Introduction
As one of the most frequent types of cancer in women
worldwide, cervical cancer is the main cause of mortality from
tumors among women, particularly in developing countries (1). There are >500,000 patients with
cervical cancer and the majority of these (80%) are in developing
countries (2). It is a challenge
in China to prevent cervical cancer; surgical excision is primarily
used for the treatment of cervical cancer at the early stage and
radiotherapy is also an effective way to cure local advanced
cervical cancer, particularly to control the disease from a
distance (3). Although there has
been progress in diagnostic and therapeutic strategies, the
survival rate for patients with cervical cancer remains poor
(4). Increasing evidence indicates
that microRNAs (miRNAs) are implicated in the pathogenesis of
cervical cancer, including miR-182 and miR-494 (5–7).
miRNAs can mediate gene expression through inducing
mRNA cleavage and suppressing translation (8). miRNAs have significant impacts on
cancer and have emerged as important factors in tumorigenesis
(9). Certain miRNAs are regarded
as oncogenes or tumor suppressor genes, therefore, miRNAs can be
used potentially as biomarkers for the diagnosis and prognosis of
various types of cancer, including cervical cancer (10,11).
For example, miR-150 contributes to cell proliferation in cervical
cancer by downregulating forkhead box O4, and miR-302 inhibits
cervical cancer proliferation and invasion by decreasing levels of
AKT1 (12). miR-411 is a member of
the miR-379 family, which is located in the miR-379/miR-656 cluster
in the DLK-DIO3 region on human chromosome 14 (13). A previous study suggested that
miR-379 may act as a tumor suppressor in cervical cancer through
directly targeting CRKL, and miR-379 may be considered as an
effective strategy for the treatment of cervical cancer (14). Additionally, miR-411 has been
reported to be involved in proliferation and differentiation in
rhabdomyosarcoma, hepatocellular carcinoma and lung cancer
(15–17). A previous study also demonstrated
that miR-411 acts as a tumor suppressor in renal cell carcinoma
(18). The microRNA.org website resource predicts that the STK17A
3′-untranslated region (UTR) contains miR-411 binding sites. It has
been shown that STK17A has low expression in acquired resistance
phenotypes of cancer cells, which are resistant to oxaliplatin and
5-fluorouracil (19). The
overexpression of STK17A is correlated with lower survival rates in
patients with glioma (20).
Furthermore, STK17A is a target gene of p53 (21). It has been reported that
abnormality of the p53 tumor suppressor gene belongs to the most
common molecular events in neoplasia of humans and animals
(22). p53 can suppress cancer
development by inducing cell-cycle arrest, cell repair/death or
anti-angiogenesis (23). In
addition, the p53 signaling pathway has been reported to be
involved in the pathology of cervical cancer (24–26).
The conclusion from the above data is that miR-411 is involved in
cervical cancer by mediating STK17A through the p53 axis. The
purpose of the present study was to determine the roles of miR-411
and STK17A in cervical cancer radiotherapy and their functions on
the cellular processes of cervical cancer cells through the p53
pathway.
Patients and methods
Study subjects
Cervical cancer tissues and adjacent normal tissues
(5-cm) were collected from 141 patients with cervical cancer, who
were pathologically diagnosed (27) and underwent cervical biopsy and
initial radiotherapy between October 2010 and October 2012 at
Jining No. 1 People's Hospital (Jining, China). All specimens were
fixed with 10% formalin and embedded with paraffin. Additionally, 5
ml of venous peripheral blood was collected from fasting patients
for polymerase chain reaction (PCR) detection. Among the enrolled
patients, there were 82 patients ≥45 years old and 59 patients
<45 years; 55 patients had stage I cancer according to the FIGO
2012 clinical staging criteria (28), 62 patients had stage II cancer and
24 patients had stage III cancer; 57 patients had a maximum local
tumor diameter of ≥4 cm and 84 patients had a maximum local tumor
diameter of <4 cm; 126 patients had squamous cell carcinoma
(SCC) and 15 patients had adenocarcinoma (pathological types); 35
patients had lymph node metastasis and 106 patients were without
lymph node metastasis; 81 patients were postmenopausal and 60
patients were premenopausal; 96 patients had a hemoglobin level
≥110 g/L and 45 patients had a level <110 g/L; 64 patients had
an SCC antigen level pre-radiotherapy of ≥2 ng/ml and 77 patients
had a level of <2 ng/ml; 33 patients were treated with
four-field conformal radiotherapy, 57 patients with
intensity-modulated radiotherapy and 51 patients with pelvic
hexagonal field radiotherapy (radiotherapy methods). The study
included patients with complete pathological and clinical data and
follow-up records. The exclusion criteria were as follows: Patients
without complete pathological and clinical data in addition to
follow-up records; patients with a history of malignant tumor in
other body regions, severe cardiovascular disease, severe liver and
kidney dysfunction or other diseases not tolerated radiotherapy.
The present study was approved by the Ethics Committee of Jining
No. 1 People's Hospital and informed consent was obtained from all
patients.
Radiotherapy methods and therapeutic
evaluation
External radiotherapy included four-field conformal
radiotherapy, intensity-modulated radiotherapy and pelvic hexagonal
field radiotherapy. Four-field conformal radiotherapy involved the
following: The upper and lower boundary of the radiation field was
radiated in the same manner as pelvic field radiation, and 6-MV
X-rays were used to irradiate in the box. When irradiation was
performed up to 17 times, the anterior and prior field conformal
radiotherapy was initiated with an irradiation dose of 45–50 Gy/1.8
Gy each time, 25–28 times every 5–6 weeks. For the
intensity-modulated radiotherapy, the upper and lower bounds of the
organs at risk were generally located at the upper and lower 2 cm
of the clinical target area and radiotherapy was performed by
irradiation with 6-MV X-rays. The prescription dose was 45–50.4 Gy
and, following synchronization of lymph node metastasis, the dose
was up to 55–65 Gy, 25–28 times/5–6 weeks. The pelvic hexagonal
field radiotherapy involved the application of irradiation in the
anterior-posterior direction with 6-MV X-rays, and an irradiation
dose of 45–50 Gy/1.8 Gy each time, 25–28 times/5–6 weeks.
Each radiotherapy method was repeated every 2–3
weeks. The efficacy was evaluated according to the response
evaluation criteria in solid tumors (RECIST) (29) guidelines issued by the World Health
Organization in 2000. A complete response (CR) was defined as the
disappearance of all target lesions; a partial response (PR) as
≥30% or reduction in the sum of the longest diameters of target
lesions compared with the baseline; progressive disease (PD) was
defined as ≥20% increase in the sum of the longest diameter of
target lesions or the appearance of any new lesions compared with
the sum of the shortest diameter recorded since treatment started;
stable disease (SD) was between PD and PR. A total of 92 CR and PR
cases were recruited into the response group (CR + PR), and a total
of 98 PD and SD cases were included in the non-response group (PD +
SD).
Follow-up
All patients were followed up via outpatient records
and telephone interviews for 3 years until October 31st 2015, and
the follow-up rate was 90%. The number of cases of survival was
recorded between grouping and end of follow-up, and the 3-year
survival rate was equal to the number of cases of survival for
>3 years accounting for the total cases followed up for >3
years. The progression-free survival rate was considered the rate
of disease with no progression (no deterioration) in a certain
period of time, including CR patients without recurrence in a
certain period of time and PR patients or patients without
deterioration of disease. The 3-year overall survival and
progression-free survival rates were calculated.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted from the tissues, peripheral
blood and cells using a TRIzol kit (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and reverse transcribed into
the cDNA template with a PCR amplification instrument. Following
this reaction, the cDNA was placed on ice or stored at −20°C. The
target gene and GAPDH (internal reference) underwent real-time qPCR
analysis using the ABI7300 PCR instrument. The reaction mixture was
2X SYBR-Green qPCR Mix for 10 µl, forward primers (10 µmol/L) for 1
µl, revised primers (10 µmol/l) for 1 µl, cDNA for 1 µl and ddwater
for 7 µl. The reaction conditions were as follows: Pre-denaturation
at 95°C for 10 min, and a total of 40 cycles of denaturation at
95°C for 15 sec, annealing at 56°C for 30 sec and extension at 72°C
for 32 sec. The primers (Table I)
for the reaction were synthesized by Applied Biosystems; Thermo
Fisher Scientific, Inc.). Each experimental sample underwent
experimental verification of complex parallel pumping station with
centrifugal pumps and each experiment was repeated three times to
obtain the mean value. Then cycle quantification (Cq) value was
calculated. The Cq value refers to the number of PCR cycles when
the fluorescence value reaches the threshold, and is a parameter
with no unit. GAPDH was used as the internal reference. The
2−ΔΔCq method (30) was
used to determine the ratio of target gene expression in the
experiment group to that in the control group, and the formula was
as follows: ΔΔCq=ΔCqexperimentgroup-ΔCqcontrol
group, and ΔCq=Cqtarget
gene-CqGAPDH.
 | Table I.Reverse transcription-quantitative
polymerase chain reaction primer sequences. |
Table I.
Reverse transcription-quantitative
polymerase chain reaction primer sequences.
| Gene | Sequence |
|---|
| miR-411 | F:
5′-GGGGTAGTAGACCGTATAG-3′ |
|
| R:
5′-TGCGTGTCGTGGAGTC-3′ |
| STK17A | F:
5′-GAACACCATGATCCCTTTGG-3′ |
|
| R:
5′-GTGCCTTTTCCATCCTGAAA-3′ |
| p53 | F:
5′-CAGCACATGACGGAGGTTG-3′ |
|
| R:
5′-TCATCCAAATACTCCACACGC-3′ |
|
p21WAF1 | F:
5′-CACTCCAAACGCCGGCTGATCTTC-3′ |
|
| R:
5′-TGTAGAGCGGGCCTTTGAGGCCCTC-3′ |
| TAp63 | F:
5′-GACCTGAGTGACCCCATGTG-3′ |
|
| R:
5′-TCTGGATGGGGCATGTCTTTGC-3′ |
| GAPDH | F:
5′-GGAGCGAGATCCCTCCAAAAT-3′ |
|
| R:
5′-GGCTGTTGTCATACTTCTCA-3′ |
Western blot analysis
The protein and tissue homogenates of the
transfected cells were prepared and transferred into a 1.5-ml
centrifuge tube, incubated on ice for 25 min, lysed by
ultrasonication for 25 sec, and centrifuged at 12,000 × g for 20
min at 4°C. The supernatant was collected and transferred into a
centrifuge tube and protein concentration was measured using a
bicinchoninic acid kit (BCA1-1KT, Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). The sodium dodecyl sulfate-polyacrylamide gel
electrophoresis separation gel (8%) and spacer gel (5%) were
prepared, and the loading quantity of each lane was 50 µg total
proteins. Following electrophoresis (spacer gel at 80 V for 30 min,
separation gel at 100 V for 80 min), the separated protein was
transferred onto polyvinylidene fluoride membranes for incubation
at room temperature for 1 h with the sealing liquid removed. The
membranes were incubated with phosphorylated (p)-STK17A antibody
(cat. no. 14433-1-AP; Proteintech, Wuhan, China; 1:1,000),
p-p21WAF1 antibody (cat. no. AP01654PU-N; Origene;
1:10,000), p-p53 antibody (cat. no. MABE518; Merck KGaA; 1:1,000),
TAp63 antibody (cat. no. TA311397; Origene; 1:1,000), and GAPDH
antibody (cat. no. 10494-1-AP; Proteintech; 1:2,000) at 4°C
overnight. All the above antibodies were purchased from Abcam
(Cambridge, MA, USA). The membranes were washed with Tris-buffered
saline with Tween-20 (TBST) times (5 min each time), incubated with
the Goat Anti-Mouse, (cat. no. SA00001-1) and Goat Anti-Rabbit,
(cat. no. SA00001-2; Proteintech; 1:10,000) for 1 h at room
temperature and washed with TBST three times (each time for 5 min).
Following scanning and developing, Image Pro Plus 6.0 software
(Media Cybernetics, Inc., Silver Spring, MD, USA) was used to
analyze the gray values of the protein bands. The experiment was
repeated three times.
Luciferase reporter gene assay
The microRNA.org
website (http://www.microrna.org) and TargetScan
website (http://www.targetscan.org/vert_71/) were used to
predict the potential target genes of miR-27a and obtain the
sequences of fragments containing binding sites. The
psiCheck-2-STK17A-MU plasmid and psiCheck-2-STK17A-WT plasmid were
constructed by inserting mutant and wild-type fragments in the
3′-UTR of the STK17A gene into the psiCheck-2 luciferase reporter
vector (Promega Corporation, Madison, WI, USA), respectively. The
CaSki cervical cancer cells were seeded into a 24-well plate with
1×105 cells per well and transfected when the cell
density was up to 80% on the following day. The miR-411 mimic and
mimic control were respectively co-transfected with
psiCheck-2-STK17A-WT and psiCheck-2-STK17A-MU into CaSki cells
using Lipofectamine 2000. After 48 h, the luciferase activity was
analyzed using a Dual-Luciferase Reporter Assay system (DLR,
Promega Corporation). The target/reference value was taken as the
relative luciferase activity and the relative luciferase activity
was accessed using a fluorescence instrument (Promega
Corporation).
Cell culture and transfection
The HeLa, CaSki, SiHa and C33A cervical cancer cell
lines were purchased from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China). The cells were cultured in Dulbecco's
modified Eagle's medium (DMEM, Gibco; Thermo Fisher Scientific,
Inc.) containing 10% fetal bovine serum (FBS, Gibco; Thermo Fisher
Scientific, Inc.) in a humidified incubator with 5% CO2
at 37°C. STK17A small interfering (si)RNA was obtained from Santa
Cruz Biotechnology, Inc. (Santa Cruz, CA, USA), and miR-411 mimic,
miR-411 inhibitor and negative control (NC) were obtained from
Rainbow Chemistry Co., Ltd. (Shanghai, China). Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to
respectively transfect the NC plasmid, miR-411 mimic plasmid,
miR-411 inhibitor plasmid, siRNA-STK17A plasmid, miR-411 inhibitor
+ siRNA-STK17A plasmid into the CaSki cell line, which had the
highest expression of miR-411. The CaSki cells were assigned into a
blank group (without any treatment), and NC, miR-411 mimic, miR-411
inhibitor, siRNA-STK17A, and miR-411 inhibitor + siRNA-STK17A
groups. Each group, with the exception of the blank group, was
respectively dissolved in Opti-MEM medium, followed by the addition
of Lipofectamine 2000 for transfection, according to the
manufacturer's protocol. The compounds prepared were directly added
into a 6-well plate containing cells and medium, agitated and mixed
gently. The cells were incubated in an incubator with 5%
CO2 at 37°C. After 6 h, the cells were cultured with
conventional culture media for the following experiments.
Colony formation assay
The CaSki cells in the logarithmic growth phase were
obtained, washed once with phosphate-buffered saline (PBS), and
treated with 0.25% trypsin (2 ml). The cells were centrifuged at
1,000 × g at room temperature for 5 min, collected, and prepared
into cell suspension with the cell density adjusted to
2.5×102 cells/ml. The cells were seeded into a 6-well
plate with 2 ml of liquid in each well and cultured in a humidified
incubator with 5% CO2 at 37°C. After 4 h, cell adhesion
was observed. The cells were respectively irradiated with a dose of
0, 2, 4, 6, 8 and 10 Gy, and the irradiation was finished within 2
h. The culture medium was removed and the cells were rinsed twice
with PBS, fixed with 500 µl of 4% polyformaldehyde for 2 h and
stained with 0.1% crystal violet for 3 h. When the dishes had been
dried out in air, the clone number was counted under a low power
light microscope (magnification, ×40). The average number of cloned
cells and survival fraction (SF) were calculated. The experiment
was repeated three times. The plating efficiency (PE) was
calculated as the number of colonies formed as a percentage of the
number of viable cells plated at the 0 Gy dose. Cell survival
rate=clone number at a given dose of irradiation/(number of cells
at the same dose xPE).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Following 48 h of transfection, the density of a
single cell suspension was adjusted to 5×105 cells/ml,
and the cells were seeded into a 96-well plate with three parallel
wells in each group, and cultured in an incubator with 5%
CO2 at 37°C. Three wells were randomly selected form
each group at 12, 24 and 48 h. To each well 20 µl of 5 mg/ml MTT
fluid (Sigma-Aldrich; Merck KGaA) was added and cultured for 4 h,
and the culture medium was discarded. To each well, 150 µl of
dimethyl sulfoxide was added. The plate was placed on the
microplate oscillator to dissolute crystal for 10 min. The optical
density (OD) of each well at 490 nm was measured by an
enzyme-linked immunometric meter (Elx800, Bio-Tek Instruments,
Inc., Winooski, VT, USA). The experiment was repeated three times.
Cell proliferation rate=(mean OD of the experiment group)/(mean OD
of the control group) ×100%.
Transwell assay
The cells in the logarithmic growth phase were
obtained to prepare the cell suspension, and seeded into the apical
chamber of the Transwell (Corning Costar, Cambridge, MA, USA).
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing
10% FBS was added into the basolateral chamber. The cells were
cultured in an incubator with 5% CO2 at 37°C for 30 h,
washed twice with PBS, fixed in 10% formaldehyde for 15 min and
stained with crystal violet for 20 min. The cells were counted and
images were captured at high magnification using an inverted
microscope. A total of 5 fields of visions were selected to obtain
the mean number of cells.
The transfected cells were cultured for 24 h and
treated with 0.25% trypsin, and cell density was modulated. The
cell suspension was seeded into the Transwell chamber. The apical
chamber was coated evenly with Matrigel with all microwells at the
bottom of the apical chamber covered. DMEM containing 10% FBS was
added to the bottom of the chamber. After 24 h, the number of cells
that invaded through the Matrigel was used to assess the invasive
ability.
Flow cytometry
Following transfection for 30 h, the CaSki cells in
each group were centrifuged at 1,000 × g for 5 min at room
temperature and the culture medium was discarded. The cells were
then washed once with PBS and incubated with 6 µl Annexin V-FITC at
room temperature for 15 min in the dark. Subsequently, 10 µl
propidium iodide (PI) was added to the cells, followed by immediate
analysis with flow cytometry (BD Biosciences, Franklin Lakes, NJ,
USA). FlowJo 7.0 software (FlowJo LLC, Ashland, OR, USA) was used
to analyze the results, and the experiments were repeated three
times. The apoptosis of CaSki cells was analyzed by flow cytometry.
The Annevin V (−)/PI (−) cells (left lower lattice) represent
normal living cells, the Annexin V (+)/PI (−) cells (right lower
lattice) represent apoptotic cells, Annevin (+)/PI (+) (left upper
lattice) cells represent necrotic cells, and Annevin V (−)/PI(+)
cells (right upper lattice) represent cells with completely
impaired membranes during cell digestion and collection.
Statistical analysis
SPSS 21.0 (IBM Corp., Armonk, NY, USA) was used for
data analysis. Measurement data are presented as the mean ±
standard deviation. The comparison between two groups was analyzed
with a Students t-test and comparison among multiple groups was
analyzed using one-way analysis of variance, and checked with
Tukey's post hoc test. Enumeration data are presented as a ratio or
percentage and were analyzed by χ2 test. The receiver
operating characteristic (ROC) curve was drawn to analyze the
predictive value of STK17A and miR-411 in the efficacy of
radiotherapy. Kaplan-Meier survival analysis and the log-rank test
were used to analyze the correlation between the overall survival
rate and the progression-free survival rate of patients with
cervical cancer. Risk factors for the prognosis of cervical cancer
were evaluated using Cox's proportional hazards regression model.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Efficacy of radiotherapy is not
associated with baseline characteristics
Initially, to understand the factors affecting the
efficacy of radiotherapy, the baseline characteristics were
compared. As shown in Table II,
age, FIGO stage, maximum tumor diameter, pathological type, lymph
node metastasis, menopause, hemoglobin level, SCC antigen level
pre-radiotherapy and radiotherapy method did not differ
significantly between the response group and the non-response group
(P>0.05).
 | Table II.Baseline characteristics of cervical
cancer between the response and non-response groups. |
Table II.
Baseline characteristics of cervical
cancer between the response and non-response groups.
|
|
| Response group | Non-response
group |
|
|---|
|
|
|
|
|
|
|---|
| Group | N=141 | (N=92) | (N=49) | P-value |
|---|
| Age (years) |
|
|
|
|
|
≥45 | 82 | 50 | 32 | 0.209 |
|
<45 | 59 | 42 | 17 |
|
| FIGO stage |
|
|
|
|
| I | 55 | 40 | 15 | 0.070 |
| II | 62 | 41 | 21 |
|
|
III | 24 | 11 | 13 |
|
| Maximum tumor
diameter (cm) |
|
|
|
|
| ≥4 | 57 | 36 | 21 | 0.668 |
|
<4 | 84 | 56 | 28 |
|
| Pathological
type |
|
|
|
|
|
SCC | 126 | 81 | 45 | 0.487 |
|
Adenocarcinoma | 15 | 11 | 4 |
|
| Lymph node
metastasis |
|
|
|
|
|
Yes | 35 | 20 | 15 | 0.246 |
| No | 106 | 72 | 34 |
|
| Menopause |
|
|
|
|
|
Yes | 81 | 52 | 29 | 0.761 |
| No | 60 | 40 | 20 |
|
| Hemoglobin level
(g/l) |
|
|
|
|
|
≥110 | 96 | 64 | 32 | 0.605 |
|
<110 | 45 | 28 | 17 |
|
| SCC antigen level
pre-radiotherapy (ng/ml) |
|
|
|
|
| ≥2 | 64 | 40 | 24 | 0.532 |
|
<2 | 77 | 52 | 25 |
|
| Radiotherapy
method |
|
|
|
|
|
Four-field conformal
radiotherapy | 33 | 21 | 12 | 0.080 |
|
Intensity-modulated
radiotherapy | 57 | 43 | 14 |
|
| Pelvic
hexagonal field radiotherapy | 51 | 28 | 23 |
|
Low expression of miR-411 or
overexpression of STK17A contributes to cervical cancer and poor
efficacy of radiotherapy
A target relationship between miR-411 and
serine/threonine kinase 38 like (STK38L) was found via the
TargetScan website (http://www.targetscan.org/vert_71/) and microRNA.org. The expression changes of STK17A and
STK38L were compared following miR mimic treatment (Fig. 1). The results showed that the
changes of STK17A were more marked, therefore, this target gene was
selected for further experiments. The expression levels of miR-411
and STK17A in the tissues and peripheral blood were detected to
examine their association with radiotherapy efficacy in cervical
cancer. As is shown in Fig. 2A, in
the cervical cancer tissues, the expression of miR-411 was markedly
decreased compared with that in the adjacent normal tissues
(P<0.05); the expression of STK17A was markedly increased
compared with that in the adjacent normal tissues (P<0.05). As
is shown in Fig. 2B, in the
response group, the expression of miR-411 was high and increased
compared with that in the non-response group (P<0.05); however,
the expression of STK17A was low and reduced significantly compared
with that in the non-response group (P<0.05). As shown in
Fig. 2C, the expression of miR-411
in the peripheral blood of the response group of cervical cancer
was 5.48±4.04 and in the peripheral blood of the non-response group
was 4.57±1.09; the expression of STK17A in the peripheral blood of
the response group of cervical cancer was 3.21±0.64 and in the
peripheral blood of the non-response group was 4.64±0.82 (all
P<0.001).
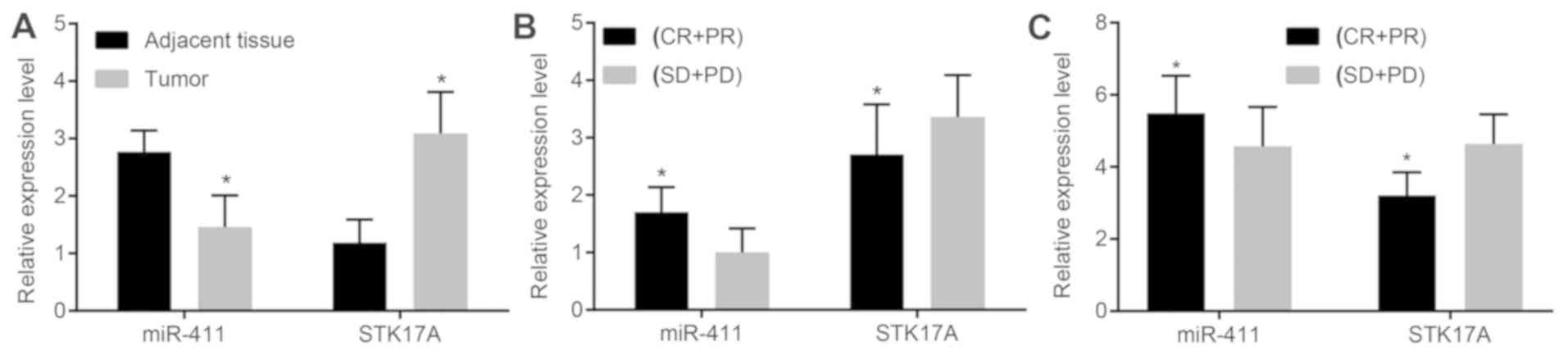 | Figure 2.miR-411 is decreased in cervical
cancer but increased following response to radiotherapy, with
STK17A changing reciprocally. (A) Cervical cancer tissues exhibited
decreased expression of miR-411 but increased expression of STK17A
compared with the adjacent normal tissues (n=141, data analyzed by
paired t-test), (B) miR-411 was increased and STK17A was decreased
in the CR + PR group (n=92) compared with the SD + PD group (n=49),
data were analyzed by independent t-test. (C) miR-411 was increased
and STK17A was decreased in the peripheral blood of the CR + PR
group (n-92) compared with the SD + PD group (n=49), data were
analyzed by independent t-test. *P<0.05, vs. adjacent
normal tissues or SD + PD group. CR, complete remission; PR,
partial remission; PD, progressive disease; SD, stable disease;
miR-411, microRNA-411; STK17A, serine/threonine kinase 17a. |
Expression of miR-411 and STK17A have
predictive value for radiotherapy efficacy in cervical cancer
The expression of miR-411 and STK17A in tissues and
peripheral blood was determined to predict radiotherapy efficacy.
When the cut-off value was 1.755 in tissues for the expression of
miR-411, the area under the ROC curve (AUC) was 0.871, and the
sensitivity and specificity were 70.7 and 91.8%, respectively (95%
confidence interval, 0.814–0.920); when the cut-off value was 4.360
in the peripheral blood, the AUC was 0.729, and the sensitivity and
specificity were 89.1 and 51.0%, respectively (95% confidence
interval, 0.641–0.817), suggesting that miR-411 had good predictive
value for radiotherapy efficacy in tissues and peripheral blood in
cervical cancer (Fig. 3A). When
the cut-off value was 2.605 in tissues for the expression of
STK17A, the AUC was 0.723, and the sensitivity and specificity were
89.8 and 50.0%, respectively (95% confidence interval,
0.641–0.806); when the cut-off value was 3.890 in the peripheral
blood for the expression of STK17A, the AUC was 0.907, and the
sensitivity and specificity were 83.7 and 91.3%, respectively (95%
confidence interval, 0.849–0.966), indicating that STK17A had good
predictive value for the efficacy of radiotherapy in tissues and
peripheral blood in cervical cancer (Fig. 3B). Furthermore, the expression of
HPV16 and HPV18 in tissues were also determined to predict
radiotherapy efficacy. In tissues positive for HPV16, the AUC was
0.969, and the sensitivity and specificity were 93.5 and 100.0%,
respectively (95% confidence interval, 0.936–1.000), indicating
that HPV16 had good predictive value for the efficacy of
radiotherapy in tissues in cervical cancer (Fig. 3C). In tissues positive for HPV18,
the AUC was 0.994, and the sensitivity and specificity were 98.9
and 98.8%, respectively (95% confidence interval, 0.000–1.000),
indicating that HPV18 had good predictive value for the efficacy of
radiotherapy in tissues in cervical cancer (Fig. 3D).
miR-411 correlates with FIGO stage,
lymph node metastasis and radiotherapy method, and STK17A
correlates with FIGO stage and radiotherapy method
The expression levels of miR-411 and STK17A were
detected to examine their correlation with the clinical
characteristics of cervical cancer. There was no significant
association between the expression of miR-411 and age, maximum
tumor diameter, pathological type, menopause, hemoglobin level, SCC
antigen level pre-radiotherapy or radiotherapy method (all
P>0.05). However, the expression of miR-411 was significantly
lower in patients at FIGO stage II and III, with lymph node
metastasis or treated with four-field conformal radiotherapy,
compared with patients at FIGO I, without lymph node metastasis or
treated with intensity-modulated radiotherapy and pelvic hexagonal
field radiotherapy (all P<0.05). The expression of STK17A was
higher in patients at FIGO stage III and treated with four-field
conformal radiotherapy and pelvic hexagonal field radiotherapy,
compared with patients at FIGO stage I and II and treated with
intensity-modulated radiotherapy (all P<0.05), whereas no
difference was found between the expression of STK17A and age,
maximum tumor diameter, pathological type, lymph node metastasis,
menopause, hemoglobin level, SCC antigen level pre-radiotherapy or
radiotherapy method (all P>0.05; Table III).
 | Table III.Correlation between the expression
levels of miR-411 and STK17A and clinical features of cervical
cancer. |
Table III.
Correlation between the expression
levels of miR-411 and STK17A and clinical features of cervical
cancer.
|
|
| miR-411
expression | STK17A
expression |
|---|
|
|
|
|
|
|---|
| Group | N=141 | High/low | χ2 | P-value | High/low | χ2 | P-value |
|---|
| Age (years) |
|
|
|
|
|
|
|
|
≥45 | 82 | 39/43 | 0.148 | 0.700 | 25/57 | 1.030 | 0.310 |
|
<45 | 59 | 30/29 |
|
| 13/46 |
|
|
| FIGO stage |
|
|
|
|
|
|
|
| I | 55 | 35/20 | 8.006 | 0.018 | 12/43 | 10.890 | 0.004 |
| II | 62 | 27/35 |
|
| 13/49 |
|
|
|
III | 24 | 7/17 |
|
| 13/11 |
|
|
| Maximum tumor
diameter (cm) |
|
|
|
|
|
|
|
| ≥4 | 57 | 24/33 | 1.787 | 0.171 | 17/40 | 0.402 | 0.526 |
|
<4 | 84 | 45/39 |
|
| 21/63 |
|
|
| Pathological
type |
|
|
|
|
|
|
|
|
SCC | 126 | 62/64 | 0.035 | 0.852 | 34/92 | 0.001 | 0.980 |
|
Adenocarcinoma | 15 | 7/8 |
|
| 4/11 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
|
Yes | 35 | 9/26 | 10.051 | 0.002 | 12/23 | 1.272 | 0.259 |
| No | 106 | 60/46 |
|
| 26/80 |
|
|
| Menopause |
|
|
|
|
|
|
|
|
Yes | 81 | 39/42 | 0.047 | 0.828 | 21/60 | 0.102 | 0.750 |
| No | 60 | 30/30 |
|
| 17/43 |
|
|
| Hemoglobin level
(g/l) |
|
|
|
|
|
|
|
|
≥110 | 96 | 50/46 | 1.192 | 0.275 | 26/70 | 0.003 | 0.959 |
|
<110 | 45 | 19/26 |
|
| 12/33 |
|
|
| SCC antigen level
pre-radiotherapy (ng/ml) |
|
|
|
|
|
|
|
| ≥2 | 64 | 32/32 | 0.053 | 0.818 | 18/46 | 0.082 | 0.774 |
|
<2 | 77 | 37/40 |
|
| 20/57 |
|
|
| Radiotherapy
method |
|
|
|
|
|
|
|
|
Four-field conformal
radiotherapy | 33 | 9/24 | 8.901 | 0.012 | 11/22 | 12.085 | 0.034 |
|
Intensity-modulated
radiotherapy | 57 | 34/23 |
|
| 10/47 |
|
|
| Pelvic
hexagonal field radiotherapy | 51 | 26/25 |
|
| 17/34 |
|
|
Low expression of miR-411 n and
overexpression of STK17A contribute to poor survival rate
Kaplan-Meier survival analysis and log-rank test
were used to analyze the association between overall survival rate,
progression-free survival rate and expression levels of miR-411 and
STK17A. The patients with a high expression of miR-411 had higher
3-year overall survival and progression-free survival rates,
compared with those with a low expression of miR-411 (P<0.05).
The patients with a high expression of STK17A had lower 3-year
overall survival and progression-free survival rates, compared with
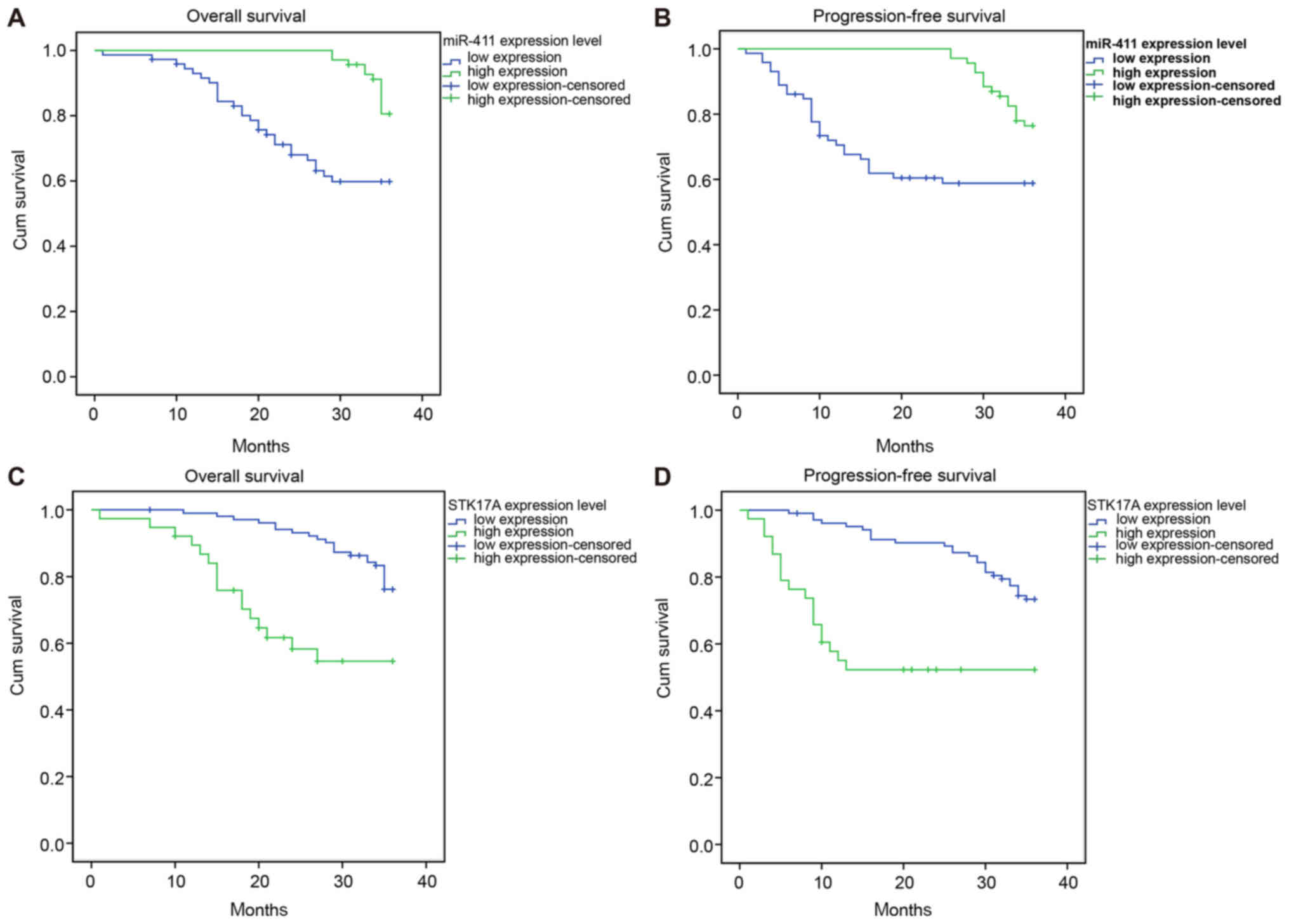
those with a low expression of STK17A (P<0.05; Fig. 4A-D and Table IV). Patients with
intensity-modulated radiotherapy and no lymph node metastasis had
higher survival rates than those with lymph node metastasis,
four-field conformal radiotherapy and pelvic hexagonal field
radiotherapy (P<0.05). No significant association was found
between survival rate and age, FIGO stage, maximum tumor diameter,
pathological type, menopause, hemoglobin level, SCC antigen level
pre-radiotherapy and radiotherapy method (all P>0.05; Table IV).
 | Table IV.Correlation between the clinical
features of cervical cancer and survival rate in patients. |
Table IV.
Correlation between the clinical
features of cervical cancer and survival rate in patients.
| Group | N=141 | 3-year overall
survival rate (%) | χ2 | P-value | 3 year
progression-free survival rate (%) | χ2 | P-value |
|---|
| miR-411 |
|
|
|
|
|
|
|
| High
expression | 69 | 56 (81.2) | 11.421 | 0.001 | 53 (76.8) | 9.421 | 0.002 |
| Low
expression | 72 | 45 (62.5) |
|
| 43 (59.7) |
|
|
| STK17A |
|
|
|
|
|
|
|
| High
expression | 38 | 22 (57.9) | 13.337 | <0.001 | 20 (52.6) | 14.515 | <0.001 |
| Low
expression | 103 | 79 (76.7) |
|
| 76 (73.8) |
|
|
| Age (years) |
|
|
|
|
|
|
|
|
≥45 | 82 | 56 (68.3) | 1.255 | 0.263 | 52 (63.4) | 1.974 | 0.160 |
|
<45 | 59 | 45 (76.3) |
|
| 44 (74.6) |
|
|
| FIGO stage |
|
|
|
|
|
|
|
| I | 55 | 43 (78.2) | 3.544 | 0.170 | 39 (70.9) | 3.625 | 0.163 |
| II | 62 | 44 (71.0) |
|
| 44 (71.0) |
|
|
|
III | 24 | 14 (58.3) |
|
| 13 (54.2) |
|
|
| Maximum tumor
diameter (cm) |
|
|
|
|
|
|
|
| ≥4 | 57 | 37 (64.9) | 2.504 | 0.114 | 35 (61.4) | 2.156 | 0.142 |
|
<4 | 84 | 64 (76.2) |
|
| 61 (72.6) |
|
|
| Pathological
type |
|
|
|
|
|
|
|
|
SCC | 126 | 89 (70.6) | 0.499 | 0.480 | 86 (68.3) | 0.007 | 0.933 |
|
Adenocarcinoma | 15 | 12 (80.0) |
|
| 10 (66.7) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
|
Yes | 35 | 20 (57.1) | 4.973 | 0.026 | 18 (51.4) | 6.721 | 0.010 |
| No | 106 | 81 (76.4) |
|
| 78 (73.6) |
|
|
| Menopause |
|
|
|
|
|
|
|
|
Yes | 81 | 56 (69.1) | 0.417 | 0.519 | 51 (63.0) | 1.609 | 0.205 |
| No | 60 | 45(75.0) |
|
| 45 (75.0) |
|
|
| Hemoglobin level
(g/L) |
|
|
|
|
|
|
|
|
≥110 | 96 | 69 (71.9) | 0.076 | 0.783 | 64 (66.7) | 0.107 | 0.744 |
|
<110 | 45 | 32 (71.1) |
|
| 32 (71.1) |
|
|
| SCC antigen level
pre-radiotherapy (ng/ml) |
|
|
|
|
|
|
|
| ≥2 | 64 | 45 (70.3) | 0.190 | 0.663 | 43 (67.2) | 0.071 | 0.791 |
|
<2 | 77 | 56 (72.7) |
|
| 53 (68.8) |
|
|
| Radiotherapy
method |
|
|
|
|
|
|
|
|
Four-field conformal
radiotherapy | 33 | 24 (72.7) | 11.946 | 0.003 | 23 (69.7) | 6.071 | 0.048 |
|
Intensity-modulated
radiotherapy | 57 | 48 (84.2) |
|
| 44 (77.2) |
|
|
| Pelvic
hexagonal field radiotherapy | 51 | 29 (56.9) |
|
| 29 (56.9) |
|
|
Risk factors for prognosis include
lymph node metastasis, radiotherapy method and expression of
STK17A
Cox's regression model was used to analyze the
association between the expression of miR-411 and STK17A and the
prognosis of patients with cervical cancer, and for the analysis of
prognostic risk factors. Cox's proportional hazards regression
model suggested that the 3-year survival rate was associated with
radiotherapy method and expression levels of miR-411 and STK17A,
and 3-year progression-free survival rate was correlated with lymph
node metastasis, radiotherapy method and expression levels of
miR-411 and STK17A (all P<0.05). miR-411 was a protective
factor, and lymph node metastasis, radiotherapy method and
expression of STK17A were risk factors for the prognosis of
patients with cervical cancer (Table
V).
 | Table V.Risk factors for prognosis of
patients with cervical cancer. |
Table V.
Risk factors for prognosis of
patients with cervical cancer.
|
| 3-year overall
survival rate | 3-year
progression-free survival rate |
|---|
|
|
|
|
|---|
| Risk factor | P-value | EXP | 95% CI | P-value | EXP | 95% CI |
|---|
| miR-411
expression | 0.013 | 0.37 | 0.17–0.81 | 0.049 | 0.506 | 0.26–0.98 |
| STK17A
expression | 0.018 | 1.9 | 1.18–3.08 | 0.001 | 3.22 | 1.63–6.36 |
| FIGO stage | 0.257 | 1.31 | 0.82–2.09 | 0.452 | 1.2 | 0.75–1.90 |
| Lymph node
metastasis | 0.411 | 1.35 | 0.66–2.73 | 0.041 | 2.01 | 1.03–3.93 |
| Radiotherapy
method | 0.009 | 1.9 | 1.17–3.08 | 0.024 | 1.67 | 1.07–2.60 |
Expression of mir-411 is higher and
expression of STK17A is lower in cervical cancer cells
The expression levels of miR-411 and STK17A in each
cervical cancer cell line were detected to identify the most
appropriate cell line for the investigation. As is shown in
Fig. 5A, the expression of miR-411
was highest in the C33A cell line, followed by the HeLa and SiHa
cell lines, and was lowest in the CaSki cells. As shown in Fig. 5B, the expression of STK17A was
lowest in the C33A cell line, followed by the HeLa and SiHa cell
lines, and was highest in the CaSki cells. Therefore, the CaSki
cell line was selected to analyze the function of miR-411 in
cervical cancer.
miR-411 targets STK17A
The microRNA.org
website was used to predict the target relationship between miR-411
and STK17A, and it was found that the STK17A 3′-UTR contained
miR-411 binding sites (Fig. 6A).
It was found that the miR-411 mimic had no significant effects on
the luciferase activity of the Mut-miR-411/STK17A plasmid. There
was a marked reduction in the luciferase activity of the
Wt-miR-411/STK17A plasmid compared with the Mut-miR-411/STK17A
plasmid (P<0.05, Fig. 6B),
which suggested that miR-411 directly downregulated STK17A.
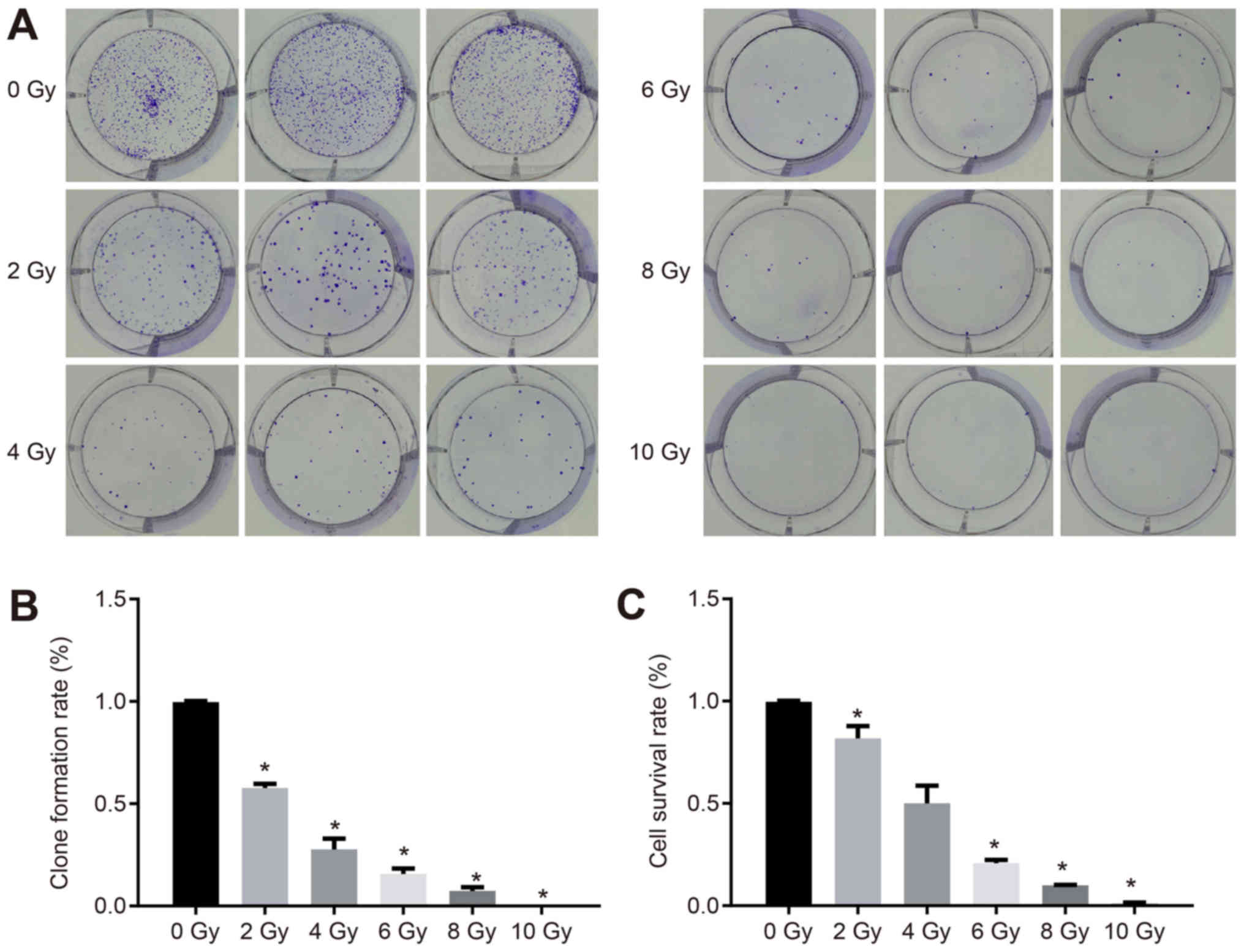
Increased radiotherapy dose decreases
the proliferation of cervical cancer cells
The proliferation of CaSki cells under different
doses of radiotherapy was measured. As is shown in Fig. 7A, the CaSki cell distribution was
dense, irregular and flat at the dose of 0 Gy. When the dose was 10
Gy, there was no colony formation, which indicated that different
doses of X-ray irradiation affected the colony forming ability and
viability of CaSki cells. The clone formation rate and survival
rate of the CaSki cells were reduced when the X-ray dose was
increased (Fig. 7B and C).
miR-411 suppresses functional STK17A
and mediates the p53 signaling pathway in cervical cancer
cells
The expression levels of p53 signaling
pathway-related genes were detected by RT-qPCR and western blot
analyses to examine the role of miR-411 in the p53 signaling
pathway. Following CaSki cell line transfection, compared with the
NC group, the expression of miR-411 was markedly increased in the
miR-411 mimic group and markedly decreased in the miR-411 inhibitor
and miR-411 inhibitor + siRNA-STK17A groups (all P<0.05).
Compared with the NC group, the mRNA and protein expression levels
of STK17A were decreased, but the mRNA and protein expression
levels of p53, p21WAF1 and TAp63 were increased in the
miR-411 mimic and siRNA-STK17A groups (all P<0.05). The mRNA and
protein expression levels of STK17A were elevated, whereas the mRNA
and protein expression of p53, p21WAF1 and TAp63 were
reduced in the miR-411 inhibitor group (all P<0.05). Compared
with the miR-411 inhibitor group, the mRNA and protein expression
levels of STK17A were lower in the miR-411 inhibitor + siRNA-STK17A
group, whereas the mRNA and protein expression levels of p53,
p21WAF1 and TAp63 were higher (all P<0.05). The mRNA
and protein expression levels of STK17A were lower, whereas those
of p53, p21WAF1 and TAp63 were higher in the
siRNA-STK17A group than in the miR-411 inhibitor + siRNA-STK17A
group (all P<0.05). No significant differences were observed in
the expression of miR-411, or the mRNA and protein expression
levels of STK17A, p53, p21WAF1 and TAp63 between the NC group and
blank group (all P>0.05). In general, the inhibition of miR-411
decreased the expression of p53, p21WAF1 and TAp63
through upregulating STK17A (Fig.
8A-C).
 | Figure 8.miR-411 activates the p53 signaling
pathway and negatively regulates STK17A in cervical cancer cells.
(A) Determination by reverse transcription-quantitative polymerase
chain reaction analysis demonstrated that ectopic expression of
miR-411 and siRNA-mediated knockdown of STK17A decreased the mRNA
expression of STK17A, but increased the mRNA expression of p53,
p21WAF1 and TAp63; miR-411 inhibitor increased the mRNA
expression of STK17A, but decreased the mRNA expression of p53,
p21WAF1 and TAp63. (B) Determination by western blot
analysis and (C) quantification demonstrated that ectopic
expression of miR-411 and siRNA-mediated knockdown of STK17A
decreased the protein expression of STK17A, but increased the
protein expression of p53, p21WAF1 and TAp63; miR-411
inhibitor increased the protein expression of STK17A, but decreased
the protein expression of p53, p21WAF1 and TAp63.
*P<0.05, vs. NC group; #P<0.05, vs. miR-411
inhibitor + siRNA-STK17A group. The experiment was repeated three
times and data were compared by one-way analysis of variance and
analyzed by Tukey's post hoc test. NC, negative control; miR-411,
microRNA-411; STK17A, serine/threonine kinase 17a; siRNA, small
interfering RNA. |
STK17A is responsible for the
inhibitory effect of miR-411 on the proliferation of cervical
cancer cells
In order to investigate the effect of miR-411 and
STK17A on cell proliferation of cervical cancer, an MTT assay was
performed. As is shown in Fig.
9A-D, compared with the NC group, the cell viability of the
miR-411 inhibitor group exhibited a sharp increase, whereas the
rates in the miR-411 mimic group and siRNA-STK17A group exhibited a
sharp decrease (all P<0.05). The cell viability in the NC group
and miR-411 inhibitor + siRNA-STK17A group were almost the same as
that in the blank group (all P>0.05), indicating that the
increase of miR-411 and decrease of STK17A inhibited the
proliferation of cervical cancer cells.
STK17A is responsible for the
inhibitory effect of miR-411 on cervical cancer cell migration and
invasion
In order to investigate the effects of miR-411 and
STK17A on cell migration and invasion of cervical cancer, a
Transwell assay was performed. CaSki cell migration and invasion in
the NC group did not differ significantly from that in the blank
group (both P>0.05). CaSki cell migration and invasion were
reduced following transfection with the miR-411 mimic or
siRNA-STK17A, but enhanced following transfection with miR-411
inhibitor, compared with the NC group (all P<0.05). CaSki cell
migration and invasion were reduced following transfection with
miR-411 inhibitor + siRNA-STK17A, compared with the miR-411
inhibitor group (both P<0.05). This indicated that the
inhibition of miR-411 or STK17A inhibited the migration and
invasion of CaSki cells (Fig.
10A-D).
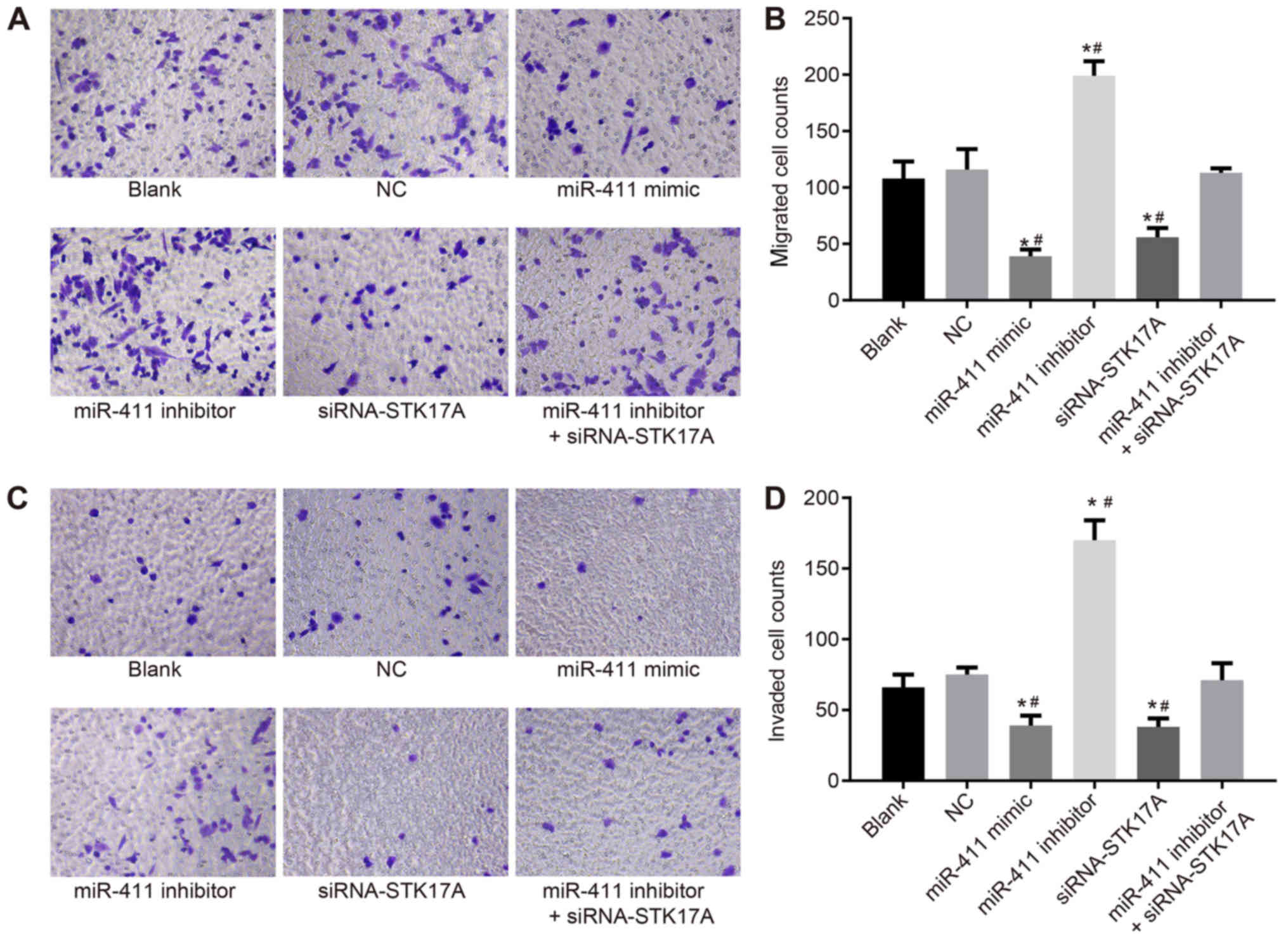 | Figure 10.Transwell assay shows that miR-411
suppresses cervical cancer cell migration and invasion through the
STK17A-dependent p53 signaling pathway. (A) Under the inverted
microscope (magnification, ×200), the number of migrated cells
reduced following upregulation of miR-411 or siRNA-mediated
knockdown of STK17A, but increased following inhibitor-mediated
knockdown of miR-411. (B) Numbers of migrated cells are indicated
in a representative histogram. (C) Under the inverted microscope
(magnification, ×200), the number of cells in the basolateral
chamber decreased following upregulation of miR-411 or
siRNA-mediated knockdown of STK17A, but increased following
inhibitor-mediated knockdown of miR-411. (D) Numbers of cells
passing through the Matrigel from the apical chamber to the
basolateral chamber are indicated in a representative histogram.
*P<0.05, vs. NC group; #P<0.05, vs. miR-411
inhibitor + siRNA-STK17A group. The experiment was repeated three
times and data were compared by one-way analysis of variance and
analyzed by Tukey's post hoc test. NC, negative control; miR-411,
microRNA-411; STK17A, serine/threonine kinase 17a; siRNA, small
interfering RNA. |
STK17A is responsible for the
promoting effect of miR-411 on cervical cancer cell apoptosis
To further investigate the effect of miR-411 and
STK17A on the apoptosis of cervical cancer cells, a Transwell assay
was performed. As is shown in Fig.
11A and B, the apoptotic rate of CaSki cells was decreased
following transfection with the miR-411 inhibitor compared with
that in the NC group, but the rate was increased following
transfection with the miR-411 mimic or siRNA-STK17A (both
P<0.05). Compared with the miR-411 inhibitor group, the
apoptotic rate was increased in the miR-411 inhibitor +
siRNA-STK17A group, however, the rate remained lower in the miR-411
inhibitor group than in the siRNA-STK17A group (all P<0.05). The
apoptotic rate in the NC group did not differ significantly to that
in the blank group (P>0.05). These results suggested that the
apoptotic rate of CaSki cells was increased by the increase of
miR-411 or decrease of STK17A.
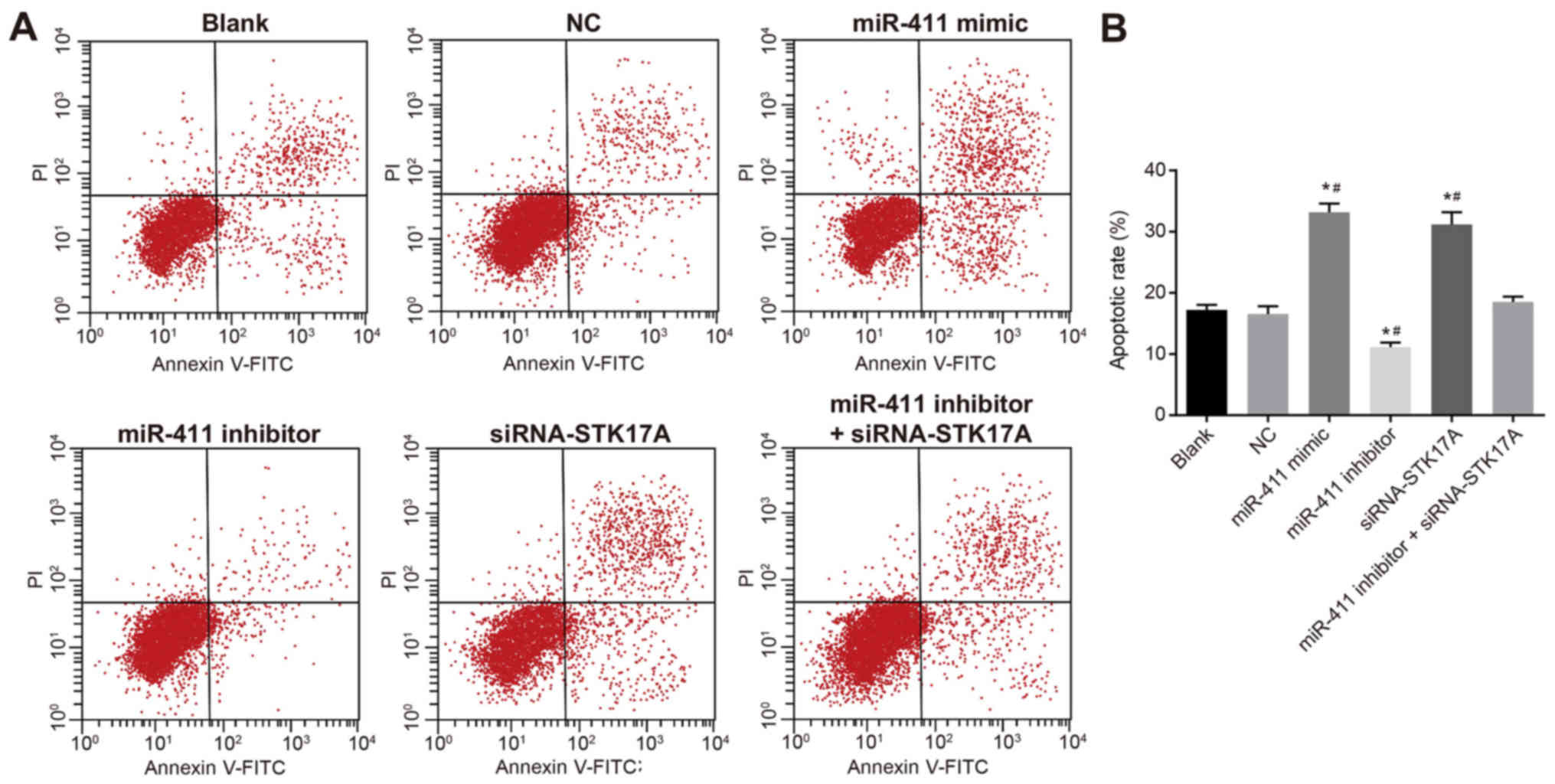 | Figure 11.Flow cytometric analysis indicates
that miR-411 promotes cervical cancer cell apoptosis through the
STK17A-dependent p53 signaling pathway. (A) CaSki cells in the
scatter plots, in which the upper left quadrant identifies necrotic
cells (Annexin V−/PI+), the upper right
quadrant identifies late apoptotic cells (Annexin
V+/PI+), the lower left quadrant identifies
live cells (Annexin V−/PI−), and the lower
right quadrant identifies early apoptotic cells (Annexin
V+/PI−). (B) Percentages of apoptotic cells
are indicated in a representative histogram. *P<0.05 vs. NC
group; #P<0.05, vs. miR-411 inhibitor + siRNA-STK17A
group. The experiment was repeated three times and data were
compared by one-way analysis of variance and analyzed by Tukey's
post hoc test. NC, negative control; miR-411, microRNA-411; STK17A,
serine/threonine kinase 17a; siRNA, small interfering RNA; PI,
propidium iodide. |
Discussion
Cervical cancer, one of the most common types of
cancer in women worldwide, requires improvements in therapy due to
the lack of defined biomarkers and targets for the development of
this disease (31). In terms of
etiology, the factors influencing cervical cancer are various,
including nutritional factors, smoking, reproductive factors,
genetic factors and sex factors (2). The objective of the present study was
to examine the correlation of miR-411 and STK17A with radiotherapy
efficacy and prognosis of cervical cancer. The main conclusion of
the study was that the upregulation of miR-411 can inhibit the
expression of STK17A, activate the p53 signaling pathway, suppress
proliferation, migration and invasion, and promote the apoptosis of
cervical cancer cells.
Firstly, it was found that, compared with adjacent
normal tissues, miR-411 exhibited decreased expression whereas
STK17A exhibited increased expression in cervical cancer tissues.
It is reported that >50% of miRNAs are present in fragile sites
and cancer-associated genomic regions, which indicates that miRNAs
are important in cancer formation and are significant regulators
for diverse types of cancer, including cervical cancer (26,32).
miR-411 has also been demonstrated to be downregulated in breast
cancer (33) and in osteoarthritis
cartilage (34). STK17A, a novel
target gene of p53, has been found to be a factor affecting the
functions of cancer cells and exhibits increased expression in
cancer cells (21,35,36).
P53 has been revealed to be a suppressive factor in cervical cancer
and its activation can inhibit the proliferation, migration and
invasion of cervical cancer cells (37). It has been observed that the
expression of miR-411 had a close association with FIGO stage and
lymph node metastasis; two previous studies have demonstrated that
miRNAs have a correlation with FIGO stage and that miR-411 is
associated with lymph node metastasis in patients with breast
cancer (33,38). Furthermore, in the present study,
it was found that the overexpression of miR-411 and low expression
of STK17A were correlated with the high efficacy of radiotherapy
and favorable prognosis. miR-411 has been identified as a
prognostic biomarker and serves as a tumor promotor for non-small
cell lung cancer (39). In a study
by Yanaihara et al (40),
the expression of miRNA was shown to be correlated with the
diagnosis and prognosis of cancer, therefore miRNAs can act as
biomarkers for cancer. The high expression of miR-411 in patients
with lung cancer is found to be correlated with poor prognosis
(41). miR-411 and STK17A have
been identified as factors influencing ovarian cancer (19,42).
The expression of STK17A has been confirmed to affect the prognosis
of patients with cervical cancer, and patients with overexpression
of STK17A are more likely to have poor outcomes (31).
Additionally, a luciferase reporter gene assay
confirmed that STK17A is a target gene of miR-411. In addition, it
was detected that the expression levels of p53, p21WAF1
and TAp63 were increased by upregulating miR-411 or downregulating
STK17A, leading to the suppression of proliferation, migration and
invasion, and the promotion of apoptosis in cervical cancer cells.
It has been identified that p53 exhibits increased expression in
cancer cells, and the p53 signaling pathway can inhibit the
progression of cancer by coordinating transcription programs when
activated by diverse stress signals (43,44).
STK17A is a novel gene found in p53 and confirmed to be a modulator
in various types of cancer, including colon cancer and testicular
cancer (21,45). It was previously revealed that
miR-411 can function as a factor suppressing the proliferation and
invasion but promoting the apoptosis of colorectal cancer cells by
directly targeting phosphoinositide-3-kinase regulatory subunit 3
(46). Another study found that
increased expression of miR-411 promoted osteosarcoma cell
proliferation and migration through inhibiting the expression of
metastasis suppressor protein 1 (47). One of the direct and DNA
damage-inducible p53 target genes is STK17A, which is involved in
cellular processes, and a functional and consensus p53 pathway
response element is located upstream of STK17A (21,48).
STK17A is regarded as a factor causing apoptosis due to a variety
of apoptotic stimuli, including certain drugs, UV light FasL and
tumor necrosis factor-α, and in a study investigating the
correlation between miR-411 and hepatocellular carcinoma cells,
miR-411 was confirmed to be involved in cell proliferation
(16,19).
In conclusion, the present study provides evidence
that miR-411 and its target STK17A are therapeutic biomarkers for
efficacy and prognosis in patients with cervical cancer treated
with radiotherapy, and that miR-411 downregulating STK17A can
inhibit the proliferation, migration and invasion, and promote the
apoptosis of cervical cancer cells by activating the p53 signaling
pathway. However, the mechanisms of miR-411 in the development and
prognosis of cervical cancer require further investigation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WW and CL designed the study. WW performed the
experiment and statistical analysis of the results. CL wrote the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Jining No. 1 People's Hospital and informed consent
was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xiong Y, Sun F, Dong P, Watari H, Yue J,
Yu MF, Lan CY, Wang Y and Ma ZB: iASPP induces EMT and cisplatin
resistance in human cervical cancer through miR-20a-FBXL5/BTG3
signaling. J Exp Clin Cancer Res. 36:482017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li M, Feng YM and Fang SQ: Overexpression
of ezrin and galectin-3 as predictors of poor prognosis of cervical
cancer. Braz J Med Biol Res. 50:e53562017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang M, Zhang H, Yu Y, Huang H, Li G and
Xu C: Synergistic effects of a novel lipid-soluble extract from
Pinellia pedatisecta Schott and cisplatin on human cervical
carcinoma cell lines through the regulation of DNA damage response
signaling pathway. Oncol Lett. 13:2121–2128. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang L, Huang Z, Fan Y, He L, Ye M, Shi
K, Ji B, Huang J, Wang Y and Li Q: FOXC1 promotes proliferation and
epithelial-mesenchymal transition in cervical carcinoma through the
PI3K-AKT signal pathway. Am J Transl Res. 9:1297–1306.
2017.PubMed/NCBI
|
|
5
|
Pedroza-Torres A, López-Urrutia E,
García-Castillo V, Jacobo-Herrera N, Herrera LA, Peralta-Zaragoza
O, López-Camarillo C, De Leon DC, Fernández-Retana J, Cerna-Cortés
JF and Pérez-Plasencia C: MicroRNAs in cervical cancer: Evidences
for a miRNA profile deregulated by HPV and its impact on
radio-resistance. Molecules. 19:6263–6281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang T, Wong HK, Gu W, Yu MY, To KF, Wang
CC, Wong YF, Cheung TH, Chung TK and Choy KW: MicroRNA-182 plays an
onco-miRNA role in cervical cancer. Gynecol Oncol. 129:199–208.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen B, Hou Z, Li C and Tong Y: MiRNA-494
inhibits metastasis of cervical cancer through Pttg1. Tumour Biol.
36:7143–7149. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cavalleri T, Angelici L, Favero C, Dioni
L, Mensi C, Bareggi C, Palleschi A, Rimessi A, Consonni D, Bordini
L, et al: Plasmatic extracellular vesicle microRNAs in malignant
pleural mesothelioma and asbestos-exposed subjects suggest a
2-miRNA signature as potential biomarker of disease. PLoS One.
12:e01766802017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cho WC: MicroRNAs: Potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lam CS, Ng L, Chow AK, Wan TM, Yau S,
Cheng NS, Wong SK, Man JH, Lo OS, Foo DC, et al: Identification of
microRNA 885-5p as a novel regulator of tumor metastasis by
targeting CPEB2 in colorectal cancer. Oncotarget. 8:26858–26870.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu Z, Zhou Y, Shi F, Cao Y, Dinh TLA, Wan
J and Zhao M: Investigation of differentially-expressed microRNAs
and genes in cervical cancer using an integrated bioinformatics
analysis. Oncol Lett. 13:2784–2790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang W, Li Y, Liu N, Gao Y and Li L:
MiR-23b controls ALDH1A1 expression in cervical cancer stem cells.
BMC Cancer. 17:2922017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Harafuji N, Schneiderat P, Walter MC and
Chen YW: miR-411 is up-regulated in FSHD myoblasts and suppresses
myogenic factors. Orphanet J Rare Dis. 8:552013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi X, Xiao X, Yuan N, Zhang S, Yuan F and
Wang X: MicroRNA-379 suppresses cervical cancer cell proliferation
and invasion by directly targeting V-crk avian sarcoma virus CT10
oncogene homolog-like (CRKL). Oncol Res. 26:987–996. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun M, Huang F, Yu D, Zhang Y, Xu H, Zhang
L, Li L, Dong L, Guo L and Wang S: Autoregulatory loop between
TGF-β1/miR-411-5p/SPRY4 and MAPK pathway in rhabdomyosarcoma
modulates proliferation and differentiation. Cell Death Dis.
6:e18592015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia K, Zhang Y, Cao S, Wu Y, Guo W, Yuan W
and Zhang S: miR-411 regulated ITCH expression and promoted cell
proliferation in human hepatocellular carcinoma cells. Biomed
Pharmacother. 70:158–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Z, Qin L and Li S: miR-411
contributes the cell proliferation of lung cancer by targeting
FOXO1. Tumour Biol. 37:5551–5560. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang X, Zhang M, Cheng J, Lv Z, Wang F
and Cai Z: MiR-411 functions as a tumor suppressor in renal cell
cancer. Int J Biol Markers. 32:e454–e460. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao J, Liu D, Li J, Song Q and Wang Q:
Effect of STK17A on the sensitivity of ovarian cancer cells to
paclitaxel and carboplatin. Oncol Lett. 12:1107–1112. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mao P, Hever-Jardine MP, Rahme GJ, Yang E,
Tam J, Kodali A, Biswal B, Fadul CE, Gaur A, Israel MA and Spinella
MJ: Serine/threonine kinase 17A is a novel candidate for
therapeutic targeting in glioblastoma. PLoS One. 8:e818032013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mao P, Hever MP, Niemaszyk LM, Haghkerdar
JM, Yanco EG, Desai D, Beyrouthy MJ, Kerley-Hamilton JS, Freemantle
SJ and Spinella MJ: Serine/threonine kinase 17A is a novel p53
target gene and modulator of cisplatin toxicity and reactive oxygen
species in testicular cancer cells. J Biol Chem. 286:19381–19391.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Prives C and Hall PA: The p53 pathway. J
Pathol. 187:112–126. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang S, Zhou L, Hong B, van den Heuvel
AP, Prabhu VV, Warfel NA, Kline CL, Dicker DT, Kopelovich L and
El-Deiry WS: Small-molecule NSC59984 restores p53 pathway signaling
and antitumor effects against colorectal cancer via p73 activation
and degradation of mutant p53. Cancer Res. 75:3842–3852. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Muthusami S, Prabakaran DS, An Z, Yu JR
and Park WY: EGCG suppresses Fused Toes Homolog protein through p53
in cervical cancer cells. Mol Biol Rep. 40:5587–5596. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu PP, Chung HW, Liu KC, Wu RS, Yang JS,
Tang NY, Lo C, Hsia TC, Yu CC, Chueh FS, et al: Diallyl sulfide
induces cell cycle arrest and apoptosis in HeLa human cervical
cancer cells through the p53, caspase- and mitochondria-dependent
pathways. Int J Oncol. 38:1605–1613. 2011.PubMed/NCBI
|
|
26
|
Au Yeung CL, Tsang TY, Yau PL and Kwok TT:
Human papillomavirus type 16 E6 induces cervical cancer cell
migration through the p53/microRNA-23b/urokinase-type plasminogen
activator pathway. Oncogene. 30:2401–2410. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Waggoner SE: Cervical cancer. Lancet.
361:2217–2225. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meva J, Chaudhary RK, Bhaduri D, Bhatia M,
Hatti S and Ba R: Lacunae in International Federation of Gynecology
and Obstetrics (FIGO) classification for cervical carcinoma:
Observational study using TNM classification as comparator. Int J
Gynecol Cancer. 23:1071–1077. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thomas A, Mahantshetty U, Kannan S,
Deodhar K, Shrivastava SK, Kumar-Sinha C and Mulherkar R:
Expression profiling of cervical cancers in Indian women at
different stages to identify gene signatures during progression of
the disease. Cancer Med. 2:836–848. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao S, Yao D, Chen J and Ding N:
Circulating miRNA-20a and miRNA-203 for screening lymph node
metastasis in early stage cervical cancer. Genet Test Mol
Biomarkers. 17:631–636. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo L, Yuan J, Xie N, Wu H, Chen W, Song S
and Wang X: miRNA-411 acts as a potential tumor suppressor miRNA
via the downregulation of specificity protein 1 in breast cancer.
Mol Med Rep. 14:2975–2982. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang G, Zhang Y, Zhao X, Meng C, Ma L and
Kong Y: MicroRNA-411 inhibited matrix metalloproteinase 13
expression in human chondrocytes. Am J Transl Res. 7:2000–2006.
2015.PubMed/NCBI
|
|
35
|
Ozeki M, Salah A, Aini W, Tamaki K, Haga H
and Miyagawa-Hayashino A: Abnormal localization of STK17A in Bile
Canaliculi in liver allografts: An early sign of chronic rejection.
PLoS One. 10:e01363812015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park Y, Kim W, Lee JM, Park J, Cho JK,
Pang K, Lee J, Kim D, Park SW, Yang KM and Kim SJ: Cytoplasmic
DRAK1 overexpressed in head and neck cancers inhibits TGF-β1 tumor
suppressor activity by binding to Smad3 to interrupt its complex
formation with Smad4. Oncogene. 34:5037–5045. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu Y, Li L, Liu Y, Geng P, Li G, Yang Y
and Song H: RECK inhibits cervical cancer cell migration and
invasion by promoting p53 signaling pathway. J Cell Biochem.
119:3058–3066. 2017. View Article : Google Scholar
|
|
38
|
Shi C and Zhang Z: MicroRNA-362 is
downregulated in cervical cancer and inhibits cell proliferation,
migration and invasion by directly targeting SIX1. Oncol Rep.
37:501–509. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lever J, Gakkhar S, Gottlieb M, Rashnavadi
T, Lin S, Siu C, Smith M, Jones M, Krzywinski M, Jones SJM and Wren
J: A collaborative filtering based approach to biomedical knowledge
discovery. Bioinformatics. 34:652–659. 2017. View Article : Google Scholar
|
|
40
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nadal E, Zhong J, Lin J, Reddy RM, Ramnath
N, Orringer MB, Chang AC, Beer DG and Chen G: A MicroRNA cluster at
14q32 drives aggressive lung adenocarcinoma. Clin Cancer Res.
20:3107–3117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim YW, Kim EY, Jeon D, Liu JL, Kim HS,
Choi JW and Ahn WS: Differential microRNA expression signatures and
cell type-specific association with Taxol resistance in ovarian
cancer cells. Drug Des Devel Ther. 8:293–314. 2014.PubMed/NCBI
|
|
43
|
Brucker J, Mayer C, Gebauer G, Mallmann P,
Belau AK, Schneeweiss A, Sohn C and Eichbaum M: Non-pegylated
liposomal doxorubicin for patients with recurrent ovarian cancer: A
multicentric phase II trial. Oncol Lett. 12:1211–1215. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Muller PA and Vousden KH: p53 mutations in
cancer. Nat Cell Biol. 15:2–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang H, Liu YJ, Liu M and Li X:
Establishment and gene analysis of an oxaliplatin-resistant colon
cancer cell line THC8307/L-OHP. Anticancer Drugs. 18:633–639. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao J, Xu J and Zhang R: MicroRNA-411
inhibits malignant biological behaviours of colorectal cancer cells
by directly targeting PIK3R3. Oncol Rep. 39:633–642.
2018.PubMed/NCBI
|
|
47
|
Xu N, Yang W, Liu Y, Yan F and Yu Z:
MicroRNA-411 promoted the osteosarcoma progression by suppressing
MTSS1 expression. Environ Sci Pollut Res Int. 25:12064–12071. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cekirge HS, Peynircioglu B and Saatci I:
Endovascular treatment of an ‘anterior cerebral artery’ aneurysm in
a patient with ‘embryonic unfused middle cerebral artery’ anomaly:
A case report. Neuroradiology. 47:690–694. 2005. View Article : Google Scholar : PubMed/NCBI
|