Introduction
Low back pain is a common and chronic disease
(1). In total, >80% of adults
will suffer from back pain at a certain stage in their lives
(2). Intervertebral disc
degeneration (IDD) is a common cause of lower back pain, and 20–33%
of the patients with this disorder will not be able to work
(3,4). Moreover, IDD is also a prevalent
musculoskeletal disorder, which may serve as a social and economic
burden worldwide (5). At present,
endoscopic lumbar discectomy is the common treatment method for
IDD. However, this method may lead to recurrence of the disease
following surgery (6,7). At present, no effective preventative
strategy and therapeutic method has been reported for treating IDD,
due to the diversity and the unclear cause of low back pain.
Therefore, it is important to identify the mechanism responsible
for the development of IDD.
MicroRNAs (miRNAs) are endogenous short non-coding
RNAs, which consist of ~20 nucleotides (8). miRNAs play vital roles in the
regulation of gene expression by controlling related physiological
processes (9). Previous studies
reported that miRNAs are involved in cell growth, proliferation and
cell death in different diseases (10–12).
It has been shown that some miRNAs exert an important role in the
development of IDD, such as miR-194, miR-3150a and miR-125b-1
(13–15).
miR-222 acts as a tumor promoter in different cancer
types, such as pancreatic and breast cancer (16,17).
miR-222 regulates the inflammatory response of macrophages in
bacterial pneumonia (18). In
addition, miR-222 regulates marrow niche-supported tumor cells,
which promote survival of residual cells (19). A previous study indicated that the
levels of miR-222 were upregulated in patients with rheumatoid
arthritis (20). However, although
various previous studies have been conducted on the biological
functions of miR-222, the role of miR-222 in IDD remains unclear.
In the present study, evidence is presented supporting the role of
miR-222 in inducing the apoptosis of NP cells via Bcl-2
regulation.
Materials and methods
Patient samples
A total of 20 lumbar nucleus pulposus specimens were
collected from patients with IDD between April 2017 and May 2018;
the patients included 12 females and 8 males (ranging 43–65 years
old) in the First Affiliated Hospital of the Jinzhou Medical
University. Patients with diabetes mellitus, spondylolisthesis,
serious systemic disease, ankylosing spondylitis, trauma history of
the spine, or back surgery were excluded. In total, 20 matched
healthy controls, recruited from public schools, were included in
the present study. The present study was approved by the Ethics
Committee of the First Affiliated Hospital of the Jinzhou Medical
University, and informed consent was obtained from each participant
prior to enrollment.
Cell culture and transfection
Human intervertebral disc nucleus pulposus (NP)
cells were obtained from the Tongpai Biological Technology Co.,
Ltd. (http://www.shtpbio.com/tongpai-Products-3469763/).
These cells are primary cells. NP cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and 100
U/ml penicillin. The cells were incubated at 37°C in a humidified
atmosphere with 5% CO2.
The 10 nM miR-222 mimics, 10 nM miR-222 inhibitor
and 10 nM mimics negative control (NC) were obtained from Shanghai
GenePharma Co., Ltd. Briefly, the cells (3×105
cells/well) were plated in 6-well plates overnight. The sequence
for miR-222 mimics was: 5′-AGCUACAUCUGGCUACUGGGU-3′. The sequence
for miR-222 inhibitor was: 5′-ACCCAGUAGCCAGAUGUAGCU-3′. The
sequence for NC was 5′-UUUGUACUACACAAAAGUACUG-3′. Subsequently,
miR-222 mimics or miR-222 inhibitor were transfected into the cells
using Lipofectamine® 2000 (Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. The transfection
efficiency of miR-222 mimics in NP cells was assessed using reverse
transcription-quantitative PCR (RT-qPCR) following 72 h of growth
after cell transfection.
RT-qPCR
Total RNA extraction from the lumbar nucleus
pulposus specimens and cells was performed using an RNA extraction
kit (Total RNA Purification kit; Sigma Aldrich; Merck KGaA). Total
RNA was reverse transcribed into cDNA using the TaqMan®
MicroRNA RT kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The RT reaction
conditions were as follows: 37°C for 60 min, followed by 85°C for 5
min and 4°C. The cDNA samples were subjected to real-time PCR using
a SYBR Green One-Step RT-qPCR kit (Thermo Fisher Scientific, Inc.)
on an ABI 7900HT instrument (ABI; Thermo Fisher Scientific, Inc.).
The qRCR reaction conditions were as follows: 95°C for 3 min,
followed by 40 cycles at 95°C for 10 sec, 58°C for 30 sec and 72°C
for 30 sec. The primer sequences for miR-222 were as follows:
Forward primer: 5′-AGCUACAUCUGGCUACUGGGU-3′ and reverse primer:
5′-CCAGUAGCCAGAUGUAGCUUU-3′. The primer sequences for U6 were as
follows: Forward primer: 5′-CTCGCTTCGGCAGCACAT-3′; reverse primer:
5′-AACGCTTCACGAATTTGCGT-3′. All samples were tested in triplicate.
The relative expression was measured using the 2−ΔΔCq
method (21). Gene expression
results were normalized by the internal control U6.
Cell Counting Kit-8 assay
Cell viability was assessed using CCK-8 (Beyotime
Institute of Biotechnology). NP cells (2×104 cells/well)
were seeded into 96-well plates overnight. Subsequently, miR-222
mimics or miR-222 inhibitor were added into each well for 0, 24, 48
and 72 h. A total of 10 µl CCK-8 reagent was added into each well
for 2 h. The absorbance of each well was determined at 450 nm with
a microplate reader (Molecular Devices, LLC).
Immunofluorescence assay
NP cells (1×105 cells/well) were plated
into 24-well plates overnight. Subsequently, miR-222 mimics,
miR-222 inhibitor or mimics control were added into each well. The
cells were fixed in 4% paraformaldehyde for 20 min at room
temperature, and in ice-cold 100% methanol for an additional 20 min
at −20°C. After 30 min of blocking with 3% BSA in TBST at room
temperature, the cells were incubated with the following
antibodies: Anti-proliferation marker protein Ki-67 (Ki67; Abcam;
cat. no. ab15580; 1:1,000), anti-Bcl-2 (Abcam; cat. no. ab32124;
1:1,000) and DAPI antibody (cat. no. ab104139; 1:1,000) at 4°C
overnight. The following morning, horseradish peroxidase-conjugated
Goat anti-rabbit IgG secondary antibodies (Abcam; cat. no.
ab150081; 1:5,000) were added for 1 h to the cells at room
temperature. The cells were observed with a Leica confocal
microscopy (Leica; magnification, ×400) and the images were
analyzed using the Leica LAS-X software.
Flow cytometry analysis of cell
apoptosis
NP cells (5×105 cells/well) were seeded
into 6-well plates overnight. miR-222 mimics, or mimics control
were added into each well for 72 h. The cells were collected,
washed 3 times with cold PBS and resuspended in 1X binding buffer.
The cells were stained with dual-staining Annexin V-FITC-propidium
iodide (PI; Thermo Fisher Scientific, Inc.) for 20 min at room
temperature according to the manufacturer's protocol. The results
were analyzed using a FACS Calibur flow cytometer (BD Biosciences)
on a BD FACSCalibur system (FACScan, BD Biosciences).
Western blot analysis
The proteins were extracted from the cells using
RIPA lysis buffer (Thermo Fisher Scientific, Inc.) and their
concentration levels were quantified using a bicinchoninic acid
protein assay kit (Beyotime Institute of Biotechnology). The
proteins (40 µg per lane) were separated by SDS-PAGE on 10% gels,
and were transferred onto 0.45 µm PVDF membranes (Thermo Fisher
Scientific, Inc.). Following blocking with 5% non-fat milk at room
temperature for 1 h, the membranes were incubated with primary
antibodies overnight at 4°C. The following primary antibodies were
used: Anti-Bax (Abcam; cat. no. ab32503; 1:1,000), anti-Bcl-2
(Abcam; cat. no. ab32124; 1:1,000), anti-cleaved caspase 3 (Abcam;
cat. no. ab2302; 1:1,000), anti-cytochrome c (Cyto C; Abcam; cat.
no. ab13575; 1:1,000), anti-apoptotic protease activating factor-1
(Apaf 1; Abcam; cat. no. ab2001; 1:1,000), anti-cleaved caspase 9
(Abcam; cat. no. ab2324; 1:1,000) and anti-β-actin (Abcam; cat. no.
ab8227; 1:1,000). Subsequently, the membranes were incubated with
horseradish peroxidase-conjugated secondary antibody (Abcam; cat.
no. ab150077; 1:200) for 1 h at room temperature. Finally, an image
of the protein band was detected using ECL reagent (Santa Cruz
Biotechnology, Inc.). Image-Pro Plus software (version 7, Media
Cybernetics, Inc.) was used for the targets that were normalized to
β-actin.
Luciferase reporter assay
The potential target genes of miR-222 were predicted
using miRanda (http://www.microrna.org), miRDB (http://www.mirdb.org/) and TargetScan (http://www.targetscan.org/). The target gene sequence
and the mutation gene sequence were synthesized by Shanghai
GenePharma Co., Ltd. The 3′-untranslated region (3′-UTR) of
Bcl-2 was amplified from genomic DNA and inserted into the
psiCHECK-2 vector (Promega Corporation) using the XhoI and
NotI sites. NP cells were co-transfected with
psiCHECK2-Bcl-2-wild-type + miR-222 mimics, and
psiCHECK2-Bcl-2-mutated type + miR-222 mimics using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.).
The cells were incubated in 5% CO2 at 37°C for 48 h. The
dual luciferase reporter assay system (Promega Corporation) was
used to measure luciferase activity. The firefly luciferase
activity was normalized, according to Renilla luciferase
activity.
Statistical analysis
Each sample was assessed for at least three
independent determinations. Data are presented as mean ± standard
error. Graphs were generated using GraphPad Prism software (version
7.0, GraphPad Software, Inc.). The comparison between the two
groups was analyzed by the Student's t-test. The comparisons among
multiple groups were performed with one-way ANOVA followed by the
Dunnett's test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-222 expression levels are
increased in IDD tissues and NP cells
To investigate the role of miR-222 in the
development of IDD, RT-qPCR was used to detect the levels of
miR-222 in IDD and normal disc tissues. A total of 20 human IDD
tissues were used with corresponding control samples. The mean
levels of miR-222 were significantly increased compared with the
normal group (Fig. 1A). In
addition, RT-qPCR was used to detect miR-222 levels in NP cells,
following transfection with miR-222 mimics for 0, 24, 48 or 72 h.
The expression levels of miR-222 in NP cells were significantly
increased following transfection with miR-222 mimics for 48 and 72
h (Fig. 1B). miR-222 mimics were
further used in the present study to successfully increase the
levels of miR-222 in NP cells. The levels of miR-222 were
significantly upregulated in NP cells following transfection with
miR-222 mimics for 72 h (Fig. 1C).
These results indicated that the levels of miR-222 were increased
in IDD tissues and NP cells.
miR-222 overexpression inhibits
proliferation of NP cells
To study the effects of miR-222 on NP cells, a CCK-8
assay was used to detect cell viability. Overexpression of miR-222
inhibited cell proliferation (Fig.
2A). Similarly, the results of the immunofluorescence assay
demonstrated that the overexpression of miR-222 significantly
decreased the number of Ki67 positive cells (Fig. 2B and C). The data suggested that
miR-222 overexpression inhibited proliferation of NP cells.
 | Figure 2.miR-222 overexpression inhibits
proliferation of NP cells. (A) Cell viability of NP cells following
transfection with NC and miR-222 mimics was determined by a CCK-8
assay at 0, 24, 48 and 72 h. Relative fluorescence expression
levels were observed by (B) Ki67 and DAPI staining (magnification,
×400), and (C) subsequent analysis. **P<0.01 vs. the NC group.
miR, microRNA; NP, nucleus pulposus; CCK-8, Cell Counting Kit-8;
Ki67, proliferation marker protein Ki-67; NC, negative control; OD,
optical density. |
miR-222 overexpression induces
apoptosis of NP cells
To further determine whether miR-222 was responsible
for the induction of apoptosis in NP cells, flow cytometry was
employed to analyze the extent of apoptosis. The cell apoptotic
rate was markedly increased in the miR-222 mimics group compared
with the NC group (Fig. 3A and B).
In addition, the expression levels of the apoptotic proteins Bax
and cleaved caspase 3 were significantly increased, while the level
of Bcl-2 was reduced in the miR-222 mimics group (Fig. 3C-F). These data s that miR-222
overexpression induced apoptosis of NP cells.
Bcl-2 is a target gene of miR-222
As predicted by miRanda, miRDB and TargetScan, there
was complementary binding between miR-222 and Bcl-2. The
miR-222 sequence exhibited a binding site for the 3′-UTR of
wild-type Bcl-2 (Fig. 4A).
To further confirm the functional interaction between miR-222 and
Bcl-2, immunofluorescence and luciferase reporter assays
were used to verify the binding of miR-222 to its target. The
expression levels of Bcl-2 were markedly decreased in the
miR-222 mimics group compared with the NC group (Fig. 4B). In addition, overexpression of
miR-222 markedly decreased the luciferase activity of the reporter
gene with regard to the wild-type genotype, whereas the
transfection of the cells with miR-222 mimics exhibited no
significant effect on the mutant genotype, indicating that miR-222
directly targeted the 3′-UTR of Bcl-2 (Fig. 4C). In addition, the expression
levels of the mitochondrial apoptotic pathway-associated proteins,
Cyto C, Apaf 1 and cleaved caspase 9, were significantly increased
in the miR-222 mimics group compared with the NC group. These data
suggested that miR-222 induced cell apoptosis by targeting
Bcl-2 in NP cells.
 | Figure 4.Bcl-2 is the target gene of miR-222.
(A) Sequence alignment of human miR-222 with Bcl-2. (B) NP
cells were transfected with NC and miR-222 mimics, and the relative
fluorescence expression levels were observed by Bcl-2 detection and
DAPI staining (magnification, ×400). (C) Luciferase reporter assays
were used to detect the luciferase activity. The cells in the WT
group were treated with psiCHECK2-Bcl-2-WT+miR-NC and
psiCHECK-2-Bcl-2-WT+miR-222 mimics separately. The cells in the MT
group were treated with psiCHECK-2-Bcl-2-MT+miR-NC and
psiCHECK2-Bcl-2-MT+miR-222 mimics separately. Both firefly and
Renilla luciferase activities were measured in the same
sample. The firefly luciferase activity was normalized, according
to Renilla luciferase activity. (D) Protein expression
levels of Cyto C, Apaf 1 and cleaved caspase 9 detected in the
intervertebral disc NP cells following transfection with NC and
miR-222 mimics. Relative expression levels of (E) Cyto C, (F) Apaf
1 and (G) cleaved caspase 9 were quantified by normalizing them to
the levels of β-actin. **P<0.01 vs. the NC group. miR, microRNA;
NP, nucleus pulposus; NC, negative control; WT, wild-type; MT,
mutant type; Cyto C, cytochrome C; Apaf 1, apoptotic protease
activating factor-1. |
Downregulation of miR-222 promotes
proliferation of NP cells
The data derived from the previous experiments
demonstrated that miR-222 overexpression could inhibit
proliferation of NP cells. To further investigate the biological
roles of miR-222 on NP cells, the cells were transfected with an
miR-222 inhibitor. The levels of miR-222 were significantly
downregulated in NP cells following transfection of the miR-222
inhibitor (Fig. 5A). In addition,
downregulation of miR-222 promoted cell proliferation (Fig. 5B). Similarly, the results of the
immunofluorescence assay suggested that downregulation of miR-222
increased Ki67 positive cells compared with the NC group (Fig. 5C and D). The present data suggested
that downregulation of miR-222 promoted proliferation of NP
cells.
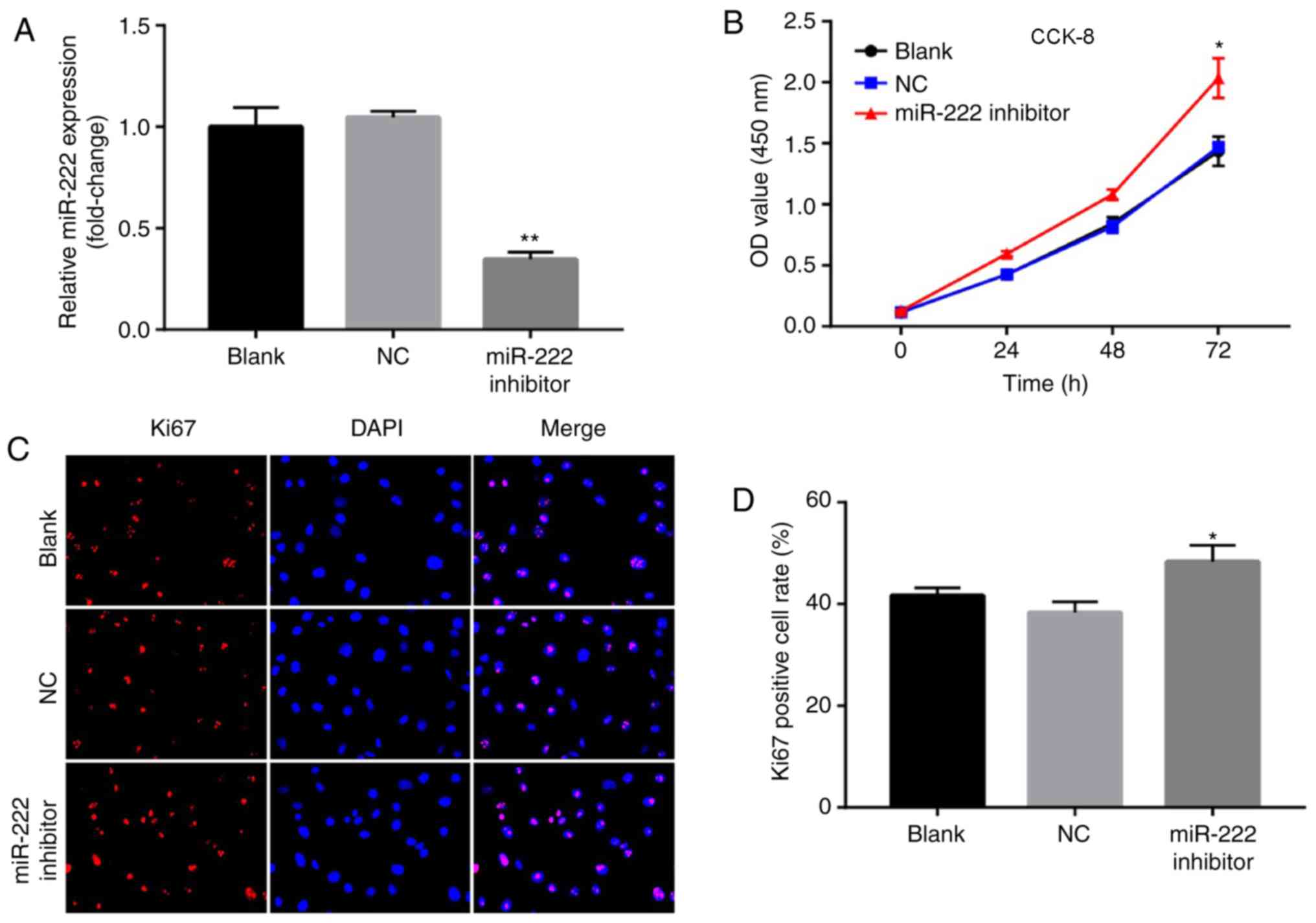 | Figure 5.Downregulation of miR-222 promotes
proliferation of NP cells. (A) Relative expression of miR-222 in NP
cells following transfection with NC and miR-222 inhibitor for 72
h. (B) Cell viability of NP cells following transfection with NC
and miR-222 inhibitor was determined by a CCK-8 assay at 0, 24, 48
and 72 h. Relative fluorescence expression levels were observed by
(C) Ki67 detection and DAPI staining (magnification, ×400), and (D)
subsequent analysis. *P<0.05, **P<0.01 vs. the NC group. miR,
microRNA; NP, nucleus pulposus; NC, negative control; CCK-8, Cell
Counting Kit-8; Ki67, proliferation marker protein Ki-67; OD,
optical density. |
Discussion
Despite the recent advances of experimental
medicine, the therapeutic applications for IDD are still limited.
Therefore, new therapeutic strategies are required for the
treatment of IDD. In the present study, it was demonstrated that
miR-222 levels were upregulated in IDD tissues. In addition,
upregulation of miR-222 could inhibit proliferation and induce
apoptosis of NP cells. Abo ElAtta et al (20) suggested that the expression levels
of miR-222 were increased in patients with rheumatoid arthritis. In
addition, Nie and Tian (22)
indicated that miR-222 was upregulated in patients with uterine
cancer. Therefore, these results indicated that miR-222 may play a
role in inhibiting cell proliferation.
Apoptosis is a physiological process, which can
remove harmful or seriously injured cells and organelles (23). A previous study indicated that the
inhibition of apoptosis in NP cells could retard disc degeneration
(24). A decrease in the number of
NP cells was associated with the development of IDD, and inhibition
of cell apoptosis was used as a strategy to reverse IDD (25). In the present study, the data
demonstrated that miR-222 induced apoptosis in NP cells by reducing
the levels of Bcl-2, and increasing the levels of Bax and cleaved
caspase 3. Zhang et al (26) further demonstrated that miR-222
induced apoptosis in glioma cells by decreasing the levels of Bcl-2
and increasing the levels of Bax. These data suggested that miR-222
could induce apoptosis in NP cells.
In the present study, a novel mechanism of the
miR-222-associated therapeutic action for the treatment of IDD was
proposed. The present results identified that Bcl-2 is a
target gene of miR-222 in NP cells, as determined by a luciferase
reporter assay. Bcl-2 is an anti-apoptotic protein, which belongs
to the Bcl-2 family of proteins, and is considered a classical
biomarker of the mitochondrial pathway of apoptosis (27). In apoptosis, external stimuli, such
as cytotoxic agents, could result in mitochondrial dysfunction, and
then result in the release of pro-apoptotic protein Cyt c and
activation of caspases to induce apoptosis (28). Previous studies demonstrated that
the dissociation of the Bcl-2/Bax complex can lead to the release
of Cyto C (29,30). The proteins Cyto C, Apaf-1 and
caspase-9 form the apoptosome, which increases the levels of
cleaved caspase-9 and leads to induction of apoptosis (30). In the present study, it was
demonstrated that overexpression of miR-222 increased the levels of
Cyto C, Apaf 1 and caspase 9 by targeting Bcl-2. The present data
suggested that miR-222 induced apoptosis of NP cells via the
mitochondrial-dependent apoptotic pathway. A decrease in the levels
of Bcl-2 and a concomitant increase in the levels of cleaved
caspase 3, cleaved caspase 9 and Bax were detected in NP cells,
which was consistent with the data reported in a previous study
(31). In addition, it is
important to note that downregulation of miR-222 promoted
proliferation of NP cells. The present findings demonstrated that
overexpression of miR-222 induced apoptosis of NP cells via the
mitochondrial-mediated apoptotic pathway. However, further studies
are needed to investigate the role of miR-222 in animal model of
intervertebral disc nucleus pulposus.
In conclusion, the present study demonstrated that
miR-222 induces apoptosis in human NP cells by directly targeting
Bcl-2. In addition, miR-222 may play a role in inhibiting
cell proliferation and inducing cell apoptosis in human NP cells.
Therefore, miR-222 may act as a potential therapeutic target for
the treatment of IDD.
Acknowledgements
Not applicable.
Funding
The present study was funded by The Department of
Science and Technology of the Liaoning Preparation of Animal Models
and Animal Experimental Research for Major Diseases (grant no.
2012408002).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
WW and JW analyzed and interpreted the patient data,
and were major contributors in the development of the first draft
of the present manuscript. JZ and WT participated in the
experimental design, tissue collection and experimental execution.
ZZ participated in the experimental design and experimental
execution, and reviewed and approved the final draft of the
manuscript prior to submission.
Ethics approval and consent to
participate
Ethics approval for the present study was provided
by The First Affiliated Hospital of The Jinzhou Medical University
Ethics Committee, and informed consent was obtained from each
participant prior to enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kadow T, Sowa G, Vo N and Kang JD:
Molecular basis of intervertebral disc degeneration and
herniations: What are the important translational questions? Clin
Orthop Relat Res. 473:1903–1912. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Andersson GB: Epidemiological features of
chronic low-back pain. Lancet. 354:581–585. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen Y, Wu Y, Shi H, Wang J, Zheng Z, Chen
J, Chen X, Zhang Z, Xu D, Wang X and Xiao J: Melatonin ameliorates
intervertebral disc degeneration via the potential mechanisms of
mitophagy induction and apoptosis inhibition. J Cell Mol Med.
23:2136–2148. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vergroesen PP, Kingma I, Emanuel KS,
Hoogendoorn RJ, Welting TJ, van Royen BJ, van Dieën JH and Smit TH:
Mechanics and biology in intervertebral disc degeneration: A
vicious circle. Osteoarthritis Cartilage. 23:1057–1070. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheung KM, Karppinen J, Chan D, Ho DW,
Song YQ, Sham P, Cheah KS, Leong JC and Luk KD: Prevalence and
pattern of lumbar magnetic resonance imaging changes in a
population study of one thousand forty-three individuals. Spine
(Phila Pa 1976). 34:934–940. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yorimitsu E, Chiba K, Toyama Y and
Hirabayashi K: Long-term outcomes of standard discectomy for lumbar
disc herniation: A follow-up study of more than 10 years. Spine
(Phila Pa 1976). 26:652–657. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Jiang L, Dai G, Li S and Mu X:
Bioinformatics analysis reveals different gene expression patterns
in the annulus fibrosis and nucleus pulpous during intervertebral
disc degeneration. Exp Ther Med. 16:5031–5040. 2018.PubMed/NCBI
|
|
8
|
Takashima Y, Kawaguchi A, Iwadate Y,
Hondoh H, Fukai J, Kajiwara K, Hayano A and Yamanaka R: MicroRNA
signature constituted of miR-30d, miR-93, and miR-181b is a
promising prognostic marker in primary central nervous system
lymphoma. PLoS One. 14:e02104002019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gagliardi D, Comi GP, Bresolin N and Corti
S: MicroRNAs as regulators of cell death mechanisms in amyotrophic
lateral sclerosis. J Cell Mol Med. 23:1647–1656. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang P, Li S, Lv C, Si J, Xiong Y, Ding
L, Ma Y and Yang Y: BPI-9016M, a c-Met inhibitor, suppresses tumor
cell growth, migration and invasion of lung adenocarcinoma via
miR203-DKK1. Theranostics. 8:5890–5902. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li M, Ding W, Tariq MA, Chang W, Zhang X,
Xu W, Hou L, Wang Y and Wang J: A circular transcript of ncx1 gene
mediates ischemic myocardial injury by targeting miR-133a-3p.
Theranostics. 8:5855–5869. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang L, Cai Y, Zhang D, Sun J, Xu C, Zhao
W, Jiang W and Pan C: miR-195/miR-497 regulate CD274 expression of
immune regulatory ligands in triple-negative breast cancer. J
Breast Cancer. 21:371–381. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kong L, Sun M, Jiang Z, Li L and Lu B:
MicroRNA-194 inhibits lipopolysaccharide-induced inflammatory
response in nucleus pulposus cells of the intervertebral disc by
targeting TNF receptor-associated factor 6 (TRAF6). Med Sci Monu.
24:3056–3067. 2018. View Article : Google Scholar
|
|
14
|
Zhang B, Guo W, Sun C, Duan HQ, Yu BB, Mu
K, Guan YY, Li Y, Liu S, Liu Y, et al: Dysregulated MiR-3150a-3p
promotes lumbar intervertebral disc degeneration by targeting
aggrecan. Cell Physiol Biochem. 45:2506–2515. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meng X, Zhu Y, Tao L, Zhao S and Qiu S:
MicroRNA-125b-1-3p mediates intervertebral disc degeneration in
rats by targeting teashirt zinc finger homeobox 3. Exp Ther Med.
15:2627–2633. 2018.PubMed/NCBI
|
|
16
|
Li Z, Tao Y, Wang X, Jiang P, Li J, Peng
M, Zhang X, Chen K, Liu H, Zhen P, et al: Tumor-secreted exosomal
miR-222 promotes tumor progression via regulating P27 expression
and Re-localization in pancreatic cancer. Cell Physiol Biochem.
51:610–629. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zong Y, Zhang Y, Sun X, Xu T, Cheng X and
Qin Y: miR-221/222 promote tumor growth and suppress apoptosis by
targeting lncRNA GAS5 in breast cancer. Biosci Rep. 39(pii):
BSR201818592019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu Z, Zhang D, Lee H, Menon AA, Wu J, Hu
K and Jin Y: Macrophage-derived apoptotic bodies promote the
proliferation of the recipient cells via shuttling
microRNA-221/222. J Leukoc Biol. 101:1349–1359. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moses BS, Evans R, Slone WL, Piktel D,
Martinez I, Craig MD and Gibson LF: Bone marrow microenvironment
niche regulates miR-221/222 in acute lymphoblastic leukemia. Mol
Cancer Res. 14:909–919. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abo ElAtta AS, Ali YBM, Bassyouni IH and
Talaat RM: Upregulation of miR-221/222 expression in rheumatoid
arthritis (RA) patients: Correlation with disease activity. Clin
Exp Med. 19:47–53. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nie X and Tian H: Correlation between
miR-222 and uterine cancer and its prognostic value. Oncol Lett.
16:1722–1726. 2018.PubMed/NCBI
|
|
23
|
Elmore S: Apoptosis: A review of
programmed cell death. Toxicol Pathol. 35:495–516. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu W, Fu J, Liu Y, Wu Y and Jiang D:
Osteogenic protein-1 inhibits nucleus pulposus cell apoptosis
through regulating the NF-κB/ROS pathway in an inflammation
environment. Biosci Rep. 38(pii): BSR201815302018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu Y, Yao H, Wang Q, Xu W, Liu K, Zhang J,
Zhao H and Hou G: Aquaporin-3 attenuates oxidative stress-induced
nucleus pulposus cell apoptosis through regulating the P38 MAPK
pathway. Cell Physiol Biochem. 50:1687–1697. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang CZ, Kang CS, Pu PY, Wang GX, Jia ZF,
Zhang AL, Han L and Xu P: Inhibitory effect of knocking down
microRNA-221 and microRNA-222 on glioma cell growth in vitro and in
vivo. Zhonghua Zhong Liu Za Zhi. 31:721–726. 2009.(In Chinese).
PubMed/NCBI
|
|
27
|
Nguyen KC, Willmore WG and Tayabali AF:
Cadmium telluride quantum dots cause oxidative stress leading to
extrinsic and intrinsic apoptosis in hepatocellular carcinoma HepG2
cells. Toxicology. 306:114–123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li X, Fang F, Gao Y, Tang G, Xu W, Wang Y,
Kong R, Tuyihong A and Wang Z: ROS induced by KillerRed targeting
mitochondria (mtKR) enhances apoptosis caused by radiation via Cyt
c/caspase-3 pathway. Oxid Med Cell Longev.
2019:45286162019.PubMed/NCBI
|
|
29
|
Eskes R, Antonsson B, Osen-Sand A,
Montessuit S, Richter C, Sadoul R, Mazzei G, Nichols A and Martinou
JC: Bax-induced cytochrome C release from mitochondria is
independent of the permeability transition pore but highly
dependent on Mg2+ ions. J Cell Biol. 143:217–224. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sinha K, Das J, Pal PB and Sil PC:
Oxidative stress: The mitochondria-dependent and
mitochondria-independent pathways of apoptosis. Arch Toxicol.
87:1157–1180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Miyazaki S, Kakutani K, Yurube T, Maeno K,
Takada T, Zhang Z, Kurakawa T, Terashima Y, Ito M, Ueha T, et al:
Recombinant human SIRT1 protects against nutrient
deprivation-induced mitochondrial apoptosis through autophagy
induction in human intervertebral disc nucleus pulposus cells.
Arthritis Res Ther. 17:2532015. View Article : Google Scholar : PubMed/NCBI
|



















