Introduction
Notable developments have been made in the systemic
therapy and early diagnosis of breast cancer; however, this disease
remains to be the second leading cause of cancer-associated
mortality in females (1,2). Distant metastasis represents as the
major cause of mortality in patients with breast cancer (3). Metastasis is a multistep process
characterized by the progressive accumulation of genetic
alterations that results in aberrant cell growth, malignant
transformation, vascular invasion and ultimately, metastasis
(4–5). Gene expression and proteomics
analyses have revealed several novel metastasis-associated factors
underlying the development and progression of breast cancer
(6–11); however, the role of these factors
requires further investigation, yet metastasis remains a serious
problem in the treatment of this disease.
Transgelin 2 (TAGLN2) is a 22-kDa protein and one of
the earliest markers of differentiated smooth muscle (12). TAGLN2 belongs to the calponin
family of proteins as it contains an N-terminal calponin homology
domain and a C-terminal calponin-like domain (13). TAGLN2 binds to actin to facilitate
the formation of cytoskeletal structures, such as stress fibers
(14). The dysregulated expression
of TAGLN2 has been observed in various types of cancer (7,15–18).
A previous study indicated that TAGLN2 inhibited the motility of
hepatocarcinoma cells by suppressing actin polymerization (16). In endometrial and ovarian cancers,
TAGLN2 was downregulated in metastatic tumors compared with primary
tumors (17). Downregulated TAGLN2
expression has been reported to be a prognostic indicator for
shorter disease-free survival in Barrett's adenocarcinoma (15). A notable correlation was observed
between reduced TAGLN2 expression and regional lymph node
metastasis.
Reactive oxygen species (ROS) serve important roles
in the migration and invasion of cancer cells (19–23).
The stimulation of cell surface receptors with growth factors and
integrin induces the production of ROS. The dynamic nature of actin
and mitochondrial dysfunction have been associated with ROS
production (24). ROS act within
cells to activate downstream signaling pathways, including the
nuclear factor-κB (NF-κB), and signaling transducer and activator
of transcription 3 pathways that promote migration and invasion
(25–29). Peroxiredoxin 1 (PRDX1) belongs to
the peroxiredoxin family of antioxidant enzymes, which serve an
important role in the maintenance of intracellular ROS levels
(30–32). The reduced expression of PRDX1
results in decreased total antioxidant capacity, inducing ROS
production (33). In breast
cancer, PRDX1 functions as a specific sensor in redox-regulated
senescence; silencing of PRDX1 increased the susceptibility for
developing Ras-induced breast cancer (34). Thus, the dysfunction of PRDX1 may
contribute to the progression of breast cancer.
Our previous results from differential proteomics
and immunohistochemical analyses revealed that TAGLN2 was
downregulated in MDA-MB-231HM cells, which has a high metastatic
potential (7). Additionally,
immunohistochemical analysis demonstrated that the expression of
TAGLN2 was suppressed in lymph node-positive invasive ductal
carcinoma tissues (7). These
findings suggested that TAGLN2 may be negatively associated with
breast cancer metastasis. Nevertheless, the biological mechanism by
which TAGLN2 downregulation induces the metastatic phenotype of
breast cancer cells remains unknown.
In the present study, it was demonstrated that
TAGLN2 knockdown promoted MDA-MB-231 cell invasion and enhanced
lung metastasis in vivo. In addition, TAGLN2 was determined
to interact with PRDX1. Knockdown of TAGLN2 resulted in reduced
PRDX1 expression, elevated ROS levels, mitochondrial redistribution
and the activation of the NF-κB pathway, which led to the induction
of metastasis-associated genes, including C-X-C chemokine receptor
4 (CXCR4), matrix metalloproteinase (MMP)1 and MMP2. The results of
the present study suggest that loss of TAGLN2 expression may
promote tumor invasion via ROS and the NF-κB pathway, PRDX1
downregulation and redistribution of the mitochondria.
Materials and methods
Cell lines and breast tumor
specimens
A panel of breast cancer cell lines were obtained
from the American Type Culture Collection in June 2015, including
BT-549, MDA-MB-231 and MCF-7. The 293T cell line was obtained from
the Cell Bank of China in 2015 and maintained in Dulbecco's
modified Eagles medium (Gibco; Thermo Fisher Scientific, Inc.)
containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen;
Thermo Fisher Scientific, Inc.). Cell lines underwent DNA profiling
of short tandem repeats in our laboratory in 2016. The cell lines
were subjected to routine cell line quality examination, via
morphological analysis and mycoplasma testing by HD Biosciences
every 3 months until used in the present study. Frozen aliquots
were stored in liquid nitrogen; cells were cultured for ≤6 months
after thawing. All human breast cancers specimens were randomly
obtained from patients aged 27–79 years old with pathologically
confirmed stage I–III primary breast cancer with no distant
metastasis; patients underwent surgery for breast cancer between
March 2003 and December 2009 at the Fudan University Shanghai
Cancer Center. The specimens were stored in liquid nitrogen until
analysis. All specimens contained >90% tumor cells. All patients
provided written informed consent. The study complied with the
Declaration of Helsinki and was approved by the Ethics Committee of
the Fudan University Shanghai Cancer Center.
Breast cancer subtypes were categorized as luminal,
or based on human epidermal growth factor receptor 2
(HER2)-positive and triple negative according to the available
estrogen receptor, progesterone receptor and HER2 status. TNM stage
and T stage were defined according to the 6th American Joint
Committee on Cancer (AJCC) TNM staging system, which include the
status of tumor, lymph node and metastasis (35).
Small interfering (si)RNA, plasmid
construction, transfection and luciferase assays
Specific siRNAs and negative controls (scrambled
siRNA) were purchased from Shanghai GenePharma Co., Ltd. Cells
cultured in 6-well plates were transfected with 100 pmol/well siRNA
using Hilymax Transfection Agent (Dojindo Molecular Technologies,
Inc.) according to the manufacturer's protocols. Scrambled siRNAs
comprised random sequences with no detectable effects in human cell
lines. Short hairpin (sh)RNAs were obtained from the RNAi
Consortium (https://www.broadinstitute.org/scientific-community/science/projects/rnai-consortium/rnai-consortium).
pLKO-shRNA vector was purchased from Addgene, Inc. (cat. no.
30,323). Green fluorescent protein-specific shRNAs were used as
unrelated negative controls. The sequences of the siRNAs, shRNA and
negative controls are listed in Table
I. The vector pGL4.32 containing NF-κB transcription responsive
elements (TREs) was from Promega Corporation (cat. no. E849A).
 | Table I.Sequences of primers, siRNA and
shRNAs. |
Table I.
Sequences of primers, siRNA and
shRNAs.
| Purposes | Sequence type | Sequence
(5′-3′) |
|---|
| TAGLN2 clone
primer | Sense |
GTCAGTGCGCTGCTCTCC |
|
| Antisense |
CCCTGACAGAAAGGAGCTTG |
| TAGLN2 FLAG clone
primer | Sense |
GCGAAGCTTGCCAACAGGGGACCTGC |
|
| Antisense |
AGAATTCTCAGAGGATCTGGCGTGGC |
| TAGLN2 shRNA
586 | Top |
CCGGGAACGTGATCGGGTTACAGATCTCGAGATCTGTAACCCGATCACGTTCTTTTTTG |
|
| Bottom |
aattCAAAAAAGAACGTGATCGGGTTACAGATCTCGAGATCTGTAACCCGATCACGTTC |
| TAGLN2 shRNA
377 | Top |
CCGGCGCTATGGCATTAACACCACTCTCGAGAGTGGTGTTAATGCCATAGCGTTTTTTG |
|
| Bottom |
aattCAAAAAACGCTATGGCATTAACACCACTCTCGAGAGTGGTGTTAATGCCATAGCG |
| TAGLN2 shRNA
252 | Top |
CCGGCGGTGCTATGTGAGCTCATTACTCGAGTAATGAGCTCACATAGCACCGTTTTTTG |
|
| Bottom |
aattCAAAAAACGGTGCTATGTGAGCTCATTACTCGAGTAATGAGCTCACATAGCACCG |
| siRNA TAGLN2
252 | Target |
CGACCAATAGCTCAGATCCTT |
| siRNA TAGLN2
377 | Target |
CAGGTGATACTATCAACCAAA |
| siRNA negative
control |
|
GUGGAUAUUGUUGCCAUCA |
The coding DNA sequence of human TAGLN2 was cloned
into the EcoRI/NotI sites of the pCDH vector
(Systembio). The clone primer sequences were listed in Table I. For the luciferase assays,
293T-pGL4.32 cells were plated in 6-well plates and transfected
with 100 pmol/well of the siRNA or a negative control. The
luciferase plasmid was purchased from Beyotime Institute of
Biotechnology. The luciferase activity was normalized to that of
Renilla. The Bright-Glo™ Luciferase Assay System was used to
analyze luciferase activity 48 h post-transfection.
Protein extraction and western blot
analysis
Cells were lysed in SDS cell lysis buffer containing
50 mM Tris (pH 8.1) and 1% SDS, supplemented with protease
inhibitors (Roche Diagnostics) for 30 min on ice. The homogenates
were centrifuged at 18,000 × g for 10 min at 4°C. Supernatants were
collected, and the protein concentration was determined by a
Bradford's assay (Bio-Rad Laboratories, Inc.); 30 µg of the
proteins was loaded for analysis. The proteins were separated via
10% SDS-PAGE and transferred to polyvinylidene fluoride membranes
(EMD Millipore). The membranes were probed with primary antibodies
against PRDX1 (1:1,000; cat. no. EPR5433; Epitomics, Abcam,
Cambridge, UK), FLAG (1:500; cat. no. F7425; Sigma-Aldrich, Merck
KGaA), TAGLN2 (1:1,000; cat. no. 60044-1-lg; ProteinTech Group,
Inc.), IκB (1:3,000; cat. no. 66418-1-lg; ProteinTech Group, Inc.),
p50 (1:200; cat. no. 15506-1-AP; ProteinTech Group, Inc.), p65
(1:1,000; cat. no 10745-1-AP; ProteinTech Group, Inc.), MMP1
(1:1,000; cat. no. 54376; Cell Signaling Technology), MMP2
(1:1,000; cat. no. 4022; Cell Signaling Technology), CXCR4
(1:1,000; cat. no. ab181020; Abcam, Cambridge, UK), Lamin B1
(1:3,000; cat. no. 12987-1-AP; ProteinTech Group, Inc.) and GAPDH
(1:3,000; cat. no. 60004-1-lg; ProteinTech Group, Inc.) on the
rocking table at 4°C overnight. After washing, the membranes were
incubated with appropriate horseradish peroxidase-conjugated
secondary antibodies at room temperature for 60 min: Goat
anti-Rabbit IgG (1:3,000; SA00001-2; ProteinTech Group, Inc.), Goat
anti-Mouse IgG (1:3,000; SA00001-1; ProteinTech Group, Inc.). The
bands were developed via enhanced chemiluminescence (EMD
Millipore). Quantity One software version 4.6.2 (Bio-Rad
Laboratories, Inc.) was used to quantify protein expression.
Immunoprecipitation
Cells were harvested and lysed in IP buffer (50 mM
Tris-Cl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100)
supplemented with proteinase inhibitors 1 mM PMSF, 10 µg/ml
aprotonin, 5 µg/ml leupeptin, 0.5 µg/ml pepstatin and 5 mM NaF on
ice for 30 min, centrifuged at 12,000 × g for 15 min at 4°C and the
supernatants were collected in fresh tube. For immunoprecipitation,
2 mg lysate was incubated and rotated with 20–30 µl Anti-FLAGM2
Magnetic Beads (cat. no. M8823; Sigma-Aldrich; Merck KGaA) or
complex of protein A/G magnetic beads (cat. no. B23202; Bimake.com) and primary antibody overnight at 4°C. The
resulting immunoprecipitates were washed at least three times in IP
buffer, before boiling with SDS sample buffer. Immunoprecipitated
material was analyzed by mass spectrometry and western
blotting.
Liquid chromatography/mass
spectrometry (LC/MS) and LC/MS-MS platforms for protein
identification
LC-MS/MS experiments were performed on LTQ-Orbitrap
hybrid mass spectrometer (Thermo Fisher Scientific, Inc.) coupled
to a Shimadzu LC-20AD LC system (Shimadzu Corporation) and SIL-20AC
autosampler (Shimadzu Corporation). Tryptic peptides were separated
by a PICOFRIT C18 reverse-phase column (0.075×100 mm, New Objective
Inc.) at a flow rate of 300 nl/min with a 110 min-gradient. Each
peptide mixture was loaded in solvent A [95% H2O, 5%
acetonitrile (ACN), 0.1% formic acid (FA)] and eluted by 5% solvent
B (5% H2O, 95% ACN, 0.1% FA) for 5 min followed by a
linear gradient to 45% solvent B in the next 90 min, then the
column was re-equilibrated at initial condition for 15 min. The
entire eluant was sprayed into the LTQ-Orbitrap mass spectrometer
via a dynamic nanospray probe (Thermo Fisher Scientific, Inc.) and
analyzed in positive mode. The 3 most abundant precursor ions
detected in the full MS survey scan (m/z range of 400–2000,
R=60,000) by Orbitrap were isolated for further MS/MS analyzing in
the linear ion trap. The automatic gain control target was set to
1,000,000 and 10,000 for MS scans and MS/MS scans, respectively.
Tandem mass spectra were extracted by Bioworks (version 3.3.1 SP1,
Thermo Fisher Scientific, Inc.) and submitted to Human
UniProtKB/Swiss-Prot database (Proteome ID: UP000005640, 20199
entries) using TurboSequest v.28 (36) search engine.
ROS detection
Cell suspensions of MDA-MB-231 and MCF-7 were
obtained in PBS and flow cytometry was performed. A total of
1×106 cells were stained with
2′,7′-dichlorodihydrofluorescein diacetate (DCFDA; 1 µM) at 37°C
for 30 min (Thermo Fisher Scientific, Inc.) and then analyzed with
a FACSCalibur™ flow cytometer (BD Biosciences). The percentage of
positively-stained cells was quantified using CELLQuest™ software
v5.1 (BD Biosciences).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The tissues were homogenized with Polytron PT100
(Kinematica AG). Total RNA was extracted using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA, isolated
from cell lines or tissues, was reverse-transcribed using the
ReverTra Ace® qPCR RT Kit (Toyobo Life Science). RT was
conducted for 15 min at 42°C followed by 5 min at 98°C. The mRNA
expression levels were quantified by qPCR using the
SYBR® Green Real-time PCR Master Mix (Toyobo Life
Science). qPCR was performed using an Applied Biosystems 7900HT
real-time PCR system, and data were collected and analyzed using
ABI SDS version 2.3. The thermocycling conditions were: 95°C for 30
sec followed by 40 cycles at 95°C for 5 sec and 65°C for 30 sec.
The mRNA expression levels of GAPDH were used as the internal
control for gene-specific mRNA analysis. The normalized expression
of each sample was designated as the quantification cycle (Cq) and
obtained by dividing the Cq value by the Cq value of GAPDH of the
same sample. The relative amount of mRNA in each sample was
calculated using the comparative Cq method (37). The results are presented as the
fold change of expression in cells or cancer tissues. The sequences
of primers were listed in Table
II.
 | Table II.List of primers employed reverse
transcription-quantitative polymerase chain reaction. |
Table II.
List of primers employed reverse
transcription-quantitative polymerase chain reaction.
| Gene | Sequence type | Sequence
(5′-3′) |
|---|
| TAGLN2 | Sense |
ACCCAGTGCCGAAAGGATG |
|
| Antisense |
TCAGTGGTGTTAATGCCATAGC |
| VCAM1 | Sense |
GCTGCTCAGATTGGAGACTCA |
|
| Antisense |
CGCTCAGAGGGCTGTCTATC |
| ICAM1 | Sense |
ATGCCCAGACATCTGTGTCC |
|
| Antisense |
GGGGTCTCTATGCCCAACAA |
| CX3CL1 | Sense |
ACCACGGTGTGACGAAATG |
|
| Antisense |
CTCCAAGATGATTGCGCGTTT |
| CCL2 | Sense |
CAGCCAGATGCAATCAATGCC |
|
| Antisense |
TGGAATCCTGAACCCACTTCT |
| CCR4 | Sense |
ATGAACCCCACGGATATAGCA |
|
| Antisense |
CACCACAGAATTTCCAAGCAGA |
| CXCR4 | Sense |
TACACCGAGGAAATGGGCTCA |
|
| Antisense |
AGATGATGGAGTAGATGGTGGG |
| IL6 | Sense |
AACCTGAACCTTCCAAAGATGG |
|
| Antisense |
TCTGGCTTGTTCCTCACTACT |
| MMP1 | Sense |
ACACATCTGACCTACAGGATTGA |
|
| Antisense |
GTGTGACATTACTCCAGAGTTGG |
| MMP2 | Sense |
CCCACTGCGGTTTTCTCGAAT |
|
| Antisense |
CAAAGGGGTATCCATCGCCAT |
| MMP9 | Sense |
TGTACCGCTATGGTTACACTCG |
|
| Antisense |
GGCAGGGACAGTTGCTTCT |
| β-actin | Sense |
TGACGTGGACATCCGCAAAG |
|
| Antisense |
CTGGAAGGTGGACAGCGAGG |
Invasion assay
The invasive potential of cells was determined by
plating 40,000 cells in the upper chamber of Matrigel (BD
Biosciences)-coated Transwell invasion chambers according to the
manufacturer's instructions. The cells were seeded in DMEM without
growth factors; DMEM with 10% FBS was added to the lower chamber.
The cells were incubated in 37°C for 24–48 h. The invasive cells
were fixed in 95% ethanol for 20 min, and stained in hematoxylin
for 2 min followed by eosin staining for 1 min in room temperature.
The invasive cells were imaged under a light microscope
(magnification, ×10) and counted using ImageJ 1.50i software
(National Institutes of Health); 5 fields per view were
analyzed.
Migration assay
Migration assay was conducted with Transparent PET
Membrane 24 well 8.0 µm migration insert (Falcon; Corning Inc.) by
seeding 36,000 cells into the top chamber in DMEM without growth
factors and adding DMEM with 10% FBS to the bottom chamber. The
cells were incubated at 37°C for 6 h. Cells were stained with 0.1%
Crystal Violet Staining Solution for 30 min at room temperature and
imaged under a light microscope (magnification, ×10). The number of
migrated cells was counted with ImageJ 1.50i software (National
Institutes of Health) and 5 fields per view were analyzed. SN50, an
NF-κB inhibitor, from MedChemExpress (cat. no. HY-P0151) was
dissolved in dimethyl sulfoxide (DMSO). MDA-MB-231 cells
transfected with scramble or shTAGLN2 were treated with 18 µM SN50
or DMSO for 24 h before migration assay.
Mitochondrial and F-actin
staining
Cells were cultured on coverslips and treated as
aforementioned. Cells were fixed in 4% paraformaldehyde for 10 min
at room temperature prior to permeabilization in PBS containing
0.1% Triton X-100 for 2 min. MitoTracker Red CMXRos (Thermo Fisher
Scientific, Inc.) was used for mitochondrial staining at 37°C for
30 min. Phalloidin-fluorescein isothiocyanate (Sigma-Aldrich; Merck
KGaA) was used for F-actin staining at 37°C for 30 min. Cells were
rinsed 3 times with PBS and incubated with DAPI (1 µg/ml; cat. no.
9542; Sigma-Aldrich; Merck KGaA) at 37°C for 30 min. Slides were
analyzed by Leica DMi6000 B inverted microscope (Leica
Microsystems, Inc.) according to manufacturer's instruction and 3
fields per view were analyzed.
In vivo metastasis assay
Female athymic mice (Shanghai SLAC Laboratory Animal
Co., Ltd.), 4–6 weeks of age, 18–22 g in weight were employed in
the present study. A total of 5×106 MDA-MB-231 cells
stably expressing scramble shRNA or shRNA-TAGLN2 were intravenously
injected into mice via the tail vein (6 mice per group). At 6 weeks
post-injection, mice in each group were sacrificed and the lungs
were dissected for the analysis of metastasis.
In addition, 1×107 MDA-MB-231HM cells
transfected with TAGLN2 or control vectors were injected into the
mammary fat pad of mice (6 mice per group). The growth of tumors
was monitored twice per week for ~2 weeks, when tumors were 1
cm3, the primary tumor was surgically removed under
anesthesia. After 4 weeks, mice were sacrificed under anesthesia
and the lungs were collected, fixed in 10% phosphate-buffered
formalin solution for 12 h at room temperature and embedded in
paraffin. Sections of the lungs were subjected to H&E staining
for 3 min at room temperature; three slides per lung section were
imaged under a light microscope (magnifications, ×10 and 20). The
presence of metastatic nodes in the lung were determined by two
independent pathologists, who were blinded to the experimental
conditions.
Statistical analysis
The results of at least three experiments are
expressed as the mean ± standard deviation. One-way ANOVA was used
for comparisons between multiple groups and post hoc multiple
comparisons were performed using Student-Newman-Keuls test. A
Student's t-test was used for the test and control samples,
respectively. A Fisher's exact test and a χ2 test were
used to analyze categorical patient variables. P<0.05 was
considered to indicate a statistically significant difference.
Statistical calculations were performed using STATA software
version 14.0 (StataCorp LLC); statistical analysis of the
clinicopathological factors was performed with GraphPad Prism 6.0
software (GraphPad Software, Inc.) and SPSS Software version 17.0
(SPSS, Inc.).
Results
TAGLN2 is a suppressor of metastasis
in human breast cancer
TAGLN2 was downregulated in MDA-MB-231HM cells,
which are a highly metastatic variant of the parental MDA-MB-231
cell line (7). To further
understand the link between TAGLN2 and breast cancer metastasis, we
analyzed the expression of TAGLN2 in 158 primary tumor samples from
patients with breast cancer at the Fudan University Shanghai Cancer
Center. Associations between TAGLN2 expression and the
clinicopathological parameters of the breast cancer patients were
summarized in Table III. The
results revealed that no significant association was observed
between TAGLN2 expression and age, histological type, tumor size,
stage, and ER, PR or HER2 status. However, TAGLN2 expression was
upregulated by >4.9-fold in the tumor specimens from lymph
node-negative patients compared with that of lymph node-positive
patients. Additionally, TAGLN2 expression was upregulated by
11.4-fold in the primary tumors of metastasis-negative patients
compared with those exhibiting metastasis (Table III). These findings suggested
that low levels of TAGLN2 were associated with the metastatic
behavior in breast cancer.
 | Table III.Association between TAGLN2 expression
and clinicopathologic parameters in breast cancer. |
Table III.
Association between TAGLN2 expression
and clinicopathologic parameters in breast cancer.
| Clinicopathological
parameters | Number of
cases | Median expression
of TAGLN2 (normalized Cq)a | P-value |
|---|
| Age, years |
|
| 0.676 |
|
≤40 | 26 | 10.158±3.124 |
|
|
>40 | 132 | 9.972±3.962 |
|
| Histological
type |
|
| 0.887 |
|
DCIS | 20 | 10.67±3.951 |
|
|
IDC | 138 | 9.973±3.542 |
|
| TNM stage |
|
| 0.523 |
|
I+II | 146 | 9.986±3.734 |
|
|
III | 12 | 9.831±3.894 |
|
| Lymph node
status |
|
| <0.001 |
|
Negative | 76 | 8.780±3.751 |
|
|
Positive | 82 | 11.081±3.926 |
|
| Tumor size |
|
| 0.322 |
| T1 +
T2 | 132 | 9.888±3.619 |
|
| T3 +
T4 | 26 | 10.412±3.820 |
|
| ER |
|
| 0.713 |
|
Negative | 61 | 9.875±3.756 |
|
|
Positive | 97 | 10.037±3.852 |
|
| PR |
|
| 0.923 |
|
Negative | 66 | 9.891±3.762 |
|
|
Positive | 92 | 10.034±3.834 |
|
| HER2 |
|
| 0.874 |
|
Negative | 125 | 9.806±3.782 |
|
|
Positive | 33 | 10.613±3.696 |
|
| Distant
metastasisb |
|
| <0.001 |
|
Absent | 117 | 9.064±3.715 |
|
|
Present | 41 | 12.573±3.786 |
|
| All cases | 158 | 9.974±3.770 |
|
Loss of TAGLN2 is associated with an
aggressive tumor phenotype
The present study reported that downregulation of
TAGLN2 was associated with breast cancer metastasis; thus, it was
proposed that knockdown of TAGLN2 in breast cancer cells could
promote cell invasion and metastasis. We established cell lines
exhibiting stable TAGLN2 silencing from the parental MDA-MB-231 and
BT-549 cell lines using the pLKO-shRNA vector (Fig. 1A). The following experiments were
conducted with shRNA252 in MDA-MB-231 and shRNA377 in BT-549 cells.
The results indicated that knockdown of TAGLN2 significantly
promoted the invasive abilities of MDA-MB-231 and BT-549 cells
compared with the corresponding control (Fig. 1B and C).
To further investigate the role of TAGLN2 in tumor
metastasis in vivo, MDA-MB-231 cells that were stably
depleted of TAGLN2 were injected into nude mice via the lateral
tail vein. Histological analysis of the mice lungs revealed that
knockdown of TAGLN2 could promote lung metastasis. The numbers and
size of the lung nodules were significantly increased in the
MDA-MB-231-shTAGLN2 group compared with that of the
MDA-MB-231-scramble group (Fig.
1D). Collectively, the results of the present study suggest
that TAGLN2 could be a negative regulator of breast cancer
metastasis.
TAGLN2 directly interacts with PRDX1
and loss of TAGLN2 promotes ROS production
TAGLN2 has been reported as an actin-binding
protein, and it may serve a role in regulating the cytoskeleton,
which cooperatively functions in a variety of cellular processes
and regulates the distribution of mitochondria (38). The present study revealed that
mitochondria were sparsely distributed in TAGLN2-knockdown cells;
however, no alterations in the morphology of F-actin were detected
(Fig. 2A). To investigate the
molecular mechanisms of TAGLN2 in breast cancer metastasis, we
examined the proteins that interact with TAGLN2. TAGLN2 protein was
immunoprecipitated from MDA-MB-231 lysates; the
co-immunoprecipitated proteins were analyzed by MS, and the
identified peptides were matched to protein databases to identify
the parent proteins (Fig. 2B;
Table IV). FLAG-tagged TAGLN2
vector was transfected in MDA-MB-231 cells and TAGLN2 was
overexpressed; PRDX1 was determined to interact with TAGLN2 by
immunoprecipitation analysis (Fig.
2C). In addition, the expression of PRDX1 was reduced in
response to TAGLN2 silencing (Fig.
2D). We also downregulated TAGLN2 in MCF-7 cells (Fig. 2E).
 | Figure 2.TAGLN2 promotes ROS production by
interacting with PRDX1 and altering actin dynamics. (A)
MDA-MB-231-scramble (left panel) and MDA-MB-231-shRNA-TAGLN2 (right
panel) cells were stained for F-actin (green), mitochondria (red)
and the nucleus (blue). Scale bar, 5 µm. (B) Liquid
chromatography-MS analysis followed by MS/MS of immunoprecipitated
FLAG-TAGLN2 from MDA-MB-231 cells revealed the interaction between
PRDX1 and TAGLN2 in MDA-MB-231 cells. (C) Overexpression of TAGLN2,
and co-immunoprecipitation of PRDX1 with TAGLN2. MDA-MB-231 cells
were transfected with FLAG-tagged TAGLN2 and subjected to
immunoprecipitation with anti-FLAG antibody followed by
immunoblotting with antibodies. (D) Representative immunoblots of
PRDX1 with TAGLN2 expression following TAGLN2 depletion in
MDA-MB-231 cells. (E) The mRNA expression levels of TAGLN2 with
TAGLN2 siRNA in MDA-MB-231 and MCF-7 cells. (F) Levels of ROS in
MDA-MB-231 and MCF-7 cells treated with siRNA TAGLN2 as determined
by flow cytometry, and presented as the fluorescence intensity of
2′,7′-dichlorodihydrofluorescein diacetate. (G) The data were
representative of five independent experiments. *P<0.05 and
****P<0.00005 vs. control. MS, mass spectrometry; PRDX1,
peroxiredoxin 1; ROS, reactive oxygen species; shRNA, short hairpin
RNA; siRNA, small interfering RNA; TAGLN2, transgelin 2. |
 | Table IV.Parent proteins. |
Table IV.
Parent proteins.
| Protein name
(Uniprot ID) | MS-identified
sequence | MH+ | z | m/z |
|---|
| Transgelin-2
(TAGL2) | NFSDNQLQEGK | 1279.59 | 2 | 640.30 |
| (P37802) | TLM*NLGGLAVAR | 1231.68 | 2 | 616.35 |
|
| TLMNLGGLAVAR | 1215.69 | 2 | 608.35 |
|
|
QM*EQISQFLQAAER | 1694.82 | 2 | 847.91 |
|
| DDGLFSGDPNWFPK | 1594.72 | 2 | 797.86 |
|
| QMEQISQFLQAAER | 1678.82 | 2 | 839.91 |
|
|
YGINTTDIFQTVDLWEGK | 2100.03 | 2 | 1050.52 |
| Peroxiredoxin-1
(PRDX1) (Q06830) | ATAVMPDGQFK | 1164.57 | 2 | 582.79 |
|
| QITVNDLPVGR | 1211.67 | 2 | 606.34 |
|
|
QGGLGPM*NIPLVSDPK | 1638.85 | 2 | 819.93 |
|
|
QGGLGPMNIPLVSDPK | 1622.86 | 2 | 811.93 |
|
| GLFIIDDKGILR | 1359.80 | 2 | 680.40 |
Mitochondria are a major cellular source of ROS
(39). PRDX1 is also involved in
cellular ROS production (30).
Flow cytometry was conducted to measure the levels of intracellular
ROS following TAGLN2 downregulation (Fig. 2F). TAGLN2-depleted MDA-MB-231 and
MCF-7 cells were stained with DCFDA; ROS production was then
analyzed by flow cytometry (40).
ROS levels were significantly increased by 21 and 12% in MDA-MB-231
and MCF-7 cells following TAGLN2 knockdown, respectively, compared
with the corresponding controls (Fig.
2F and G). These results suggest that downregulation of TAGLN2
induced the rearrangement of the mitochondria and the abnormal
expression of PRDX1, by which several mechanisms underlying ROS
production were induced.
Downregulation of TAGLN2 activates the
NF-κB pathway in breast cancer cell lines
ROS have been reported to promote cellular migration
and invasion by activating downstream signaling, including the
NF-κB signaling pathway (41).
Thus, we proposed that TAGLN2 knockdown could induce ROS
production, activating the NF-κB signaling pathway. Therefore, a
luciferase-reporter system was generated to monitor NF-κB
activation. The luciferase activity within the reporter system
increased in a dose-dependent manner following treatment with
various concentrations of tumor necrosis factor-α (Fig. S1A). Furthermore, in cells
transfected with the luciferase reporter vector and NF-κB
transcription responsive elements (TREs) were treated with
siRNA-TAGLN2 or scramble siRNA. As expected, cells with NF-κB TREs
exhibited a 2.2-fold increase in luciferase expression in response
to TAGLN2 knockdown (Fig. 3A).
Western blot analysis was performed to examine NF-κB pathway
activation. The results revealed that the expression levels of p50
and p65 were upregulated in MDA-MB-231 and MCF-7 cells transfected
with shTAGLN2. Of note, IκB expression was suppressed following
downregulation of TAGLN2 (Fig.
3B). In addition, the nuclear localization of p65 was analyzed,
and served as a marker of activated NF-κB (Fig. S1B). In 10 human breast cancer cell
lines, TAGLN2 expression was negatively correlated with the nuclear
expression of p65 (Fig. S1C).
Similar trends in the expression of p50 and p100 were observed in
ZR-75-1, MCF-7, T47D, MDA-MB-231 and MDA-MB-468 cells (Fig. S1B and C). To further investigate
the association between TAGLN2 and NF-κB activation, MDA-MB-231
cells transfected with scramble or shTAGLN2 were treated with 18 µM
SN50, an NF-κB inhibitor, for 24 h. Following inhibition of the
NF-κB signaling pathway, the pro-migratory effects of TAGLN2
downregulation were reversed (Fig.
3C). These results supported the findings of the luciferase
reporter assay and revealed that knockdown of TAGLN2 promoted the
activation of NF-κB pathway in breast cancer cells.
Downregulation of TAGLN2 promotes
microenvironment-associated gene expression in breast cancer
cells
The present study reported that knockdown of TAGLN2
promoted the formation of lung metastases by breast cancer cells.
To determine the genes that were associated with the metastatic
phenotype, RT-qPCR analysis of the microenvironment-associated
genes was conducted, including genes involved in the immune and
inflammatory responses, such as cell adhesion molecules, chemokines
and MMPs (42). The results
indicated that interleukin-6, C-X-C motif chemokine ligand 1,
CXCR4, MMP1 and MMP2 were significantly upregulated in MDA-MB-231
cells transfected with shTAGLN2 compared with the control (Fig. 4A). MMP1, MMP2 and MMP9 were
significantly upregulated in MCF-7 cells following TAGLN2 knockdown
compared with the control (Fig.
4B). Western blot analysis was performed to examine the
expression of the metastasis-associated genes. In accordance with
results of the RT-qPCR analysis, MMP1 and MMP2 were upregulated
following downregulation of TAGLN2 in MDA-MB-231 and MCF-7 cells
(Fig. 4C). Furthermore, MDA-MB-231
scramble and shTAGLN2-transfected cells were treated with SN50 to
inhibit the NF-κB signaling pathway. The results demonstrated that
the expression levels of MMP1 and MMP2 were downregulated in
response to SN50 treatment compared with shTAGLN2 transfection
alone (Fig. 4D). These findings
suggested that downregulation of TAGLN2 promoted the expression of
microenvironment-associated genes in breast cancer cells.
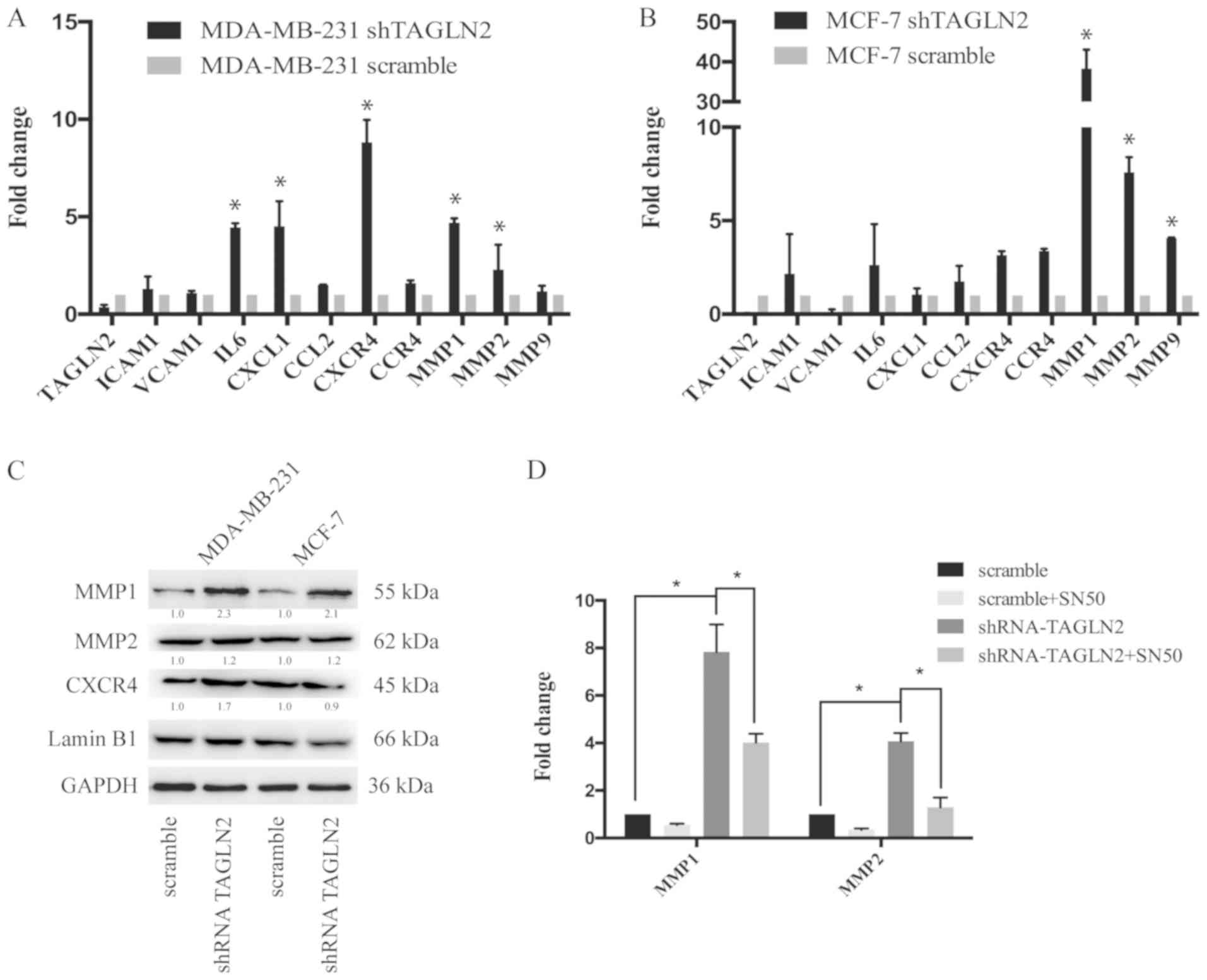 | Figure 4.Analysis of metastasis-associated
genes in TAGLN2-knockdown cancer cells. (A) RT-qPCR analysis of
ICAM1, VCAM1, IL6, CXCL1, CCL2, CXCR4, CCR4, MMP1, MMP2, and MMP9
mRNA expression in MDA-MB-231 cells treated with scramble or
shTAGLN2; results are presented relative to β-actin expression.
Data shown was representative of three independent experiments. (B)
RT-qPCR analysis of ICAM1, VCAM1, IL6, CXCL1, CCL2, CXCR4, CCR4,
MMP1, MMP2 and MMP9 mRNA expression in MCF-7 cells treated with
shControl or shTAGLN2; results are presented as relative to β-actin
expression. Data are representative of three independent
experiments. (C) Immunoblotting of CXCR4, MMP1 and MMP2 expression
in MDA-MB-231 and MCF-7 cells treated with scramble or shTAGLN2.
Western blots were representative of three independent experiments.
(D) RT-qPCR analysis of MMP1 and MMP2 in MDA-MB-231 cells
transfected with scramble or shTAGLN2 under treatment of DMSO or
SN50 (18 µM) for 24 h. Data are representative of three independent
experiments. *P<0.05 vs. scramble or as indicated. CCL2, C-C
motif chemokine ligand 2; CXCL1, C-X-C motif chemokine ligand 1;
CCR4, C-C chemokine receptor type 4; CXCR4, including C-X-C
chemokine receptor 4; DMSO, dimethyl sulfoxide; ICAM1,
intercellular adhesion molecule 1; IL6, interleukin-6; MMP, matrix
metalloproteinase; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; shRNA, short hairpin RNA; TAGLN2,
transgelin 2; VCAM1, vascular cell adhesion protein 1. |
Overexpression of TAGLN2 suppresses in
vivo breast cancer cell lung metastasis in a mouse model
To further examine the effects of TAGLN2 on
host-tumor interactions in a mouse model, MDA-MB-231HM cells stably
expressing the control or TAGLN2 overexpression vectors were
injected into the mammary fat pad of athymic mice. Compared with
the control group, the orthotropic transplantation of
pCDH-CMV-TAGLN2-transfected MDA-MB-231HM cells resulted in fewer
metastatic tubes in the lungs in vivo (Fig. 5A). Statistical analysis revealed a
significant decrease in the number of metastatic tubes derived from
the TAGLN2-overexpressing MDA-MB-231HM cells than of the control
cells (Fig. 5B). Collectively, the
findings of the present study suggested that overexpression of
TAGLN2 suppressed breast cancer cell metastasis in a mouse
model.
Discussion
In the present study, loss of TAGLN2 was reported to
promote breast cancer cell invasion and metastasis by increasing
intracellular ROS levels and activation of the NF-κB signaling
pathway. The effects of TAGLN2 may be mediated by its interaction
with PRDX1 and the redistribution of the mitochondria (Fig. 6).
TAGLN, which contains a calponin-like domain,
functions as a tumor suppressor (43,44).
Loss of TAGLN has been associated with the progression,
differentiation and metastasis of colon cancer (18). TAGLN also inhibits tumor cell
invasion via MMP9 suppression (45). Similar to TAGLN, TAGLN2 also
belongs to the calponin family of proteins and may act as a tumor
suppressor (14). Discrepancies in
the functions of TAGLN2 have been reported previously. Upregulated
TAGLN2 expression was associated with the development of gliomas
(46), uterine cervical squamous
carcinoma (47) and esophageal
squamous carcinoma (48). On the
contrary, Yoshida et al (17) reported that TAGLN2 was
downregulated in metastatic tumors of the brain than in primary
endometrial and ovarian cancers, suggesting TAGLN2 as a suppressor
of metastasis. Zheng et al (49) revealed that knockdown of TAGLN2
improved the sensitivity of paclitaxel-resistant MCF-7 cells to
paclitaxel treatment, and suppressed cellular migration and
invasion; however, the mechanisms underlying the functions of
TAGLN2 remain poorly understood. The results of the present study
indicated that TAGLN2 was downregulated in metastasis-positive
patients with breast cancer and MDA-MB-231HM cells. Therefore,
TAGLN2 may be negatively associated with breast cancer metastasis.
Depleted TAGLN2 expression by RNA interference could enhance the
metastatic properties including invasion ability in vitro
and promotion of lung metastasis in vivo of breast cancer
cells. Xenograft models indicated that TAGLN2 inhibited the
metastasis of breast cancer cells to the lungs. The findings of the
present study suggest that TAGLN2 may serve as a suppressor of
breast cancer metastasis.
Using LC/MS, we identified several
TAGLN2-interacting proteins, including oxidation-regulation protein
PRDX1, which has been reported to reduce the effects of ROS and
regulate ROS-dependent signaling pathways (32,50).
Additionally, downregulation of TAGLN2 was proposed to reduce the
expression of PRDX1 in the present study, which may partially
contribute to increases in ROS levels. Alternatively, TAGLN2 may
regulate ROS levels through the redistribution of the mitochondria.
The dynamic actin cytoskeleton and specific actin-binding proteins
are required for the regular and ordered transport of the
mitochondria (51–53). The actin cytoskeleton is a key
component involved in the regulation of intracellular ROS
production (54–55). TAGLN2 associated with the
cytoskeleton may serve as a sensor of environmental stress and
participate in the modulation of certain signaling pathways.
Consistent with this notion, TAGLN2 depletion may result in the
redistribution of mitochondria and elevated ROS levels; increased
ROS production in response to TAGLN2 depletion suggests that TAGLN2
may function as a suppressor of metastasis via signaling pathways
mediated by ROS. Activation of the NF-κB pathway induces the
expression of microenvironment-associated genes that promote the
metastasis of breast cancer cells (56). The findings of the present study
may provide insight as to how cytoskeletal proteins may actively
affect cancer metastasis. Therefore, inducing the expression of
TAGLN2 could serve as a therapeutic strategy to suppress the
expression of microenvironment-associated genes in various types of
cancer.
Colonization is a critical, rate-limiting step of
the metastatic cascade (57). The
communication between tumor cells and surrounding cells serves an
important role in colonization (58–59).
CXCR4 is an important molecule involved in the spread and
progression of breast cancer. Increased CXCR4 expression serves as
an indicator of poor prognosis in patients with breast cancer. MMPs
are crucial for the development of healthy tissue architecture, and
serve critical roles in numerous physiological and pathological
processes. Similar to CXCR4, MMPs also play key roles in tumor
invasion and the induction of metastasis. The expression of MMPs
has also been associated with the poor prognosis of patients with
breast cancer (60). Our findings
demonstrated that loss of TAGLN2 induces CXCR4, MMP1 and MMP2
expression, and promotes cancer metastasis. Additionally, TAGLN2
expression was associated with lymph node metastasis and distant
metastasis, which are important prognostic factors in breast cancer
patients. However, the clinical significance of TAGLN2 requires
further investigation, in particular the outcomes of breast cancer
patients of the same age, gender, ER/PR/HER-2 status, receiving
similar treatment, and varying expression levels of TAGLN2, which
we aim to study in the future. In addition, our in vivo
analysis of lung metastasis for comparing MDA-MB-231 cells with
TAGLN2 low/high-expression with the control cells may provide
further insight into the role of TAGLN2 in the present study.
In summary, we identified TAGLN2 as a tumor
suppressor associated with breast cancer metastasis, in which its
effects could be mediated by redox signaling, and may be a novel
role for TAGLN2 in the regulation of cancer cell metastasis.
Therefore, TAGLN2 may be considered as a critical regulator of the
ROS/NF-κB pathway in breast cancer.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Yin Chen and
colleagues at the Shanghai Institute of Materia Medica, Chinese
Academy of Science for their excellent guidance in the animal
experiments. The authors would also like to thank Dr Dongyin Guan
and Dr Dan Liu from Shanghai InnoStar Bio-Tech Co. Ltd. for their
critical analysis of the manuscript.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81302303
and 81502293). The funders had no role in study design, data
collection and analysis, decision to publish or preparation of the
manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GHD, SJY, JW and ZMS made substantial contributions
to the conception and design of the present study. LY, QH and SJY
conducted the majority of the experiments. SGX and XYK assisted in
the acquisition of data, including patient management. LY, QH and
GYL assisted with the analysis and interpretation of data. SJY and
JW wrote, reviewed and revised the manuscript. GHD and ZMS
supervised the research. All authors participated in final approval
of the version to be published and contributed to ensuing that
questions related to the accuracy or integrity of any part of the
work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Fudan University Shanghai Cancer Center. All patients provided
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TAGLN2
|
transgelin 2
|
|
PRDX1
|
peroxiredoxin 1
|
|
ROS
|
reactive oxygen species
|
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
HER2
|
human epidermal growth factor receptor
2
|
References
|
1
|
Berry DA, Cronin KA, Plevritis SK, Fryback
DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD and
Feuer EJ; Cancer Intervention and Surveillance Modeling Network
(CISNET) Collaborators, : Effect of screening and adjuvant therapy
on mortality from breast cancer. N Engl J Med. 353:1784–1792. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eccles SA and Welch DR: Metastasis: Recent
discoveries and novel treatment strategies. Lancet. 369:1742–1757.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lambert AW, Pattabiraman DR and Weinberg
RA: Emerging biological principles of metastasis. Cell.
168:670–691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Geiger T, Madden SF, Gallagher WM, Cox J
and Mann M: Proteomic portrait of human breast cancer progression
identifies novel prognostic markers. Cancer Res. 72:2428–2439.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu SG, Yan PJ and Shao ZM: Differential
proteomic analysis of a highly metastatic variant of human breast
cancer cells using two-dimensional differential gel
electrophoresis. J Cancer Res Clin Oncol. 136:1545–1556. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bertucci F, Birnbaum D and Goncalves A:
Proteomics of breast cancer: Principles and potential clinical
applications. Mol Cell Proteomics. 5:1772–1786. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Murugaesu N, Iravani M, van Weverwijk A,
Ivetic A, Johnson DA, Antonopoulos A, Fearns A, Jamal-Hanjani M,
Sims D, Fenwick K, et al: An in vivo functional screen identifies
ST6GalNAc2 sialyltransferase as a breast cancer metastasis
suppressor. Cancer Discov. 4:304–317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ooms LM, Binge LC, Davies EM, Rahman P,
Conway JR, Gurung R, Ferguson DT, Papa A, Fedele CG, Vieusseux JL,
et al: The inositol polyphosphate 5-phosphatase PIPP regulates
AKT1-dependent breast cancer growth and metastasis. Cancer Cell.
28:155–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Okita Y, Kimura M, Xie R, Chen C, Shen LT,
Kojima Y, Suzuki H, Muratani M, Saitoh M, Semba K, et al: The
transcription factor MAFK induces EMT and malignant progression of
triple-negative breast cancer cells through its target GPNMB. Sci
Signal. 10(pii): eaak93972017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stanier P, Abu-Hayyeh S, Murdoch JN,
Eddleston J and Copp AJ: Paralogous sm22alpha (Tagln) genes map to
mouse chromosomes 1 and 9: Further evidence for a paralogous
relationship. Genomics. 51:144–147. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li M, Li S, Lou Z, Liao X, Zhao X, Meng Z,
Bartlam M and Rao Z: Crystal structure of human transgelin. J
Struct Biol. 162:229–236. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shapland C, Hsuan JJ, Totty NF and Lawson
D: Purification and properties of transgelin: A transformation and
shape change sensitive actin-gelling protein. J Cell Biol.
121:1065–1073. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Elsner M, Rauser S, Maier S, Schöne C,
Balluff B, Meding S, Jung G, Nipp M, Sarioglu H, Maccarrone G, et
al: MALDI imaging mass spectrometry reveals COX7A2, TAGLN2 and
S100-A10 as novel prognostic markers in Barrett's adenocarcinoma. J
Proteomics. 75:4693–4704. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Leung WK, Ching AK, Chan AW, Poon TC, Mian
H, Wong AS, To KF and Wong N: A novel interplay between oncogenic
PFTK1 protein kinase and tumor suppressor TAGLN2 in the control of
liver cancer cell motility. Oncogene. 30:4464–4475. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoshida A, Okamoto N, Tozawa-Ono A,
Koizumi H, Kiguchi K, Ishizuka B, Kumai T and Suzuki N: Proteomic
analysis of differential protein expression by brain metastases of
gynecological malignancies. Human Cell. 26:56–66. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shields JM, Rogers-Graham K and Der CJ:
Loss of transgelin in breast and colon tumors and in RIE-1 cells by
Ras deregulation of gene expression through Raf-independent
pathways. J Biol Chem. 277:9790–9799. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luanpitpong S, Talbott SJ, Rojanasakul Y,
Nimmannit U, Pongrakhananon V, Wang L and Chanvorachote P:
Regulation of lung cancer cell migration and invasion by reactive
oxygen species and caveolin-1. J Biol Chem. 285:38832–38840. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng Y, Miyamoto DT, Wittner BS, Sullivan
JP, Aceto N, Jordan NV, Yu M, Karabacak NM, Comaills V, Morris R,
et al: Expression of β-globin by cancer cells promotes cell
survival during blood-borne dissemination. Nat Commun. 8:143442017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou G, Peng F, Zhong Y, Chen Y, Tang M
and Li D: Rhein suppresses matrix metalloproteinase production by
regulating the Rac1/ROS/MAPK/AP-1 pathway in human ovarian
carcinoma cells. Int J Oncol. 50:933–941. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen B, Liu J, Ho TT, Ding X and Mo YY:
ERK-mediated NF-κB activation through ASIC1 in response to
acidosis. Oncogenesis. 5:e2792016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao L, Chen X, Xiao X, Ma Q and Li W:
Resveratrol inhibits hyperglycemia-driven ROS-induced invasion and
migration of pancreatic cancer cells via suppression of the ERK and
p38 MAPK signaling pathways. Int J Oncol. 49:735–743. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Księżakowska-Łakoma K, Żyła M and
Wilczyński JR: Mitochondrial dysfunction in cancer. Prz
Menopauzalny. 13:136–144. 2014.PubMed/NCBI
|
|
25
|
Morgan MJ and Liu ZG: Crosstalk of
reactive oxygen species and NF-κB signaling. Cell Res. 21:103–115.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vasquez-Dunddel D, Pan F, Zeng Q,
Gorbounov M, Albesiano E, Fu J, Blosser RL, Tam AJ, Bruno T, Zhang
H, et al: STAT3 regulates arginase-I in myeloid-derived suppressor
cells from cancer patients. J Clin Invest. 123:1580–1589. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang CS, Kim JJ, Lee SJ, Hwang JH, Lee CH,
Lee MS and Jo EK: TLR3-triggered reactive oxygen species contribute
to inflammatory responses by activating signal transducer and
activator of transcription-1. J Immunol. 190:6368–6377. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yoon S, Woo SU, Kang JH, Kim K, Kwon MH,
Park S, Shin HJ, Gwak HS and Chwae YJ: STAT3 transcriptional factor
activated by reactive oxygen species induces IL6 in
starvation-induced autophagy of cancer cells. Autophagy.
6:1125–1138. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Q, Raje V, Yakovlev VA, Yacoub A,
Szczepanek K, Meier J, Derecka M, Chen Q, Hu Y, Sisler J, et al:
Mitochondrial-localized Stat3 promotes breast cancer growth via
phosphorylation of serine 727. J Biol Chem. 288:31280–31288. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Egler RA, Fernandes E, Rothermund K,
Sereika S, de Souza-Pinto N, Jaruga P, Dizdaroglu M and Prochownik
EV: Regulation of reactive oxygen species, DNA damage and c-Myc
function by peroxiredoxin 1. Oncogene. 24:8038–8050. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lv WP, Li MX and Wang L: Peroxiredoxin 1
inhibits lipopolysaccharide-induced oxidative stress in lung tissue
by regulating P38/JNK signaling pathway. Eur Rev Med Pharmacol Sci.
21:1876–1883. 2017.PubMed/NCBI
|
|
32
|
Ding C, Fan X and Wu G: Peroxiredoxin 1-an
antioxidant enzyme in cancer. J Cell Mol Med. 21:193–202. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang J, Lin D, Peng H, Huang Y, Huang J
and Gu J: Cancer-derived immunoglobulin G promotes tumor cell
growth and proliferation through inducing production of reactive
oxygen species. Cell Death Dis. 4:e9452013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cao J, Schulte J, Knight A, Leslie NR,
Zagozdzon A, Bronson R, Manevich Y, Beeson C and Neumann CA: Prdx1
inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J.
28:1505–1517. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Singletary SE and Connolly JL: Breast
cancer staging: Working with the sixth edition of the AJCC Cancer
Staging Manual. CA Cancer J Clin. 56:37–47; quiz 50-1. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lundgren DH, Han DK and Eng JK: Protein
identification using TurboSEQUEST. Curr Protoc Bioinformatics
Chapter. 13:Unit 13.3. 2005. View Article : Google Scholar
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Meng T, Liu L, Hao R, Chen S and Dong Y:
Transgelin-2: A potential oncogenic factor. Tumour Biol.
39:10104283177026502017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Collins Y, Chouchani ET, James AM, Menger
KE, Cochemé HM and Murphy MP: Mitochondrial redox signalling at a
glance. J Cell Sci. 125:801–806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Eruslanov E and Kusmartsev S:
Identification of ROS using oxidized DCFDA and flow-cytometry.
Methods Mol Biol. 594:57–72. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang F, Yang JL, Yu KK, Xu M, Xu YZ, Chen
L, Lu YM, Fang HS, Wang XY, Hu ZQ, et al: Activation of the NF-κB
pathway as a mechanism of alcohol enhanced progression and
metastasis of human hepatocellular carcinoma. Mol Cancer.
14:102015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lu H, Ouyang W and Huang C: Inflammation,
a key event in cancer development. Mol Cancer Res. 4:221–233. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Assinder SJ, Stanton JA and Prasad PD:
Transgelin: An actin-binding protein and tumour suppressor. Int J
Biochem Cell Biol. 41:482–486. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang Z, Chang YJ, Miyamoto H, Ni J, Niu Y,
Chen Z, Chen YL, Yao JL, di Sant'Agnese PA and Chang C: Transgelin
functions as a suppressor via inhibition of ARA54-enhanced androgen
receptor transactivation and prostate cancer cell growth. Mol
Endocrinol. 21:343–358. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nair RR, Solway J and Boyd DD: Expression
cloning identifies transgelin (SM22) as a Novel repressor of 92-kDa
type IV collagenase (MMP-9) expression. J Biol Chem.
281:26424–26436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Han MZ, Xu R, Xu YY, Zhang X, Ni SL, Huang
B, Chen AJ, Wei YZ, Wang S, Li WJ, et al: TAGLN2 is a candidate
prognostic biomarker promoting tumorigenesis in human gliomas. J
Exp Clin Cancer Res. 36:1552017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yakabe K, Murakami A, Kajimura T,
Nishimoto Y, Sueoka K, Sato S, Nawata S and Sugino N: Functional
significance of transgelin-2 in uterine cervical squamous cell
carcinoma. J Obstet Gynaecol Res. 42:566–572. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Du YY, Zhao LM, Chen L, Sang MX, Li J, Ma
M and Liu JF: The tumor-suppressive function of miR-1 by targeting
LASP1 and TAGLN2 in esophageal squamous cell carcinoma. J
Gastroenterol Hepatol. 31:384–393. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zheng X, Chen S, Yang Q, Cai J, Zhang W,
You H, Xing J and Dong Y: Salvianolic acid A reverses the
paclitaxel resistance and inhibits the migration and invasion
abilities of human breast cancer cells by inactivating transgelin
2. Cancer Biol Ther. 16:1407–1414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ahmed W and Lingner J: PRDX1 and MTH1
cooperate to prevent ROS-mediated inhibition of telomerase. Genes
Dev. 32:658–669. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Boldogh IR and Pon LA: Interactions of
mitochondria with the actin cytoskeleton. Biochim Biophys Acta.
1763:450–462. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Rappaport L, Oliviero P and Samuel JL:
Cytoskeleton and mitochondrial morphology and function. Mol Cell
Biochem. 184:101–105. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Moore AS, Wong YC, Simpson CL and Holzbaur
EL: Dynamic actin cycling through mitochondrial subpopulations
locally regulates the fission-fusion balance within mitochondrial
networks. Nat Commun. 7:128862016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Breitenbach M, Laun P and Gimona M: The
actin cytoskeleton, RAS-cAMP signaling and mitochondrial ROS in
yeast apoptosis. Trends Cell Biol. 15:637–639. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bhat SS, Parray AA, Mushtaq U, Fazili KM
and Khanday FA: Actin depolymerization mediated loss of SNTA1
phosphorylation and Rac1 activity has implications on ROS
production, cell migration and apoptosis. Apoptosis. 21:737–748.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Roy P, Sarkar UA and Basak S: The NF-κB
activating pathways in multiple myeloma. Biomedicines. 6:E592018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Klein CA: Cancer. The metastasis cascade.
Science. 321:1785–1787. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shahriari K, Shen F, Worrede-Mahdi A, Liu
Q, Gong Y, Garcia FU and Fatatis A: Cooperation among heterogeneous
prostate cancer cells in the bone metastatic niche. Oncogene.
36:2846–2856. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shibue T, Brooks MW and Weinberg RA: An
integrin-linked machinery of cytoskeletal regulation that enables
experimental tumor initiation and metastatic colonization. Cancer
Cell. 24:481–498. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang J, Chen J, Wo D, Yan H, Liu P, Ma E,
Li L, Zheng L, Chen D, Yu Z, et al: LRP6 ectodomain prevents
SDF-1/CXCR4-induced breast cancer metastasis to lung. Clin Cancer
Res. 25:4832–4845. 2019. View Article : Google Scholar : PubMed/NCBI
|




















