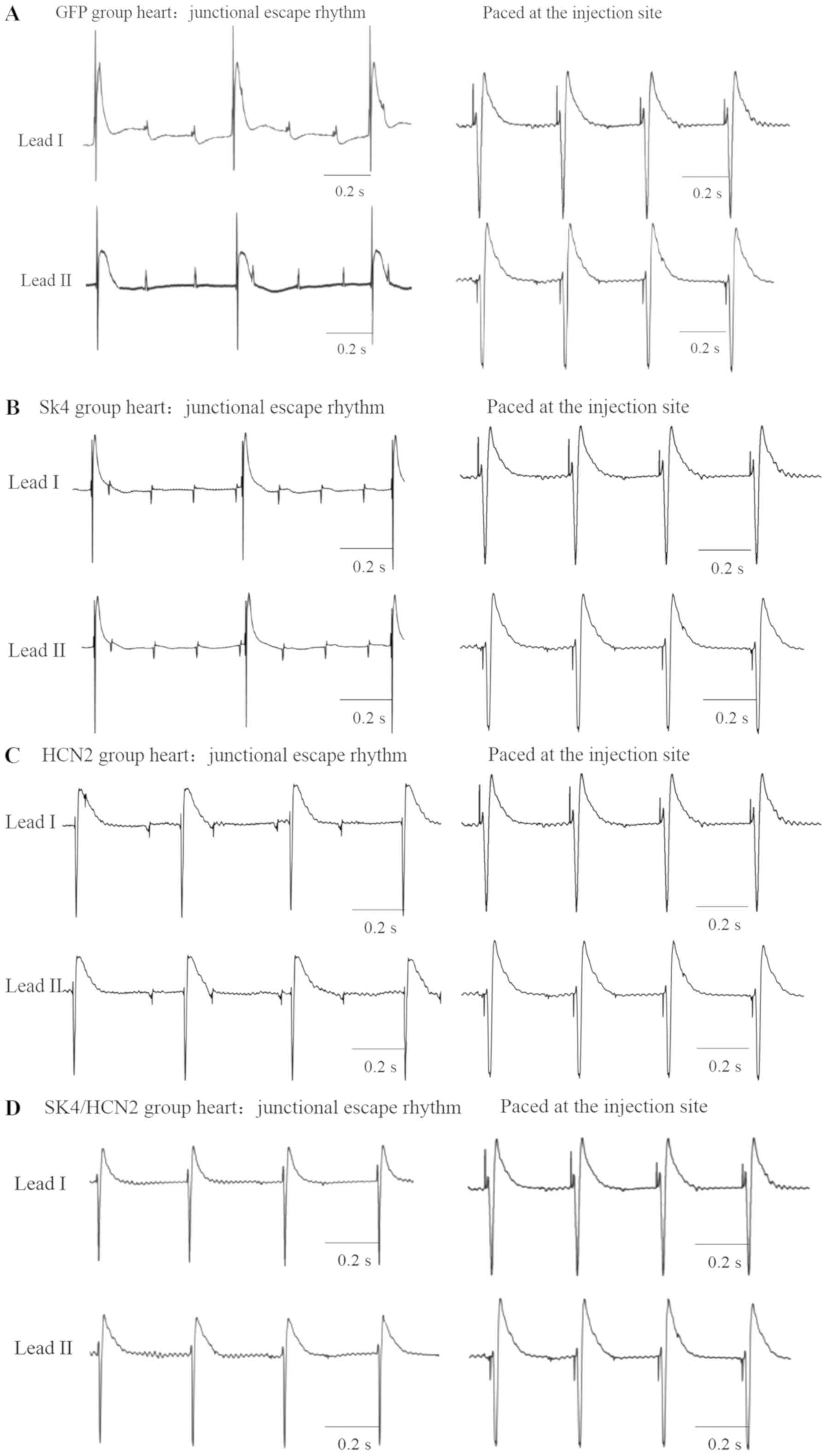
Ex vivo complete heart block model and
origin of ectopic rhythm
The atrioventricular node region was ablated with
75% alcohol using a microsyringe. After the atrioventricular node
was ablated, the electrocardiogram showed obvious atrioventricular
dissociation, and the heart rate was clearly slowed. After the
atrioventricular node was ablated, all of the isolated perfused
hearts showed junctional escape rhythm or ventricular rhythm
(Fig. 2). Compared with the
spontaneous electrocardiogram after complete heart block and the
pacing electrocardiogram of the transgene site, it was observed
that the hearts in the GFP and SK4 groups showed junctional escape
rhythms (Fig. 2A and B).
Furthermore, the pacing electrocardiogram at the transgene site was
markedly different compared with the spontaneous electrocardiogram
(Fig. 2A and B). By contrast, the
pacing electrocardiograms at the transgene sites in the HCN2 (n=8)
and the SK4/HCN2 (n=7) groups were similar to the corresponding
spontaneous electrocardiograms, and exhibited a ventricular rhythm
(Fig. 2C and D). As opposing P
wave axes were observed in the GFP group, and the SK4 and HCN2
groups, it was hypothesized that the P waves may change polarity
during the process of atrioventricular block, due to the
heterogeneity of conduction velocity in the heart. Compared with
the GFP [96.7±7.6 beats per min (BPM; maximum, 105 BPM; minimum, 80
BPM)] and SK4 groups [98.1±8.9 BPM (maximum, 105 BPM; minimum, 74
BPM)], the spontaneous heart rates in the HCN2 group were
significantly increased [111.7±5.5 BPM (maximum, 123 BPM; minimum,
103 BPM); P<0.05 vs. GFP and SK4], and the heart rates in the
SK4/HCN2 group were further increased compared with the HCN2 group
[139.9±21.9 BPM (maximum, 217 BPM; minimum, 120 BPM); P<0.05,
Table II]. To determine the
stability of heart rates after complete heart block was
established, heart rate variability with an assessment of the
standard deviation of NN intervals (SDNN). The SDNNs of the four
groups are presented in Table
III. It was determined that the SDNN was not significantly
affected by SK4 and/or HCN2 overexpression.
 | Table II.Heart rate after a complete heart
block was established between groups. |
Table II.
Heart rate after a complete heart
block was established between groups.
| Groups | GFP | SK4 | HCN2 | SK4/HCN2 |
|---|
| Heart rate
(beats/minute) | 96.7±7.6 | 98.1±8.9 |
111.7±5.5a |
139.9±21.9a,b |
 | Table III.Heart rate variability after a
complete heart block was established between groups. |
Table III.
Heart rate variability after a
complete heart block was established between groups.
| Groups | GFP | SK4 | HCN2 | SK4/HCN2 | P-value |
|---|
| SDNN (mean ±
standard deviation, msec) | 0.52±0.07 | 0.50±0.06 | 0.55±0.05 | 0.53±0.06 | P>0.05 for
all |
APD alternans and VA inducibility
To evaluate the risk of VA, APD alternans and VA
inducibility were measured. The threshold of APD alternans was not
statistically significant between the various groups. Additionally,
neither non-sustained ventricular tachycardia nor ventricular
fibrillation was induced in each group, and the incidence of
ventricular premature contraction was not significantly different
between the groups (Table
IV).
 | Table IV.APD alternans and VA inducibility
between groups. |
Table IV.
APD alternans and VA inducibility
between groups.
| Groups | GFP | SK4 | HCN2 | SK4/HCN2 | P-value |
|---|
| APD alternans
(msec) | 82.5±8.9 | 83.8±10.6 | 83.8±7.4 | 82.5±12.8 | P>0.05 for
all |
| VA
inducibility | 3/30 | 5/30 | 4/30 | 4/30 | P>0.05 for
all |
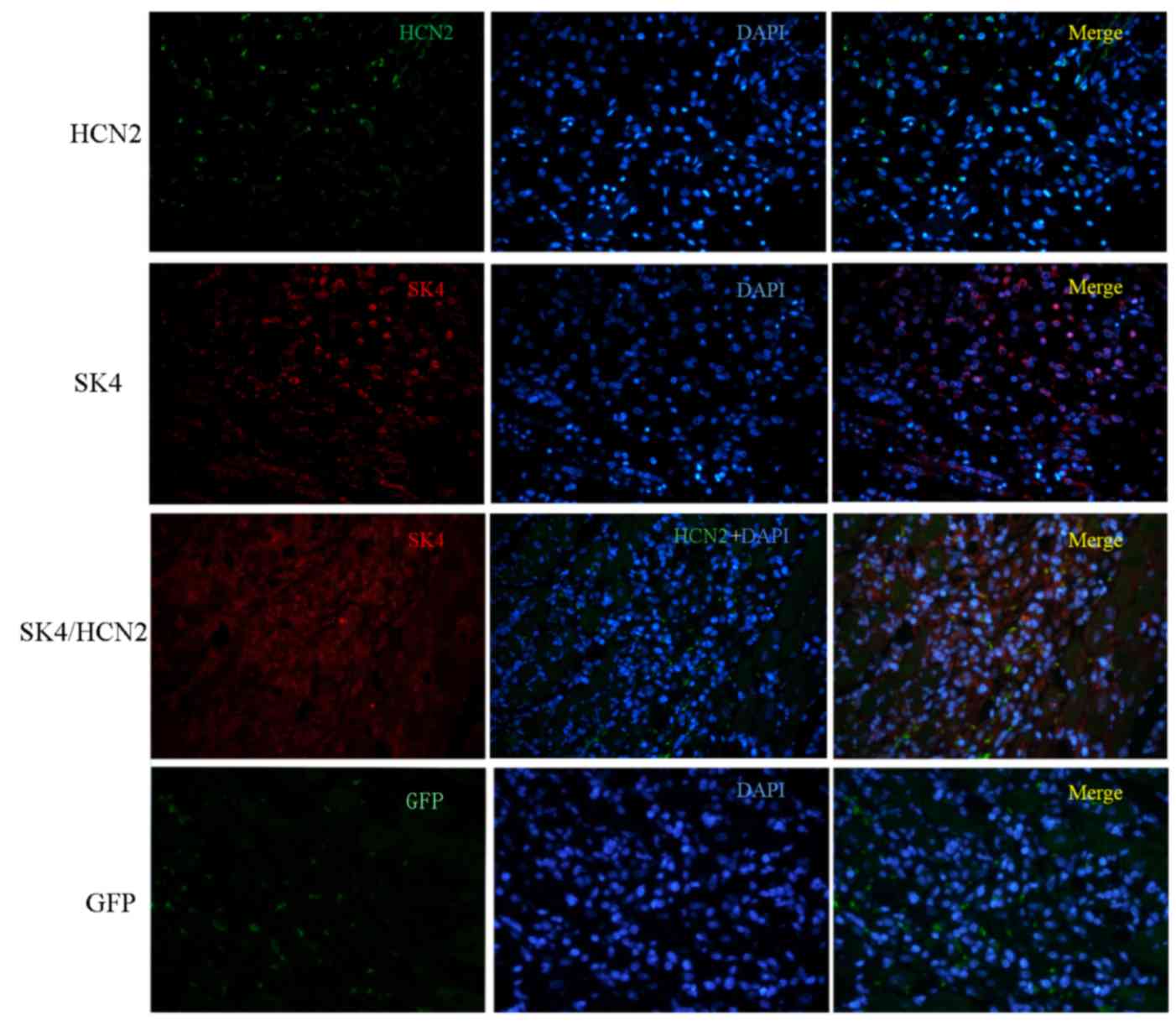
Expression levels of SK4 and HCN2
Myocardial tissue was extracted from the transgene
site to detect the expression of SK4 and HCN2 protein, and the
corresponding genes HCN2 and KCNN4 (Figs. 3 and 4). Western blotting revealed that the
expression levels of SK4 were higher in the SK4 group (0.83±0.04)
and the SK4/HCN2 group (0.84±0.04) compared with in the GFP group
(0.45±0.03; P<0.05; Fig. 3A);
however, there was no significant difference between the SK4 and
SK4/HCN2 groups (P>0.05), nor between the HCN2 and GFP groups.
Similarly, the expression levels of HCN2 in the HCN2 group
(0.65±0.03) and the SK4/HCN2 group (0.67±0.02) were higher than in
the GFP group (0.15±0.01; P<0.05; Fig. 3A). There was no significant
difference between the HCN2 and the SK4/HCN2 groups (P>0.05), or
the SK4 and GFP groups (P>0.05). The RT-qPCR results
demonstrated similar expression profiles at the mRNA level
(Fig. 3B). SK4 and HCN2
overexpression were further validated by immunohistochemistry. As
presented in Fig. 4, SK4 (red) and
HCN2 (green) were both overexpressed in tissue from the SK4, HCN2
and SK4/HCN groups compared with the GFP group.
Discussion
In the present study, biological pacemaker activity
was successfully generated via the overexpression of SK4/HCN2 in a
rat model of complete heart block. The results showed the
following: i) SK4 and HCN2 were successfully expressed in the
myocardium after the adenoviral vectors was injected; ii) SK4/HCN2
co-overexpression could generate an ectopic rhythm in the
ventricle; and iii) the pacing efficiency in the SK4/HCN2
co-overexpression group was improved compared with that in the
groups with overexpression of HCN2 or SK4 alone.
SK4 previously was found to be expressed in the
mouse SAN (6), ESC-CM (4,5),
adult human SAN (6) and right
atrium, and in ventricular biopsies (2). The present study found that SK4 was
expressed in the rat atrium, ventricle, SAN and atrioventricular
node, with the highest expression levels observed in the ventricle.
A previous study has shown that after the SK4 channel was
specifically inhibited, the MDP of cardiomyocytes was depolarized
but the duration of APD50 was unaltered, indicating that SK4
currents do not notably affect the action potential repolarization
duration and only act on the late stage of repolarization (5).
Previous studies have shown that overexpression or
activation of SK4 in embryonic stem cell-derived cardiomyocytes
(ESC-CM) can increase the APD, trigger frequency and beating area
of the embryoid body (3,4); a mathematical model predicted that
the trigger frequency of the sinoatrial node cells could be
increased by increasing SK4 current (6). Therefore, the improved biological
pacing activity induced by SK4/HCN2 co-overexpression in the
present study may be regulated by the outward SK4 potassium
current. This current is responsible for the notch of the MDP,
providing a driving force that is sufficiently strong to activate
the I(f) current at the early phase of the DD slope; SK4
synergistically interacts with I(f) by maintaining the voltage
changes within a range where HCN channels can most effectively
operate (2). It is proposed that
this phenomenon is termed SK4-induced I(f) activation. However, the
overexpression of SK4 alone cannot generate biological pacing
activity, possibly due to inadequate activation of HCN
channels.
Similar to IK1-induced I(f) activation (18), IK1 has long been considered to
hyperpolarize cell membranes and inhibit cardiac automaticity;
inhibition or knockout of IK1 can increase cardiac automaticity
(19). However, it has been
observed that co-overexpression of IK1/HCN can significantly
increase the beating frequency of left ventricular cardiomyocytes
(20) and induce the automaticity
of unexcitable 293T cells and ESC-CM cells (21). Electrophysiological data have shown
that IK1 can promote an increase in MDP hyperpolarization,
accelerate the four-phase depolarization slope and shorten the APD,
thus increasing the frequency of I(f)-induced automaticity
(18). Other studies indicated
that biological pacemaker activity can also be successfully created
by overexpression of T-box18 (TBX18) in vivo (22–24).
The hypothesized mechanisms underlying this phenomenon are as
follows: i) TBX18 can convert ventricular myocytes into sinoatrial
node-like cells; and ii) TBX18 drives upregulation of HCN4 or HCN2,
and downregulation of connexin 43, which increases automaticity and
the conduction of impulse. The similarities between the above
studies and the present study is that HCN channel is involved;
however, unlike TBX18, which affects the expression of HCN, the
present study focused on the synergistic effects of SK4 and HCN2
channel overexpression.
Previous studies have shown that the biological
heart rates generated by overexpression of HCN212, Adenylate
yclase1 (AC1) and T-box3 far exceed the physiological heart rates
and even cause ventricular tachycardia (9,10,25).
The present study found that the heart rate of the SK4/HCN2 group
increased significantly compared with other groups; however, the
fastest heart rate observed was 214 BPM, which is remains slower
than the normal heart rate of rats (400–500 BPM). This result
indicated that the co-overexpression of SK4/HCN2 can safely and
successfully generate biological pacing activity. In addition, a
study has reported on methods of introducing exogenous genes, such
as the overexpression of AC1 and HCN2/AC1, that may have
proarrhythmic effects, which can occur after depolarization;
overexpression can also induce both activity and calcium
overload-induced VAs (10). APD
alternans is closely associated with VAs, with calcium-triggered
calcium release and abnormal intracellular calcium leakage
contributing to these effects (26). In the present study, APD alternans
and burst pacing-induced VAs were measured, revealed that the
threshold of APD alternans between each group was not significantly
different. Furthermore, neither non-sustained ventricular
tachycardia nor ventricular fibrillation was induced in any group,
and the incidence of VA inducibility was not significantly
different between the groups (Table
II). These results indicated that co-overexpression of SK4/HCN2
does not increase the risk of VAs. However, there are limitations
to the present study. First, the myocardium at the transgene site
was not isolated, nor were other cellular electrophysiological
parameters measured, such as SK4 and I(f) currents, MDP or APD.
Data such as these may provide additional information regarding the
underlying mechanisms of the observed effects. Additionally, SK4
and HCN2 were not co-expressed at different ratios, as has been
performed in other studies (15);
such experiments may enable the identification of optimal
co-expression ratios for achieving biological pacemaker
activity.
In conclusion, biological pacemaker activity can be
successfully generated by co-overexpression of SK4 and HCN2 without
increasing the risk of VAs. The overexpression of SK4 alone was
insufficient to generate biological pacemaker activity. This study
provides evidence that SK4 and HCN2 combined could function as an
ectopic pacemaker, laying the groundwork for the development of
improved biological pacing strategies in the future.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant no. 81670303).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ and MY conducted the experiments, acquired and
analysed the data and drafted the manuscript, with support from QZ
and CH. HZ, FW and AY contributed to sample preparation, performed
the calculations and designed the figures. XW, YT and TW conceived
the study and supervised the project. All authors discussed the
results and contributed to the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Animal Studies
Subcommittee of Wuhan University School of Medicine and conducted
in accordance with the Guide for the Care and Use of Laboratory
Animals of the National Institutes of Health.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vergara C, Latorre R, Marrion NV and
Adelman JP: Calcium-activated potassium channels. Curr Opin
Neurobiol. 8:321–329. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weisbrod D, Khun SH, Bueno H, Peretz A and
Attali B: Mechanisms underlying the cardiac pacemaker: The role of
SK4 calcium-activated potassium channels. Acta Pharmacol Sin.
37:82–97. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kleger A, Seufferlein T, Malan D,
Tischendorf M, Storch A, Wolheim A, Latz S, Protze S, Porzner M,
Proepper C, et al: Modulation of calcium-activated potassium
channels induces cardiogenesis of pluripotent stem cells and
enrichment of pacemaker-like cells. Circulation. 122:1823–1836.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liebau S, Tischendorf M, Ansorge D, Linta
L, Stockmann M, Weidgang C, Iacovino M, Boeckers T, von Wichert G,
Kyba M and Kleger A: An inducible expression system of the
calcium-activated potassium channel 4 to study the differential
impact on embryonic stem cells. Stem Cells International.
2:4568152011.
|
|
5
|
Weisbrod D, Peretz A, Ziskind A, Menaker
N, Oz S, Barad L, Eliyahu S, Itskovitz-Eldor J, Dascal N,
Khananshvili D, et al: SK4 Ca2+ activated K+ channel is
a critical player in cardiac pacemaker derived from human embryonic
stem cells. Proc Natl Acad Sci USA. 110:E1685–E1694. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Haron-Khun S, Weisbrod D, Bueno H, Yadin
D, Behar J, Peretz A, Binah O, Hochhauser E, Eldar M, Yaniv Y, et
al: SK4 K+ channels are therapeutic targets for the treatment of
cardiac arrhythmias. EMBO Mol Med. 9:415–429. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lai MH, Wu Y, Gao Z, Anderson ME, Dalziel
JE and Meredith AL: SK4 Ca2+ activated K+channels
regulate sinoatrial node firing rate and cardiac pacing in vivo.
Bio J. 112:35a2017.
|
|
8
|
Bucchi A, Plotnikov AN, Shlapakova I,
Danilo P Jr, Kryukova Y, Qu J, Lu Z, Liu H, Pan Z, Potapova I, et
al: Wild-type and mutant HCN channels in a tandem
biological-electronic cardiac pacemaker. Circulation. 114:992–999.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Plotnikov AN, Bucchi A, Shlapakova I,
Danilo P Jr, Brink PR, Robinson RB, Cohen IS and Rosen MR:
HCN212-channel biological pacemakers manifesting ventricular
tachyarrhythmias are responsive to treatment with I(f) blockade.
Heart Rhythm. 5:282–288. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boink GJ, Nearing BD, Shlapakova IN, Duan
L, Kryukova Y, Bobkov Y, Tan HL, Cohen IS, Danilo P Jr, Robinson
RB, et al: Ca(2+)-stimulated adenylyl cyclase AC1generates
efficient biological pacing as single gene therapy and in
combination with HCN2. Circulation. 126:528–536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miake J, Marbán E and Nuss HB: Biological
pacemaker created by gene transfer. Nature. 419:132–133. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cingolani E, Yee K, Shehata M, Chugh SS,
Marbán E and Cho HC: Biological pacemaker created by percutaneous
gene delivery via venous catheters in a porcine model of complete
heart block. Heart Rhythm. 9:1310–1318. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boink GJ, Duan L, Nearing BD, Shlapakova
IN, Sosunov EA, Anyukhovsky EP, Bobkov E, Kryukova Y, Ozgen N,
Danilo P Jr, et al: HCN2/SkM1 gene transfer into canine left bundle
branch induces stable, autonomically responsive biological pacing
at physiological heart rates. J Am Coll Cardiol. 61:1192–1201.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals, . Guide for the care and use of laboratory animals, 8th
editionNational Academies Press (US); 2011
|
|
15
|
Qu J, Barbuti A, Protas L, Santoro B,
Cohen IS and Robinson RB: HCN2 overexpression in newborn and adult
ventricular myocytes: Distinct effects on gating and excitability.
Circ Res. 89:E8–E14. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baldzizhar A, Manuylova E, Marchenko R,
Kryvalap Y and Mary MG: Ventricular tachycardias: Characteristics
and management. Crit Care Nurs Clin North Am. 28:317–329. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun Y, Timofeyev V, Dennis A, Bektik E,
Wan X, Laurita KR, Deschênes I, Li RA and Fu JD: A singular role of
IK1, promoting the development of cardiac automaticity
during cardiomyocyte differentiation by Ik1-induced
activation of pacemaker current. Stem Cell Rev. 13:631–643. 2017.
View Article : Google Scholar :
|
|
19
|
Zaritsky JJ, Redell JB, Tempel BL and
Schwarz TL: The consequences of disrupting cardiac inwardly
rectifying K(+) current (I(K1)) as revealed by the targeted
deletion of the murine Kir2.1 and Kir2.2 genes. J Physiol.
533:697–710. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chan YC, Siu CW, Lau YM, Lau CP, Li RA and
Tse HF: Synergistic effects of inward rectifier (IK1) and pacemaker
(If) currents on the induction of bioengineered cardiac
automaticity. J Cardiovasc Electrophysiol. 20:1048–1054. 2010.
View Article : Google Scholar
|
|
21
|
Chen K, Zuo D, Wang SY and Chen H: Kir2
inward rectification-controlled precise and dynamic balances
between Kir2 and HCN currents initiate pacemaking activity. FASEB
J. 32:3047–3057. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu YF, Dawkins JF, Cho HC, Marbán E and
Cingolani E: Biological pacemaker created by minimally invasive
somatic reprogramming in pigs with complete heart block. Sci Transl
Med. 6:245ra942014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Choudhury M, Black N, Alghamdi A, D'Souza
A, Wang R, Yanni J, Dobrzynski H, Kingston PA, Zhang H, Boyett MR
and Morris GM: TBX18 overexpression enhances pacemaker function in
a rat subsidiary atrial pacemaker model of sick sinus syndrome. J
Physiol. 596:6141–6155. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gorabi AM, Hajighasemi S, Khori V,
Soleimani M, Rajaei M, Rabbani S, Atashi A, Ghiaseddin A, Saeid AK,
Ahmadi Tafti H and Sahebkar A: Functional biological pacemaker
generation by T-Box18 protein expression via stem cell and viral
delivery approaches in a murine model of complete heart block.
Pharmacol Res. 141:443–450. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hoogaars WM, Engel A, Brons JF, Verkerk
AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P,
et al: Tbx3 controls the sinoatrial node gene program and imposes
pacemaker function on the atria. Genes Dev. 21:1098–1112. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Orini M, Hanson B, Taggart P and Lambiase
P: Detection of transient, regional carrdiac repolarization
alternans by time-frequency analysis of synthetic eletrograms. Eng
Med Biol Soc. 2013:3773–3776. 2013.
|