Introduction
Cancer-induced bone pain (CIBP) is a common symptom
of patients with advanced cancer, which seriously affects their
quality of life (1,2). Although pain is common in patients
with bone cancer, effectively preventing and controlling CIBP
remains one of the most difficult tasks for pain management
professionals (3–5). In recent years, oxycodone (OXY) has
become the first line of treatment for CIBP (6). OXY is a semi-synthetic opioid
analgesic derived from the naturally occurring thebaine (7). An improved understanding of the
mechanism of action of OXY is required due to the increase in the
number of OXY-dependent patients and cases of mortality caused by
OXY overuse (8,9).
The phosphorylation of certain molecules in the
central and peripheral nerves can cause hyperalgesia in rats with
CIBP (10,11). For example, the phosphorylation of
calmodulin can activate a variety of signaling pathways to promote
inflammatory factors (12,13). OXY, a G protein-coupled receptor
agonist, inhibits adenylate cyclase activity, thereby inhibiting
cAMP production which affects the phosphorylation of membrane
proteins (14). In nerve cells,
the phosphorylation of membrane proteins alters the permeability of
certain ions, including calcium ions, resulting in increased
calcium influx (15). OXY exerts
analgesic effects by inhibiting adenylate cyclase activity and
regulating the phosphorylation of cellular proteins (14,15).
However, the majority of previous studies have focused on the role
of a specific molecule (16,17).
At present, little is known regarding the alterations in the
phosphorylated protein profile in spinal dorsal horn tissue from
rats with bone cancer-induced pain. Therefore, the effect of OXY on
the phosphorylated protein profile of spinal dorsal horn tissue
during the treatment of rats with CIBP requires further
investigation. In the present study, tandem mass tag (TMT)
phosphorylation proteomics was used to analyze the phosphorylated
molecular signal of OXY in rats with CIBP.
As an essential complement to the postgenomic era,
proteomics techniques have been used to study the proteome
expression levels in various animal pain models (18–20).
Quantitative phosphorylation proteomics analysis based on the
isobaric tag for relative and absolute quantitation/TMT is one of
the most commonly used methods, having numerous advantages over gel
technology, and can directly quantify and compare protein levels in
samples with greater efficiency and accuracy (21). Information concerning up- or
downregulation of phosphorylated proteins or kinases can be
obtained using TMT proteomics techniques, which aid predictions of
potential signal transduction processes (22). The purpose of the present study was
to obtain a phosphorylated protein profile of the spinal cord
dorsal horn tissue of rats with CIBP before and after OXY
administration using the TMT phosphorylation proteomics method. The
present findings may provide novel insight into the mechanism
underlying CIBP and the effects of OXY on hyperalgesia.
Materials and methods
Animals
Adult female Sprague-Dawley rats (9 weeks old,
n=99), weighing 180–220 g, were purchased from ZheJiang Academy of
Medical Sciences and were housed in a 12-h dark/light environment
at 22–24°C with a relative humidity of 40–60%. Food and water were
freely accessible. The rats were acclimated to the environment for
5 days prior to the experiments. They were 33 rats per group in the
study for the various assays; 12 rats in each group were used to
evaluate the PWT and the X-ray of the tibia in rats with CIBP, 21
rats in each group were used for protein quantitative analysis and
WB analysis, and 3 rat tibias were used for HE staining. All animal
experiments and protocols were approved by the Jiaxing University
Institutional Animal Care and Use Committee, and were performed in
accordance with the Association for Assessment and Accreditation of
Laboratory Animal Care (http://www.aaalac.org/accreditation/rules.cfm) and the
Institutional Animal Care and Use Committee (https://blink.ucsd.edu/sponsor/iacuc/links.html#Guidelines)
guidelines All rats were sacrificed by intraperitoneal injection of
excess pentobarbital (100 mg/kg). Death was verified by a lack of
cardiac pulse, and fixed and dilated pupils.
Tumor cell preparation
Tumor cells were prepared as described previously
(23–26). Walker 256 rat breast tumor cells
were donated by Nanjing University. Walker-256 cells were grown in
Dulbecco's modified Eagle medium containing 4.5 g/l glucose, 100
mg/l penicillin, 100 mg/l streptomycin, supplemented with 10% fetal
bovine serum and 5% carbon dioxide at 37°C. The cells (0.5 ml;
2×107 cells/ml) were intraperitoneally injected into the
female rats. After a week, ~5 ml of ascites was extract from the
rats, then centrifuged at 2,500 × g for 3 min at 4°C, then washed
with PBS and suspended in D-Hank solution (PB180321, Procell Life
Science & Technology Co., Ltd.) to reach a final concentration
of 1×107 cells/ml. The same concentration of heat-killed
tumor cells (obtained by heating at a high temperature) was used in
the sham group.
CIBP model
Walker 256 tumor cells were injected into the left
iliac marrow cavity of rats to simulate the development of CIBP
clinical pathophysiology as previously described (23,24,27).
Briefly, 10 µl Walker 256 cells (1×107 cells/ml) were
slowly injected into the medullary cavity of the left tibia of rats
in the CIBP group after the rats were anesthetized with sodium
pentobarbital (60 mg/kg; intraperitoneally). The same method was
used for heat-killed cells in rats in the sham group. Finally, all
rats undergoing inoculation were allowed to recover naturally for 3
days prior to experiments.
Bone X-ray examination
X-ray examination of the left tibia bone was
performed on the 12th and 21st days following cancer cell
inoculation to confirm destruction of the tibial bone caused by
tumor inoculation. The rats underwent a flat X-ray examination
following anesthesia with sodium pentobarbital (60 mg/kg;
intraperitoneal injection). Tumor cell infiltration and bone
destruction were assessed in the bone using an E-COM Technology
Digital Radiographer system (n=3 rats per group; E-COM Technology,
Ltd.).
Histological analysis of bone
Rats (n=3 per group) were sacrificed with
pentobarbital (100 mg/kg; intraperitoneal injection) on the 12th
day after tumor inoculation. The tibial tissue surrounding the
inoculation site was collected (1 cm total) and fixed for 24 h in
the 4% phosphate-buffered paraformaldehyde at 4°C. The tibia was
decalcified in 10% EDTA solution for 24 h at 55°C. Subsequently,
the tissue was dehydrated, embedded in paraffin and cut into 8-µm
sections using a microtome (Reichert-Leica RM2235; Leica
Microsystems GmbH). The extent of tumor cell infiltration and bone
destruction was verified by staining with hematoxylin and eosin.
Briefly, the section was placed in Mayer's hematoxylin dye solution
(cat. no. H9627; Sigma-Aldrich; Merck KGaA) for 5–7 min at room
temperature, and washed with tap water to blue. All images were
captured under a fluorescent microscope using a ×10 or ×20
objective lens (Olympus BX51; Olympus Corporation).
Drug administration
The administration of saline (0.9%) and OXY
hydrochloride injection (OxyNorm; 10 mg/ml; Napp Pharmaceuticals,
Ltd.) was randomized (n=21). OXY was diluted to 0.5 mg/ml with 0.9%
saline and intraperitoneally injected at a dose of 2.5 mg/kg twice
daily for 5 consecutive days from the 8th day after the injection
of tumor cells to establish the CIBP model, as described previously
(7,17,28,29).
Saline was used in the control group.
Mechanical allodynia assessment
The alterations in the pain threshold of the left
hind paw of rats, which were represented by paw withdrawal
threshold (PWT) values (in g), were evaluated using an electronic
von Frey's anesthesia meter (IITC Life Science Inc.). Prior to each
test, the rats were placed in a glass box (25×20×20 cm3)
and were allowed to move freely for 30 min to adapt to the wire
mesh platform. The test was repeated three times with a minimum of
5 min between each stimulus. The test was conducted 1 h before
(pretest) and 30 min after the second daily saline or drug
administration. The test was also conducted 0, 15, 30, 60, 120 and
240 min after last administration. The mean of three tests was used
as the PWT value of the hind paw of each rat. All tests were
performed by investigators blinded to the experimental group.
Protein preparation
The rats were sacrificed 30 min after the last drug
or saline administration on the 12th day, and the lumbar
enlargement was quickly removed and stored in liquid nitrogen
(23) (n=21). The tissue sample
was mixed with a suitable amount of SDT lysate (4% SDS, 100 mM
Tris-HCl, 1 mM DTT; pH 7.6), transferred to a 2-ml centrifuge tube
pre-packed with a suitable amount of quartz sand and homogenized
using an MP homogenizer (24×2; 6.0 M/S; 60 sec; twice).
Subsequently, the sample was sonicated (80 W; work 10 sec,
intermittent 15 sec, cycle 10 times) and kept in boiling water for
15 min. Following centrifugation at 14,000 × g for 40 min at 4°C,
the supernatant was filtered through a 0.22-µm filter and the
filtrate was collected. Protein quantification was performed using
the bicinchoninic acid method. The samples were stored at −80°C,
and 54 samples were used for liquid chromatography (LC)/mass
spectrometry (MS) proteomics analysis, and nine samples were used
for western blot analysis.
TMT labeling
Each of the six protein samples was labeled as a
sample pool, with three sample pools in each group. A total of nine
sample pools were used for TMT labeling. Protein samples (100 µg)
were labeled according to the manufacturer's protocol (TMT 6plex
Mass Tag Labeling Kits and Reagents; cat. no. 90068; Thermo Fisher
Scientific, Inc.). Briefly, the sham group was labeled with 126,
127N and 127C, and the CIBP group was labeled with 128N, 128C and
129N. The OXY group was labeled with 129C, 130N and 130C.
Phosphopeptide enrichment
The labeled peptide solution was freeze-dried, and
1X 1,5-dihydroxybenzoic acid (DHB) buffer was added (5X DHB buffer
comprised 3% DHB, 80% acetonitrile and 0.1% trifluoroacetic acid)
and diluted with water at a 1:4 ratio. The TiO2 beads
were added to the solution, agitated for 40 min and the solution
was centrifuged at 14,000 × g for 40 min at 37°C to remove the
supernatant. The beads were transferred to the stopper tip and
washed. The elution buffer (10 mM Tris-Cl; pH 8.5) was added for
elution, and phosphopeptides were collected, concentrated as
previously described (30) and
dissolved in 30 µl 0.1% formic acid. A total of 20 µl was used for
MS analysis.
LC-MS/MS analysis
A total of 3 sample pools in each group were
separated and injected using the Easy nLC1000 nanoliter flow
high-performance liquid chromatography (Thermo Scientific EASY-nLC
1000 system, Thermo Fisher Scientific, Inc.). Buffer solution A was
0.1% formic acid in water, and solution B was 0.1% formic acid in
acetonitrile (84% for acetonitrile). Furthermore, 95% of solution A
was used to balance the column. The samples were loaded from an
autosampler onto a loading column (Thermo Scientific Easy column; 2
cm ×100 µm, 5 µm-C18; Thermo Fisher Scientific, Inc.) and then
separated using an analytical column (0.075×250 mm, 3 µm-C18) at a
flow rate of 250 nl/min. The relevant liquid-phase gradients were
as follows: 0–220 min, B liquid linear gradient from 0–55%; 220–228
min, B liquid linear gradient from 55–100%; 228–240 min, B fluid
maintained at 100%. Next, the samples were analyzed using a
Q-Exactive mass spectrometer (Thermo Fisher Scientific, Inc.)
following separation using the following parameters: Analysis time,
240 min; detection method, positive ion; parent ion scanning range,
350–1,800 m/z; the primary mass spectrometry resolution, 70,000,
m/z 200; automatic gain control target, 3×106;
first-level maximum ion-trap time (IT), 20 msec; scan range, 1; and
dynamic exclusion time, 30.0 sec; the nitrogen gas temperature was
300°C, the nebulizer pressure 310.28 kpa and the flow rate, 12
l/min.
Database searching
Mass spectral analysis raw data for RAW files using
Mascot 2.2 (Matrix Science, Ltd.) and Proteome Discoverer 1.4
software (Thermo Fisher Scientific, Inc.) were used for
identification and quantitative analysis. The database used in the
present study was uniptot_Rat_35897_20170511.fasta with the
following search parameters: Enzyme, trypsin; max missed cleavages,
2; fixed modifications, carbamidomethyl I; variable modifications,
oxidation (M) phospho (ST) phospho (Y); peptide mass tolerance, ±20
ppm; fragment mass tolerance, 0.1 Da. The results of the filter
parameters were peptide false discovery rate ≤0.01, as described
previously (31). The Proteome
Discoverer 1.4 software was used to perform quantitative analysis
based on the reported peak intensity of peptide ions. The peptide
quantification result was the ratio of the signal intensity value
of the label where the reference sample was located to the signal
intensity value of other labels. The protein quantification result
was the median of the quantitative results of the identified
peptides. The final quantification results were normalized by the
median of each label to eliminate the error in the amount of sample
introduced by human factors in the experiment. Mass spectral data
were searched using the Mascot software and analyzed using Proteome
Discoverer 1.4 software for phosphopeptides. The Phospho RS score
was >50, and Phospho RS site probabilities >75% indicated
that the phosphorylation modification had higher credibility
(32). The MS data were deposited
to the Proteome X change Consortium (http://www.proteomexchange.org) with the dataset
identifier PXD011729.
Bioinformatics analysis
The distribution of individual Gene Ontology (GO)
(33,34) classifications in the target and
overall protein sets was compared using a right-tailed Fisher's
exact test when performing a GO annotation enrichment analysis on
the target protein set by Blast2GO (35). The domain enrichment analysis was
performed with a right-tailed Fisher's exact test using the Pfam
(http://pfam.xfam.org/) database. P<0.05 was
considered to indicate a statistically significant difference.
Western blot analysis
Total protein extracts were obtained from tissue
homogenates using RIPA buffer (Sigma) and were quantified using a
Bicinchoninic Acid Assay kit (Bio-Rad Laboratories, Inc.). Equal
amounts of protein (50 µg) were separated by 10% SDS-PAGE and
transferred onto a PVDF membrane (EMD Millipore). The membrane was
then blocked with 5% non-fat milk for 1 h at room temperature and
incubated with primary antibodies overnight at 4°C. The primary
antibodies used were as follows: Mouse anti-GAPDH (mouse monoclonal
antibody; 1:1,000 dilution; cat. no. ab8245; Abcam) and rabbit
anti-disks large homolog 3 (DLG3; rabbit polyclonal antibody; 1:750
dilution; cat. no. ab3438; Abcam). The membrane was incubated with
horseradish peroxidase-conjugated secondary goat anti-rabbit IgG
antibody (goat polyclonal antibody; 1:3,000 dilution; cat. no.
A0545; Sigma-Aldrich; Merck KGaA) for 1 h at room temperature after
three washes in TBS with Tween-20 (0.05%). Finally, the optical
density of the target tape was analyzed using ImageLab 3.0 software
(Bio-Rad Laboratories, Inc.). GAPDH was used as the internal
control.
Statistical analysis
SPSS 21.0 statistical software (IBM Corp.) was used
for analysis, and measurement data (n=12) are presented as the mean
± SD. One-way ANOVA was used for comparisons among groups followed
by the Bonferroni post hoc test; for time course experiments of
nociception, repeated-measures ANOVA followed by Bonferroni post
hoc test was used. P<0.05 was considered to indicate a
statistically significant difference.
Results
Verification of CIBP model
In the present study, a rat model of CIBP was
established by inoculation of Walker-256 breast cancer cells into
the medullary cavity of the tibia and the model was subsequently
verified by three methods, including X-ray imaging, hematoxylin
staining and determination of PWT of the hind paw. No significant
changes were observed in the X-ray image of the tibia of the sham
group. Cortical bone destruction occurred in the CIBP group rats as
indicated by the black arrow on the 12th days after inoculation of
tumor cells (Fig. 1A). Hematoxylin
staining of the tibia showed that the tumor cells infiltrated into
the medullary cavity on 12th days after modeling in the CIBP rats
as indicated by the black arrow (Fig.
1B); This phenomenon was not observed in the sham group. By
measuring the PWT of hind paw in rats, it was found that there was
no significant difference in the PWT of hind paw in the CIBP group
compared with the sham group on baseline (BL; P>0.05). Compared
with the control group, the PWT of hind paws was statistically
significant (P<0.05) on the fifth day of inoculation of tumor
cells in the CIBP group, and the PWT of hind paws continued to
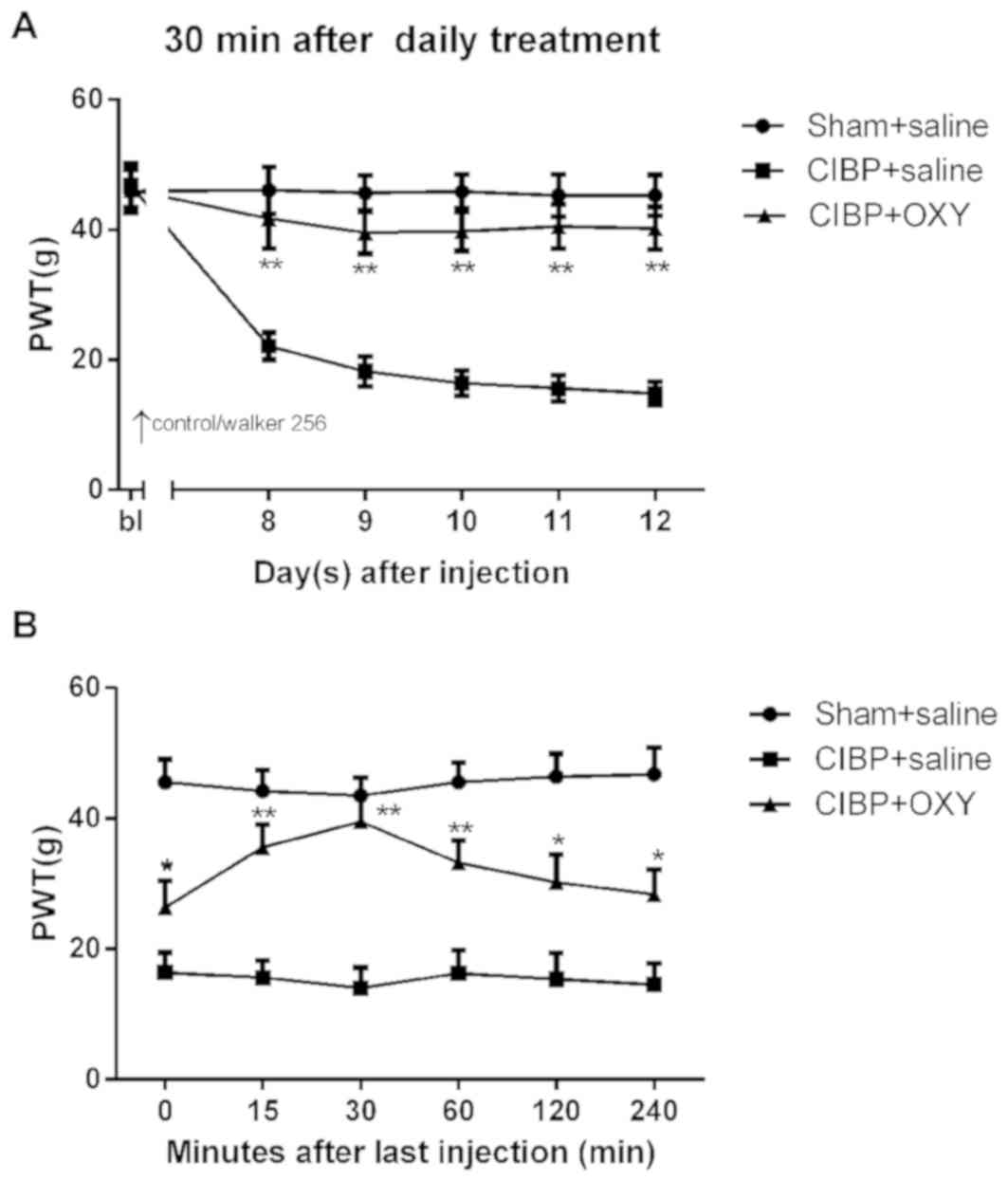
decrease, and began to stabilize after the 12th day (Fig. 1C).
Effects of intraperitoneal injection
of OXY on mechanical allodynia in CIBP
Saline or OXY were intraperitoneally administered in
the sham and CIBP groups to examine the effects of OXY on CIBP
behavior. The preliminary experiments demonstrated that the onset
time of OXY using the systemic administration route was 15 min,
with peak efficacy after 30 min and efficacy lasting for 2–4 h
(Fig. 2B), which was consistent
with the results of previous studies (7,28,36,37).
As shown in Fig. 2A, the PWTs in
the OXY group were increased significantly 30 min after daily
treatment (2.5 mg/kg; intraperitoneally). These results indicated
that OXY (2.5 mg/kg; intraperitoneally) reversed the decreased PWTs
in the CIBP model.
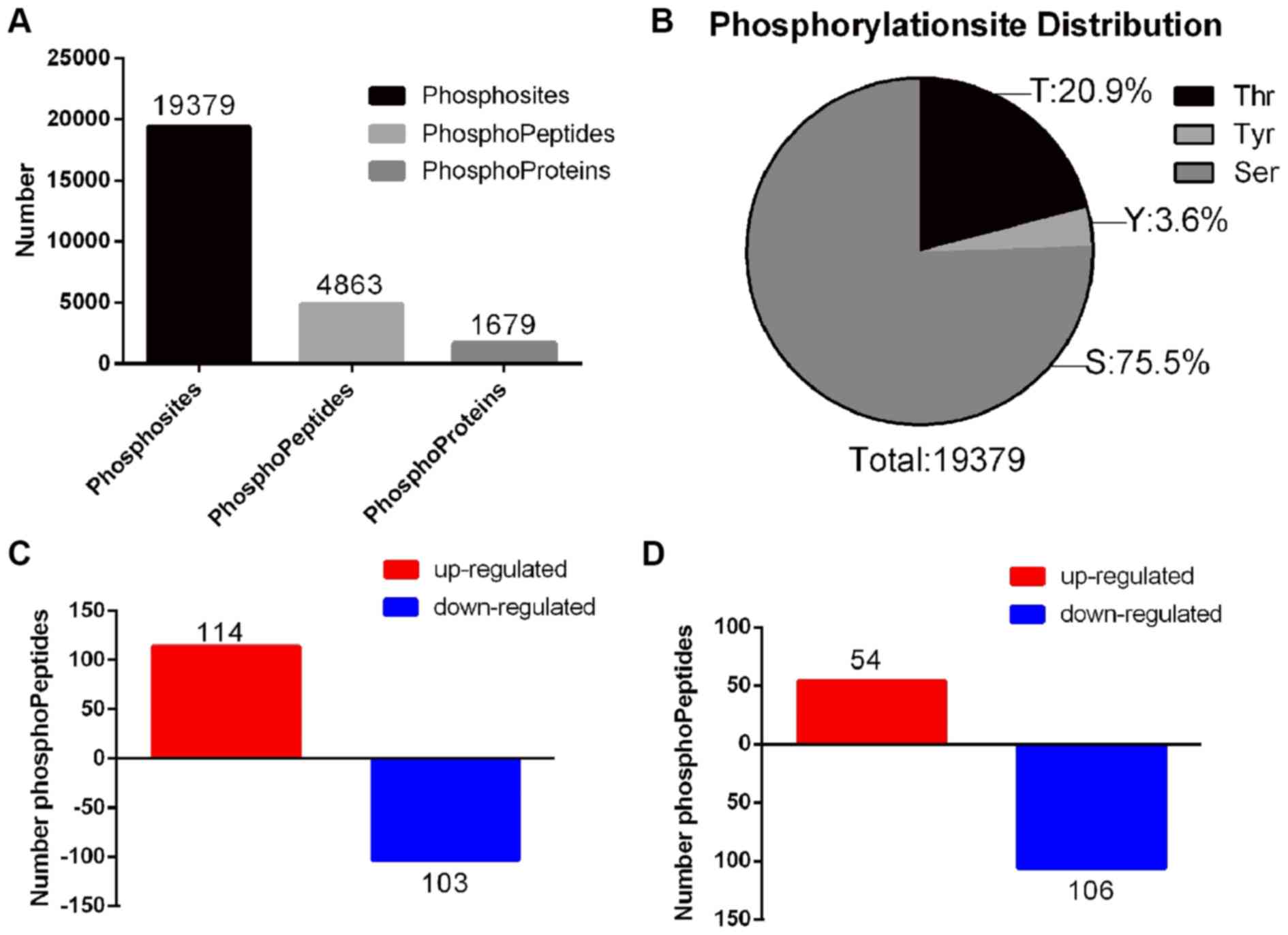
Phosphorylated protein atlas in rats
with CIBP following OXY treatment
TMT quantitative phosphoproteomic technology was
used in the present study to further investigate the alterations in
protein and phosphorylated protein profile of the spinal dorsal
horn following the intraperitoneal injection of OXY in rats with
CIBP. As a result, a total of 4,863 phosphorylated peptides and
1,679 phosphorylated proteins were identified in all three groups
(sham, CIBP and OXY). Additionally, 19,379 phosphosites were
identified (Fig. 3A; Table SI). Among all phosphosites, serine
accounted for 75.5%, threonine for 20.9% and tyrosine for 3.6%
(Fig. 3B). Using a 1.2-fold
cut-off for hypophosphorylation and hyperphosphorylation events
(P<0.05), 217 differentially abundant phosphorylated peptides,
including 160 differentially abundant phosphorylated proteins, were
identified between the CIBP and sham groups. Compared with the sham
group, 114 phosphorylated peptides were upregulated and 103
downregulated in the CIBP group (Fig.
3C; Table SII). A total of
160 differentially abundant phosphorylated peptides, including 113
differentially abundant phosphorylated proteins, were identified
between the CIBP and OXY groups. A total of 54 phosphorylated
peptides were upregulated and 106 were downregulated in the CIBP
group (Fig. 3D; Table SIII). Furthermore, 14
differentially abundant phosphorylated peptides were identified in
all three groups (Table I).
Notably, the levels of all eight upregulated phosphorylated
peptides were decreased and the levels of five downregulated
phosphorylated peptides were increased following OXY treatment in
the CIBP group compared with those in the sham group. Only the
level of one downregulated phosphorylated peptide remained low
following OXY treatment (Table
I).
 | Table I.Differentially abundant
phosphorylated peptides between the CIBP and sham groups, and
between the OXY and CIBP groups. |
Table I.
Differentially abundant
phosphorylated peptides between the CIBP and sham groups, and
between the OXY and CIBP groups.
| Sequence | Protein IDs | Protein name | Phosphorylation
site probabilities (%) site | Fold change
CIBP/Sham | P-value | Fold change
OXY/CIBP | P-value |
|---|
|
sPASVksPGEAksPAEAk | P16884; F1LRZ7 | Neurofilament heavy
polypeptide | S(1): 100.0; S(4):
100.0; S(7): 100.0; S(13): 100.0 | 0.792127 | 0.030177 | 1.301539 | 0.043963 |
|
eEITtFIDEtPLPsPtAsPGPSPRRPRPLGFSPR | D4AAS1 | G protein-coupled
receptor 162 | T(4): 37.8; T(5):
37.8; T(10): 79.9; S(14): 86.1; T(16): 86.1; S(18): 86.1; S(22):
86.1; S(32): 0.0 | 3.394491 | 0.014797 | 0.382391 | 0.029113 |
| gVTSNTsDsESSSk | Q62936 | Disks large homolog
3 | T(3): 0.0; S(4):
0.1; T(6): 96.9; S(7): 3.0; S(9): 99.7; S(11): 0.1; S(12): 0.1;
S(13): 0.1 | 1.288081 | 0.016595 | 0.665242 | 0.0195 |
|
eSPPQPPADDGsEEPGsETSDAkSTPTAEDVTAPLVEER | D4A1Q2 |
Microtubule-associated protein | S(2): 0.0; S(12):
33.0; S(17): 33.0; T(19):33.0; S(20): 33.0; S(24): 33.0; T(25):
33.0; T(27): 1.8; T(32): 0.3 | 1.262656 | 0.019150 | 0.592023 | 0.002063 |
|
eSEAEsDEssDEDsDSEETSk | Q6LDZ3 | Leukocyte common
antigen | S(2): 0.0; S(6):
100.0; S(9): 100.0; S(10):100.0; S(14): 50.0; S(16): 50.0; T(19):
0.1; S(20): 0.0 | 1.611936 | 0.025247 | 0.665296 | 0.007388 |
|
eGQGADkAsEGEEDPGNR | Q4V8H9 | Interferon-induced
protein with tetratricopeptide repeats 2 | S(9): 100.0 | 1.705975 | 0.026870 | 0.644842 | 0.002323 |
|
eALGGNAADsDTEDEDQLQNDkER | A0A0G2JXY3 | Uncharacterized
protein | S(10): 100.0;
T(12): 0.0 | 1.352634 | 0.033045 | 0.795199 | 0.009367 |
|
sQEPISNDQkDsDDDkEk | Q9Z1W6 | Protein LYRIC | S(1): 0.0; S(6):
0.0; S(12): 100.0 | 1.244283 | 0.038597 | 0.787261 | 0.026174 |
| aIEEssEsEssFsD | D3ZKX1 | RCG48807, isoform
CRA_a | S(5): 100.0; S(6):
100.0; S(8): 100.0; S(10): 100.0; S(11): 100.0; S(13): 100.0 | 1.573604 | 0.049062 | 0.807985 | 0.046741 |
| vGSEkGsTGsRDGk | Q80XF7 | Gap junction γ-2
protein | S(3): 0.0; S(7):
100.0; T(8): 0.0; S(10): 100.0 | 0.75329 | 0.004827 | 1.205399 | 0.016649 |
|
sPVPksPVEEVkPkPEAk | G3V7S2 | Neurofilament
medium polypeptide | S(1): 0.0; S(6):
100.0 | 0.811632 | 0.007093 | 1.298441 | 0.040118 |
|
sPAEAksPASVksPGEAk | P16884; F1LRZ7 | Neurofilament heavy
polypeptide | S(1): 100.0; S(7):
100.0; S(10): 2.3; S(13): 97.7 | 1.411921 | 0.003365 | 1.282388 | 0.025597 |
| fAsFIER | P19527 | Neurofilament light
polypeptide | S(3): 100.0 | 0.82836 | 0.040310 | 1.254364 | 0.003244 |
| rFsMEDLNk | O35832 | Cyclin-dependent
kinase 18 | S(3): 100.0 | 0.66579625 | 0.048207 | 0.782255 | 0.030252 |
GO enrichment analysis of cellular
components
GO analysis of the cellular components of
differentially abundant phosphorylated proteins between the CIBP
and sham groups was performed to ascertain how these differentially
abundant phosphorylated proteins contributed to analgesia in rats
with CIBP following OXY treatment. The top 10 enriched cellular
components in the CBP group are presented in Fig. 4A. The top 10 enriched cellular
components between the CIBP and OXY groups are presented in
Fig. 4B. Notably, seven GO terms
were included in both lists. Of these, six GO terms were closely
associated with synaptic function, including GO0044456 (‘synapse
part’), GO0098794 (‘postsynapse’), GO0014069 (‘postsynaptic
density’), GO0099572 (‘postsynaptic specialization’), GO0032279
(‘asymmetric synapse’) and GO0098984 (‘neuron-to-neuron synapse’).
A total of 23 differentially abundant phosphorylated proteins were
associated with these 6 GO terms between the sham and CIBP groups
(Table SIV). Additionally, 22
proteins belonged to these GO terms between the CIBP and OXY groups
(Table SV). These were further
analyzed as candidate proteins, and included eight common
phosphorylated proteins (Tables
SIV and SV) represented by a
red font. Of note, the levels of five phosphorylated proteins,
including phosphorylated microtubule-associated protein 1A,
phosphorylated microtubule-associated protein 1B (MAP1B),
phosphorylated protein bassoon, phosphorylated erythrocyte membrane
protein band 4.1-like 3 and phosphorylated disks large homolog 3
(DLG3), were increased in rats with CIBP compared with in rats in
the sham group. However, the levels of these phosphorylated
proteins decreased following OXY treatment.
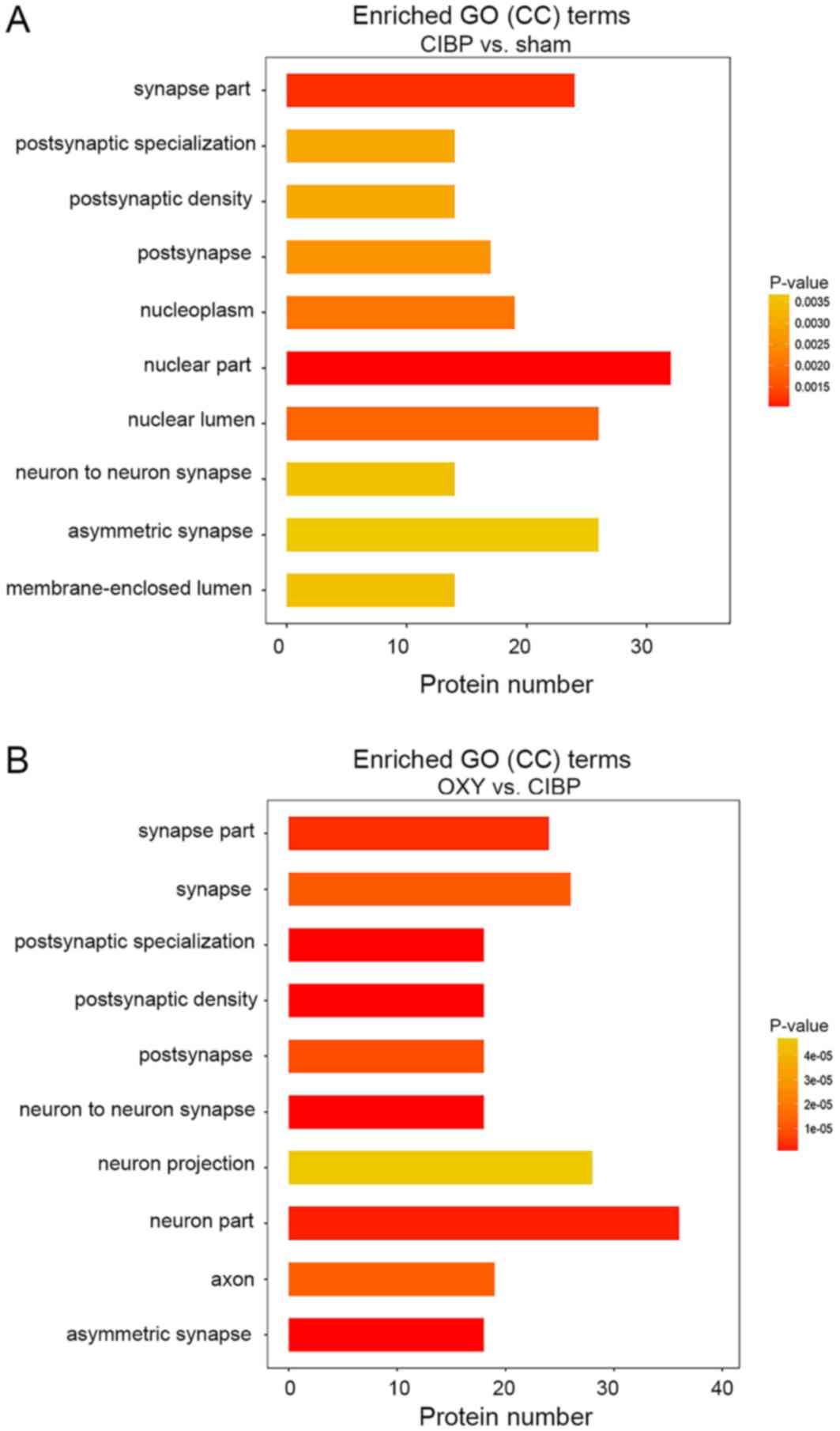 | Figure 4.GO enrichment analysis of CC terms.
(A) Top 10 CCs between the sham and CIBP groups. (B) Top 10 CCs
between the CIBP and OXY groups. Among these, six GO terms were
closely associated with synaptic function, including GO0044456
(‘synapse part’), GO0098794 (‘postsynapse’), GO0014069
(‘postsynaptic density’), GO0099572 (‘postsynaptic
specialization’), GO0032279 (‘asymmetric synapse’) and GO0098984
(‘neuron-to-neuron synapse’). The vertical coordinate is the
process name, and the horizontal coordinate is the number of
differential proteins enriched. A right-tailed Fisher's exact test
was used. The P-value is represented by a color. P<0.05 was
considered to indicate a statistically significant difference. CC,
cellular component; CIBP, cancer-induced bone pain; GO, Gene
Ontology; OXY, oxycodone. |
Domain enrichment analysis of the
differential proteins
Subsequent domain analysis of the differential
proteins compared with the sham group suggested several significant
synapse-associated domains. Furthermore, these domains were also
significantly enriched following OXY treatment of rats with CIBP.
The domains included PF04382: ‘Spectrin/actin-binding (SAB)
domain’, PF05902: ‘4.1 protein C-terminal domain’ (CTD) and
PF00038: ‘intermediate filament protein’ (Fig. 5A and B). In addition, the PF00414:
‘Neuraxin and MAP1B repeat domains of MAP1B’ were significantly
enriched in the CIBP group compared with the sham group, whereas
this domain was not significantly enriched after OXY administration
(Fig. 5A). The PF00625: ‘PDZ
[postsynaptic density-95 (PSD-95)/Disks
large/zonaoccludens-1]-associated domain of N-methyl-D-aspartic
acid (NMDA) receptors’, PF10608: ‘polyubiquitination (PEST)
N-terminal domain of membrane-associated guanylate kinase (MAGUK)’
and PF00625: ‘guanylate kinase domains’ of the bone cancer pain
group were not significantly enriched, whereas these domains were
significantly enriched after OXY treatment compared with those in
the sham group (Fig. 5B).
 | Figure 5.Domain enrichment analysis of
differential proteins. (A) Domain enrichment analysis of the
differential proteins between the sham and CIBP groups. (B) Domain
enrichment analysis of the differential proteins between the CIBP
and OXY groups. The vertical coordinate is the name of the domain
and the horizontal coordinate is the number of differential
proteins enriched. The right-tailed Fisher's exact test was used.
Different colors indicate P-values, and P<0.05 was considered to
indicate a statistically significant difference. AbLIM,
actin-binding LIM protein; CIBP, cancer-induced bone pain; FERM,
4.1 protein ezrin radixin moesin; MAGUK, membrane-associated
guanylate kinase; MAP1B, microtubule-associated protein 1B; MHC I,
major histocompatibility complex I; NMDA, N-methyl-D-aspartic acid;
OXY, oxycodone; PDZ, postsynaptic density-95/disks large
zonaoccludens-1; SAB, spectrin and actin binding; Surp module,
suppressor-of-white-apricot domain. |
Analysis of non-phosphorylated protein
levels
Combined GO and domain analyses revealed that
synapse-associated protein components exhibited differential
enrichment in spinal dorsal horn tissues of rats with CIBP
following OXY treatment, and phosphorylated DLG3 may be a key
molecule in OXY treatment of CIBP. Subsequently, the levels of
non-phosphorylated DLG3 in the spinal dorsal horn tissue were
further analyzed using western blot analysis. As shown in Fig. 6, compared with the sham group, the
DLG3 levels in the spinal dorsal horn tissues of rats with CIBP
were increased, and OXY reduced this effect. It has not been
possible to analyze the alterations in phosphorylation levels using
western blot analysis due to a lack of phosphorylation
site-specific antibodies.
Discussion
The CIBP rat model induced using Walker 256 mammary
tumor cells may simulate the pathogenesis of patients with CIBP,
and has been widely used in CIBP research (23–25).
Of course, a number of studies have also used a variety of other
animal models to study bone cancer pain (38,39).
Schwei et al (38)
established a mouse femur bone cancer (FBC) pain model by injecting
osteolytic mouse sarcoma NCTC2472 cells into the femoral bone
marrow cavity. Similar to the FBC model, the injection of NCTC2472
cells into the mouse calcaneus can establish a calcaneus bone
cancer pain model (39). However,
three types of cancer often cause bone metastases; breast, prostate
and lung cancer (2). Therefore,
breast cancer cells were used to establish a CIBP model in the
present study. In the present study, three methods were used to
validate the CIBP rat model, including radiological examination of
the left tibia bone, hematoxylin staining of the tibia and
measurement of the PWT of the left hind paw. OXY is an opioid
agonist widely used for treating moderate to severe pain in
clinical practice (7). A number of
clinical and animal studies have demonstrated that OXY reaches peak
efficacy at 30 min after systemic administration, with the effects
lasting for 2–4 h (7,36,37).
Therefore, in the present study, rats were sacrificed 30 min after
administration on the 12th day of CIBP rats, and spinal cord tissue
was collected for phosphoproteomic analysis. A total of 1,679
phosphoproteins and 4,863 phosphopeptides were identified, in three
groups using the TMT phosphorylation technique, in addition to
19,379 phosphorylation sites. Interestingly, 14 differentially
phosphorylated peptides belonging to 13 phosphorylated proteins
were identified in the three groups. It was inferred that these
proteins, including neurofilament heavy polypeptide, DLG3 and
microtubule-associated protein, were associated with the analgesic
effects of OXY.
In the present study, all differentially abundant
phosphoproteins were subjected to GO enrichment analysis. Notably,
six synapse-associated GO entries, including GO0044456: ‘synapse
part’, GO0098794: ‘postsynapse’, GO0014069: ‘postsynaptic density’,
GO0099572: ‘postsynaptic specialization’, GO0032279: ‘asymmetric
synapse’ and GO0098984: ‘neuron-to-neuron synapse’, existed in both
rankings. Synapses are the functional link between neurons and a
key part of information transmission (40,41).
Presynaptic calcium influx is a key step in synaptic transmission
(42). The neuronal activity
triggered by long-term continuous excitatory peripheral evoked
impulses can specifically alter the structure and function of
synapses, and this abnormal long-term potentiation of synaptic
transmission is crucial in the development of pain (43,44).
Phosphorylation modification can regulate synaptic function, and
the enhancement of NMDA receptor function in the glutamatergic
postsynaptic membrane is central in the formation of central
hyperalgesia (44,45). The NMDA receptor-gated channel
regulates calcium influx in the postsynaptic membrane (10,42).
Intracellular calcium influx leads to the activation of calmodulin,
further activating a variety of signaling pathways, which serve
important roles in pain transmission (12,13),
including the p38-mitogen-activated protein kinase signaling
pathway (46). Therefore, synaptic
structure and functional adaptation is an important step in the
development of central hyperalgesia (43,44,46).
MAGUKs located in the postsynaptic density, including DLG3, PSD-95,
DLG1 and PSD-93, can anchor NMDA receptors in the postsynaptic
membrane (47,48). Their protein interaction with the
NMDA receptor enhances the function of glutamatergic excitatory
synapses (47,48). The inhibition of postsynaptic
density activity can interfere with NMDA receptor activation and
produce an analgesic effect (47,49).
In the present study, DLG3 abundance was increased in the spinal
dorsal horn tissue in the CIBP group compared with that in the sham
group, and OXY administration reversed this effect. This was
consistent with the results of previous studies (14,50).
Subsequently, domain enrichment analysis was
performed to understand the functional region alterations of these
differentially abundant proteins. The results indicated that the
domains involved in synapse-associated differential proteins were
the PDZ-associated domain of NMDA, polyubiquitination (PEST)
N-terminal domain of MAGUK, intermediate filament protein, neuraxin
and MAP1B repeat, 4.1 protein-ezrin-radixin-moesin N-terminal
domain, 4.1 protein CTD and SAB domain. The differential proteins
with PDZ-associated domain of NMDA receptors and a
polyubiquitination (PEST) N-terminal domain of MAGUK domains were
DLG3 and DLG1 before and after OXY administration in rats with
CIBP, respectively. Unexpectedly, no significant difference in this
domain was identified in rats with CIBP compared with the sham
group, despite an increase in DLG3 abundance. PDZ-associated domain
binds to glutamate NMDA receptor NR2 subunit, promotes NMDA
receptor function activation and enhances glutamatergic excitatory
synaptic activity (47,48,51).
In addition, the N-terminal of DLG3 also possesses a specific NMDA
receptor N2B subunit-binding site (47). Phosphorylation of DLG3, which
depends on calcineurin activation, promotes the transport of
glutamate receptors and their anchoring on the cell membrane
(52). It is worth noting that OXY
inhibits calcium influx in cells and calmodulin activity (14). In the present study, the decrease
in abundance of phosphorylated DLG3 after OXY administration may
have been due to the inhibition of calcineurin. This would result
in the inhibition of glutamatergic synaptic function (14). Therefore, it was hypothesized that
calcineurin was a key kinase for the phosphorylation of synaptic
proteins, whereas OXY could cause analgesia by inhibiting
phosphorylation kinase activity and regulating synaptic protein
phosphorylation. However, the alterations in abundance of
phosphorylated DLG3 were not conclusively due to alterations in the
abundance of non-phosphorylated DLG. The focus of future research
should be on how OXY regulates the functional regions of synaptic
protein phosphorylation.
In conclusion, the present study revealed
alterations in the phosphorylated protein profile of spinal cord
tissues before and after intraperitoneal injection of OXY in rats
with CIBP. By combining phosphorylation proteomics and
bioinformatics analysis, the present study provided a novel
perspective for further understanding the mechanism underlying bone
pain hyperalgesia and the mechanism of action of OXY. The
phosphorylation of synapse-associated cellular component proteins
may have an important role in the development of hyperalgesia in
rats with CIBP.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Professor Renshan
Ge, the former head of the Endocrinology Lab of the Population
Council's Center for Biomedical Research (New York, NY, USA), for
assistance with the English language.
Funding
The present study was supported, in part, by grants
from the Natural Science Foundation of Zhejiang Province (grant
nos. LY17H090019 and LY16H090016), TCM Research Foundation of
Zhejiang Province (grant no. 2016ZA190), Science and Technology
Project of Jiaxing City (grant nos. 2017AY33008, 2017AY33020 and
2016AY23033), the National Natural Science Foundation of China
(grant nos. 81341035 and 81171057), the Construction Project of
Anesthesiology Discipline Special Disease Center in Zhejiang North
Region (grant no. 201524) and the Construction Project of Key
Laboratory of Nerve and Pain Medicine in Jiaxing City.
Availability of data and materials
The MS data have been deposited to the Proteome X
change Consortium with the dataset identifier PXD011729. The
datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
MY, LSX and HSD analyzed and interpreted the data.
HSD, HDN, YGW, HBL, QLH and MX performed the experiments. HSD was a
major contributor in writing the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments and protocols were approved
by the Jiaxing University Institutional Animal Care and Use
Committee (JUMC2018-015) and performed in accordance with AAALAC
and IACUC guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CIBP
|
cancer-induced bone pain
|
|
DLG3
|
disks large homolog 3
|
|
GO
|
Gene Ontology
|
|
LC
|
liquid chromatography
|
|
MAGUK
|
membrane-associated guanylate
kinase
|
|
MS
|
mass spectrometry
|
|
NMDA
|
N-methyl-D-aspartic acid
|
|
OXY
|
oxycodone
|
|
PDZ
|
postsynaptic density-95 (PSD-95)/Disks
large zonaoccludens-1
|
|
PWT
|
paw withdrawal threshold
|
|
TMT
|
tandem mass tag
|
References
|
1
|
Harding D, Giles SL, Brown MRD, Ter Haar
GR, van den Bosch M, Bartels LW, Kim YS, Deppe M and deSouza NM:
Evaluation of quality of life outcomes following palliative
treatment of bone metastases with magnetic resonance-guided high
intensity focused ultrasound: An international multicentre study.
Clin Oncol (R Coll Radiol). 30:233–242. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Buga S and Sarria JE: The management of
pain in metastatic bone disease. Cancer Control. 19:154–166. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mercadante S and Arcuri E: Breakthrough
pain in cancer patients: Pathophysiology and treatment. Cancer
Treat Rev. 24:425–432. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brown M and Farquhar-Smith P: Pain in
cancer survivors; filling in the gaps. Br J Anaesth. 119:723–736.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Frost CØ, Hansen RR and Heegaard AM: Bone
pain: Current and future treatments. Curr Opin Pharmacol. 28:31–37.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ahmad I, Ahmed MM, Ahsraf MF, Naeem A,
Tasleem A, Ahmed M and Farooqi MS: Pain management in metastatic
bone disease: A literature review. Cureus. 10:e32862018.PubMed/NCBI
|
|
7
|
Pöyhiä R and Kalso EA: Antinociceptive
effects and central nervous system depression caused by oxycodone
and morphine in rats. Pharmacol Toxicol. 70:125–130. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cooper ZD, Bedi G, Ramesh D, Balter R,
Comer SD and Haney M: Impact of co-administration of oxycodone and
smoked cannabis on analgesia and abuse liability.
Neuropsychopharmacology. 43:2046–2055. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Coe MA, Nuzzo PA, Lofwall MR and Walsh SL:
Effects of short-term oxycodone maintenance on experimental pain
responses in physically dependent opioid abusers. J Pain.
18:825–834. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hanamura K, Washburn H, Sheffler-Collins
SL, Xia NL, Henderson N, Tillu DV, Hassler S, Spellman DS, Zhang G,
Neubert TA, et al: Extracellular phosphorylation of a receptor
tyrosine kinase controls synaptic localization of NMDA receptors
and regulates pathological pain. PLoS Biol. 15:e20024572017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pareek TK, Keller J, Kesavapany S, Agarwal
N, Kuner R, Pant HC, Iadarola MJ, Brady RO and Kulkarni AB:
Cyclin-dependent kinase 5 modulates nociceptive signaling through
direct phosphorylation of transient receptor potential vanilloid 1.
Proc Natl Acad Sci USA. 104:660–665. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo N, Zhang X, Huang M, Li X, Li Y, Zhou
X and Bai J: Geranylgeranylacetone blocks the reinstatement of
morphine-conditioned place preference. Neuropharmacology.
143:63–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen SP, Sun J, Zhou YQ, Cao F, Braun C,
Luo F, Ye DW and Tian YK: Sinomenine attenuates cancer-induced bone
pain via suppressing microglial JAK2/STAT3 and neuronal CAMKII/CREB
cascades in rat models. Mol Pain. 14:17448069187932322018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vaughan CW, Ingram SL, Connor MA and
Christie MJ: How opioids inhibit GABA-mediated neurotransmission.
Nature. 390:611–614. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Scherer PC, Zaccor NW, Neumann NM, Vasavda
C, Barrow R, Ewald AJ, Rao F, Sumner CJ and Snyder SH: TRPV1 is a
physiological regulator of µ-opioid receptors. Proc Natl Acad Sci
USA. 114:13561–13566. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gaspari S, Cogliani V, Manouras L,
Anderson EM, Mitsi V, Avrampou K, Carr FB and Zachariou V: RGS9-2
modulates responses to oxycodone in pain-free and chronic pain
states. Neuropsychopharmacology. 42:1548–1556. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Largent-Milnes TM, Guo W, Wang HY, Burns
LH and Vanderah TW: Oxycodone plus ultra-low-dose naltrexone
attenuates neuropathic pain and associated mu-opioid receptor-Gs
coupling. J Pain. 9:700–713. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Wang YH, Zhang XH, Ge HY,
Arendt-Nielsen L, Shao JM and Yue SW: Proteomic analysis of
differential proteins related to the neuropathic pain and
neuroprotection in the dorsal root ganglion following its chronic
compression in rats. Exp Brain Res. 189:199–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao Y, Chen S, Xu Q, Yu K, Wang J, Qiao L,
Meng F and Liu J: Proteomic analysis of differential proteins
related to anti-nociceptive effect of electroacupuncture in the
hypothalamus following neuropathic pain in rats. Neurochem Res.
38:1467–1478. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lind AL, Emami Khoonsari P, Sjödin M,
Katila L, Wetterhall M, Gordh T and Kultima K: Spinal cord
stimulation alters protein levels in the cerebrospinal fluid of
neuropathic pain patients: A proteomic mass spectrometric analysis.
Neuromodulation. 19:549–562. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mertins P, Udeshi ND, Clauser KR, Mani DR,
Patel J, Ong SE, Jaffe JD and Carr SA: iTRAQ labeling is superior
to mTRAQ for quantitative global proteomics and phosphoproteomics.
Mol Cell Proteomics. 11:M111.014423. 2012. View Article : Google Scholar :
|
|
22
|
McAlister GC, Huttlin EL, Haas W, Ting L,
Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD
and Gygi SP: Increasing the multiplexing capacity of TMTs using
reporter ion isotopologues with isobaric masses. Anal Chem.
84:7469–7478. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mao-Ying QL, Zhao J, Dong ZQ, Wang J, Yu
J, Yan MF, Zhang YQ, Wu GC and Wang YQ: A rat model of bone cancer
pain induced by intra-tibia inoculation of Walker 256 mammary gland
carcinoma cells. Biochem Biophys Res Commun. 345:1292–1298. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
An K, Rong H, Ni H, Zhu C, Xu L, Liu Q,
Chen Y, Zheng Y, Huang B and Yao M: Spinal PKC activation-Induced
neuronal HMGB1 translocation contributes to hyperalgesia in a bone
cancer pain model in rats. Exp Neurol. 303:80–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang LX and Wang ZJ: Animal and cellular
models of chronic pain. Adv Drug Deliv Rev. 55:949–965. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Ni H, Li H, Deng H, Xu LS, Xu S,
Zhen Y, Shen H, Pan H and Yao M: Nuclear factor kappa B regulated
monocyte chemoattractant protein-1/chemokine CC motif receptor-2
expressing in spinal cord contributes to the maintenance of
cancer-induced bone pain in rats. Mol Pain.
14:17448069187886812018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu M, Yao M, Wang H, Xu L, Zheng Y, Huang
B, Ni H, Xu S, Zhou X and Lian Q: P2Y12
receptor-mediated activation of spinal microglia and p38MAPK
pathway contribute to cancer-induced bone pain. J Pain Res.
10:417–426. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kato A, Minami K, Ito H, Tomii T,
Matsumoto M, Orita S, Kihara T, Narita M and Suzuki T:
Oxycodone-induced analgesic effects in a bone cancer pain model in
mice. Oncology. 74 (Suppl 1):S55–S60. 2008. View Article : Google Scholar
|
|
29
|
Nielsen CK, Ross FB, Lotfipour S, Saini
KS, Edwards SR and Smith MT: Oxycodone and morphine have distinctly
different pharmacological profiles: Radioligand binding and
behavioural studies in two rat models of neuropathic pain. Pain.
132:289–300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Larsen MR, Thingholm TE, Jensen ON,
Roepstorff P and Jørgensen TJ: Highly selective enrichment of
phosphorylated peptides from peptide mixtures using titanium
dioxide microcolumns. Mol Cell Proteomics. 4:873–886. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wiśniewski JR, Zougman A, Nagaraj N and
Mann M: Universal sample preparation method for proteome analysis.
Nat Methods. 6:359–362. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Olsen JV, Blagoev B, Gnad F, Macek B,
Kumar C, Mortensen P and Mann M: Global, in vivo, and site-specific
phosphorylation dynamics in signaling networks. Cell. 127:635–648.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
The Gene Ontology Consortium, . The Gene
Ontology Resource: 20 years and still GOing strong. Nucleic Acids
Res. 47(D1): D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Götz S, García-Gómez JM, Terol J, Williams
TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J and Conesa A:
High-throughput functional annotation and data mining with the
Blast2GO suite. Nucleic Acids Res. 36:3420–3435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peckham EM and Traynor JR: Comparison of
the antinociceptive response to morphine and morphine-like
compounds in male and female Sprague-Dawley rats. J Pharmacol Exp
Ther. 316:1195–1201. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lemberg KK, Kontinen VK, Siiskonen AO,
Viljakka KM, Yli-Kauhaluoma JT, Korpi ER and Kalso EA:
Antinociception by spinal and systemic oxycodone: Why does the
route make a difference? In vitro and in vivo studies in rats.
Anesthesiology. 105:801–812. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schwei MJ, Honore P, Rogers SD,
Salak-Johnson JL, Finke MP, Ramnaraine ML, Clohisy DR and Mantyh
PW: Neurochemical and cellular reorganization of the spinal cord in
a murine model of bone cancer pain. J Neurosci. 19:10886–10897.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cain DM, Wacnik PW, Turner M,
Wendelschafer-Crabb G, Kennedy WR, Wilcox GL and Simone DA:
Functional interactions between tumor and peripheral nerve: Changes
in excitability and morphology of primary afferent fibers in a
murine model of cancer pain. J Neurosci. 21:9367–9376. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li H, Li Y, Lei Z, Wang K and Guo A:
Transformation of odor selectivity from projection neurons to
single mushroom body neurons mapped with dual-color calcium
imaging. Proc Natl Acad Sci USA. 110:12084–12089. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zheng R, Yi X, Lu W and Chen T: Stability
of analytic neural networks with event-triggered synaptic
feedbacks. IEEE Trans Neural Netw Learn Syst. 27:483–494. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Steinert JR, Kopp-Scheinpflug C, Baker C,
Challiss RA, Mistry R, Haustein MD, Griffin SJ, Tong H, Graham BP
and Forsythe ID: Nitric oxide is a volume transmitter regulating
postsynaptic excitability at a glutamatergic synapse. Neuron.
60:642–656. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ko HG, Choi JH, Park DI, Kang SJ, Lim CS,
Sim SE, Shim J, Kim JI, Kim S, Choi TH, et al: Rapid turnover of
cortical NCAM1 regulates synaptic reorganization after peripheral
nerve injury. Cell Rep. 22:748–759. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chirila AM, Brown TE, Bishop RA, Bellono
NW, Pucci FG and Kauer JA: Long-term potentiation of glycinergic
synapses triggered by interleukin 1β. Proc. Natl Acad Sci USA.
111:8263–8268. 2014. View Article : Google Scholar
|
|
45
|
Huang LE, Guo SH, Thitiseranee L, Yang Y,
Zhou YF and Yao YX: N-methyl D-aspartate receptor subtype 2B
antagonist, Ro 25–6981, attenuates neuropathic pain by inhibiting
postsynaptic density 95 expression. Sci Rep. 8:78482018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee JO, Kim N, Lee HJ, Lee YW, Kim JK, Kim
HI, Lee SK, Kim SJ, Park SH and Kim HS: Visfatin, a novel
adipokine, stimulates glucose uptake through the Ca2 +-dependent
AMPK-p38 MAPK pathway in C2C12 skeletal muscle cells. J Mol
Endocrinol. 54:251–262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cui H, Hayashi A, Sun HS, Belmares MP,
Cobey C, Phan T, Schweizer J, Salter MW, Wang YT, Tasker RA, et al:
PDZ protein interactions underlying NMDA receptor-mediated
excitotoxicity and neuroprotection by PSD-95 inhibitors. J
Neurosci. 27:9901–9915. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan J, Cowan CM, Zhang LY, Hayden MR and
Raymond LA: Interaction of postsynaptic density protein-95 with
NMDA receptors influences excitotoxicity in the yeast artificial
chromosome mouse model of Huntington's disease. J Neurosci.
29:10928–10938. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lee WH, Li LL, Chawla A, Hudmon A, Lai YY,
Courtney MJ and Hohmann AG: Disruption of nNOS-NOS1AP
protein-protein interactions suppresses neuropathic pain in mice.
Pain. 159:849–863. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Vickers CA, Stephens B, Bowen J,
Arbuthnott GW, Grant SG and Ingham CA: Neurone specific regulation
of dendritic spines in vivo by post synaptic density 95 protein
(PSD-95). Brain Res. 1090:89–98. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cuthbert PC, Stanford LE, Coba MP, Ainge
JA, Fink AE, Opazo P, Delgado JY, Komiyama NH, O'Dell TJ and Grant
SG: Synapse-associated protein 102/dlgh3 couples the NMDA receptor
to specific plasticity pathways and learning strategies. J
Neurosci. 27:2673–2682. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wei Z, Wu G and Chen BS: Regulation of
SAP102 synaptic targeting by phosphorylation. Mol Neurobiol.
55:6215–6226. 2018. View Article : Google Scholar : PubMed/NCBI
|