Introduction
Esophageal cancer is the sixth most commonly
diagnosed cancer and the fifth leading cause of cancer motility
globally (1). It was estimated
that esophageal cancer accounted for >4% of cancer-related
mortality in the USA in 2017 (2).
In East Asia, the risk of incidence for esophageal cancer is almost
4-fold higher compared with North America (1). The two major types of esophageal
cancer are esophageal squamous-cell carcinoma and esophageal
adenocarcinoma (3). Owing to its
aggressive nature and difficulty to diagnose, the overall 5-year
survival rate of patients with esophageal cancer is ~20% (4,5).
Therefore, there is an urgent necessity to discover the molecular
mechanisms that lead to the metastasis of esophageal cancer.
MicroRNAs (miRNAs) are a class of small, non-coding,
single-stranded RNAs that are ubiquitously expressed in eukaryotic
cells (6). Through directly
binding to the 3′ untranslated region (UTR) of target gene mRNA,
miRNAs induce degradation of mRNA or inhibit mRNA translation,
resulting in downregulation of target gene expression (7). Previous studies have revealed that
mRNA-miRNA regulatory networks are crucial for normal biological
processes, including cell differentiation, migration and apoptosis
(8–10). Dysregulation of miRNA expression
contributes to a number of human diseases such as cancer (11). A microarray study has demonstrated
that the expression levels of several miRNAs are promising
biomarkers for esophageal cancer (12). miRNA (miR)-542-3p downregulation
has been reported in several cancer types (13,14);
however, the potential role and molecular mechanism of miR-542-3p
in esophageal cancer remains to be determined.
Ovarian tumor domain-containing ubiquitin
aldehyde-binding protein 1 (OTUB1) is a hydrolase that is able to
specifically remove ubiquitin from proteins to prevent protein
degradation (15). By upregulating
the expression of oncogenes, OTUB1 facilitates tumor progression in
several types of cancers (15–17).
In esophageal cancer, OTUB1 stabilizes Snail protein to promote the
metastasis of cancer cells (18);
however, it is unknown how OTUB1 is regulated in esophageal
cancer.
In the present study, miR-542-3p and OTUB1
mRNA expression levels were examined in normal and tumor tissues
from patients with esophageal cancer. A negative correlation was
observed between miR-542-3p and OTUB1 mRNA expression in
tumor tissues. Western blotting and reverse
transcription-quantitative PCR (RT-qPCR) data indicated that
OTUB1 was negatively regulated by miR-542-3p in esophageal
cancer cells. A dual-luciferase assay validated OTUB1 as a
target gene for miR-542-3p. Furthermore, function assays
demonstrated that miR-542-3p inhibited the migration and invasion
of KYSE150 human esophageal squamous cell carcinoma cells through
repression of OTUB1 expression. The data from the present study
suggested a potential tumor suppressor role for miR-542-3p in
esophageal cancer.
Materials and methods
Tissue collection
Tumor tissues and matched normal tissues were
collected from 40 patients (mean age 56.33±7.21, male:female =
25:15) with esophageal squamous cell carcinoma or esophageal
adenocarcinoma in Sheyang People's Hospital (Sheyang, China)
between June 2015 and July 2017. Written consent was provided by
all participants before the experiments and all procedures were
approved by the Ethics Committee of Sheyang People's Hospital (IRB
no. SYPH1506). Tissues were stored in −80°C upon collection.
Cell culture
Human esophageal squamous cell carcinoma cell line
KYSE150 was purchased from American Type Culture Collection and
used within 6 months. Cells were maintained in DMEM (Sigma-Aldrich,
Merck KGaA) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C.
Overexpression and downregulation of
miR-542-3p
miR-542-3p mimic (5′-UGUGACAGAUUGAUAACUGAAA-3′),
miR-negative control (NC) mimic (5′-AAUUCUCCGAACGUGUCACTT-3′),
miR-542-3p inhibitor (5′-UUUCAGUUAUCAAUCUGUCACA-3′) and miR-NC
inhibitor (5′-GUGACACGUUCGGAGAAUUTT-3′) were synthesized by and
purchased from Shanghai GenePharma Co., Ltd. For overexpression or
downregulation of miR-542-3p, miR-542-3p mimic (50 nM) or
miR-542-3p inhibitor (50 nM) was mixed with
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in serum-free DMEM medium for 15 min. The
mixtures were added into each well in 6-well plates and incubated
for 48 h before the cells (2×105) were harvested for
subsequent experiments.
RNA extraction and RT-qPCR
Total RNA from cell lines (1×104) or
tissue samples (50–100 mg) was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. For mRNA quantification, RNA was
reverse-transcribed into first-stranded cDNA using PrimeScript RT
Master Mix (Takara Bio, Inc.). RT-qPCR was conducted using a
SYBR-Green qPCR Master Mix kit (Takara Bio, Inc.) on an ABI PRISM
7900HT Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with the followed thermocycling parameters:
Initial denaturation at 95°C for 2 min, 40 cycles of 95°C for 15
sec and 64°C for 30 sec. For miRNA quantification, RNA was
reverse-transcribed with a Mir-X miRNA First Strand Synthesis kit
(Takara Bio, Inc.) followed by qPCR with Mir-X™ miRNA qRT-PCR SYBR
kit (Takara Bio, Inc.). Relative expression levels of miRNA and
mRNA were calculated using the 2−ΔΔCq method (19). GAPDH and U6 served as internal
controls for mRNA and miRNA, respectively. The primer sequences
were as listed: miR-542-3p forward, 5′-TGTGACAGATTGATAACT-3′ and
stem-loop RT primer,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGCGGTTTCAGT-3′;
OTUB1 forward, 5′-TCGGTCCTATACAAGGAGTATGC-3′ and reverse,
5′-GGTCTTGCGGATGTACGAGT-3′; U6 forward, 5′-GTGCTCGCTTCGGCAGCACAT-3′
and reverse, 5′-AATATGGAACGCTTCACGAAT-3′, GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′. Expression level of miR-542-3p was
normalized to U6. Expression level of OTUB1 was normalized to
GAPDH.
Protein lysate preparation and western
blotting
The antibodies used were as follows: OTUB1 (cat. no.
A302-917A; 1:1,000; Bethyl Laboratories, Inc.); GAPDH (cat. no.
G8795; 1:5,000; Sigma-Aldrich; Thermo Fisher Scientific, Inc.);
Snail (cat. no. sc-393172; 1:1,000; Santa Cruz Biotechnology, Inc.,
CA, USA); horseradish peroxidase-conjugated secondary antibodies
against rabbit (cat. no. 7074, 1:10,000; Cell Signaling Technology,
Danvers, MA, USA) and mouse (cat. no. 7076, 1:10,000; Cell
Signaling Technology). Protein lysates from cells
(1×104) were prepared using RIPA lysis buffer (Roche
Diagnostics GmbH) followed by determination of protein
concentration by the BCA method. Then, protein samples (20 µg) were
separated on an 10% SDS-PAGE gel and transferred to a PVDF
membrane. The, PVDF membrane was blocked by 5% bovine serum albumin
(BSA, Beijing Solarbio Science & Technology Co., Ltd.) at room
temperature for 2 h. Following incubation with the primary antibody
at 4°C overnight and a secondary antibody at room temperature for 2
h, the bands were visualized using ECL Western Blot Substrate
(Pierce; Thermo Fisher Scientific, Inc.). Densitometric analysis
was performed by ImageJ software (version 1.8.0; National
Institutes of Health). GAPDH was used to normalize expression
data.
Dual-luciferase reporter assay
The position 107–114 of 3′UTR of OTUB1
(5′…UUCACCCCCUUCUUCCUGUCACA…) mRNA containing the putative target
site of miR-542-3p (3′AAAGUCAAUAGUUAGACAGUGU) was determined by
TargetScan version 7.1 (20) and
amplified from the cDNA of KYSE150 cells and ligated into the
pGL3-basic vector (Promega Corporation).
pGL3-OTUB1−3′UTR-mutant (Mut, 5′…UUCACCCCCUUCUUCCUGGAACA…)
was created by introducing two site mutations into miR-542-3p
potential target sites using QuickChangeSite-Directed Mutagenesis
kits (Agilent Technologies, Inc.). pGL3-OTUB1−3′UTR-WT (200
ng) or pGL3-OTUB1−3′UTR-Mut (200 ng) was co-transfected with
Renilla plasmid into KYSE150 cells using
Lipofectamine® 3000, followed by transfection of miR-NC
mimic (10 nM) or miR-542-3p mimic (10 nM) for 48 h at 37°C. The
Dual-Luciferase Reporter Assay System (Promega Corporation) was
used to measure the relative luciferase activity of each well. The
firefly luciferase expression was normalized to Renilla.
OTUB overexpression plasmid
construction and transfection
Full-length OTUB1 cDNA was amplified from
KYSE150 cells and ligated to a pcDNA3.1 vector (https://www.70dir.com/seo/report_www_youbio_cn.html,
YouBio) with the restriction sites of KpnI and XhoI.
For overexpression of OTUB1, pcDNA3.1-OTUB1 was mixed
with Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in serum-free DMEM medium for 15 min. The
mixtures were then added into each well in 6-well plates
(1×106) and incubated for 48 h at 37°C before the cells
were harvested for subsequent experiments.
Cell migration assay
A wound healing assay was used to examine the
migratory ability of transfected KYSE150 cells. Briefly,
1×106 cells were seeded in each well of 6-well plates.
Following transfection, as aforementioned, the cells were cultured
in 37°C to 90–100% confluence. A wound area was made in the center
of each well using a 10 µl pipette tip. The culture medium of the
cells was replaced with serum-free medium. Images were captured
using an inverted microscope (Nikon Corporation) at 0 and 24 h to
observe the cells (magnification, ×100) that migrated into the
wound area. The migrated areas were quantified using Image Pro Plus
and normalized to the miR-NC mimic + pcDNA3.1 group.
Cell invasion assay
For the cell invasion assay, BD Matrigel Invasion
Chambers (8-µm pore; BD Biosciences) were used. In brief, KYSE150
cells (1×105) transfected with miR-NC mimic + pcDNA3.1,
miR-542-3p mimic + pcDNA3.1 or miR-542-3p mimic + pcDNA3.1-OTUB1
were cultured in the upper chamber in serum-free DMEM at 37°C.
Following 72 h of invasion, cells on the upper side of the filter
were removed and cells that invaded to the underside of the
membranes were fixed using 8% formaldehyde at room temperature for
15 min, followed by staining with crystal violet at room
temperature for 30 min. The number of invaded cells (×400) was
counted using a light microscope (Olympus Corporation).
Statistical analysis
Data were analyzed with GraphPad Prism 7 (GraphPad
Software, Inc.) and presented as the mean ± standard error of the
mean. For in vitro experiments, the statistical differences
were evaluated using Student's t-test (two groups) or using ANOVA
followed by Newman Keuls method (three groups). For ex vivo
experiments in tumor tissues, the statistical analyses were
performed using paired Student's t-test (two groups). Pearson's
correlation analysis was performed to determine the correlation
between miR-542-3p and OTUB1 mRNA expression levels in
esophageal tumor tissues. P<0.05 was considered to indicate a
statistically significant difference. All experiments were
performed thrice.
Results
miR-542-3p is downregulated in
esophageal tumor tissues
To investigate the role of miR-542-3p in esophageal
cancer, RT-qPCR was performed to detect miR-542-3p expression
levels in tumoral and adjacent normal tissues from 40 patients with
esophageal cancer. The results demonstrated that the expression of
miR-542-3p was significantly downregulated in tumor tissues
compared with adjacent normal tissues (Fig. 1A).
OTUB1 is a newly identified oncogene in
esophageal cancer and has been reported to be regulated by
miR-542-3p in colorectal cancer (13,18).
In the present study, OTUB1 mRNA levels were
significantly increased in tumor tissues compared with adjacent
normal tissues (Fig. 1B). Western
blotting revealed that the protein expression of OTUB1 was also
significantly increased in tumor tissues compared adjacent normal
tissues (Fig. 1C). In addition,
Pearson's correlation analysis indicated that expression of
miR-542-3p was significantly negatively correlated with
OTUB1 mRNA expression levels in esophageal tumor tissues
(Fig. 1D). These results suggested
that miR-542-3p may inhibit esophageal cancer progression and act
as a tumor suppressor.
Expression of OTUB1 is repressed by
miR-542-3p in esophageal cancer cells
To further study the regulatory association between
OTUB1 and miR-542-3p, miR-542-3p mimic was used to elevate
miR-542-3p expression in KYSE150 cells. Compared with cells
transfected with miR-NC mimic, transfection with miR-542-3p mimic
significantly increased miR-542-3p expression in KYSE150 cells
(Fig. 2A). Overexpression of
miR-542-3p significantly decreased OTUB1 mRNA levels and
OTUB1 protein levels in cells (Fig. 2B
and C). Conversely, transfection with miR-542-3p inhibitor
significantly decreased miR-542-3p expression in KYSE150 cells and
led to an increased expression of OTUB1 at mRNA and protein
levels (Fig. 2D-F). These data
demonstrated that miR-542-3p may negatively regulate OTUB1
in esophageal cancer cells.
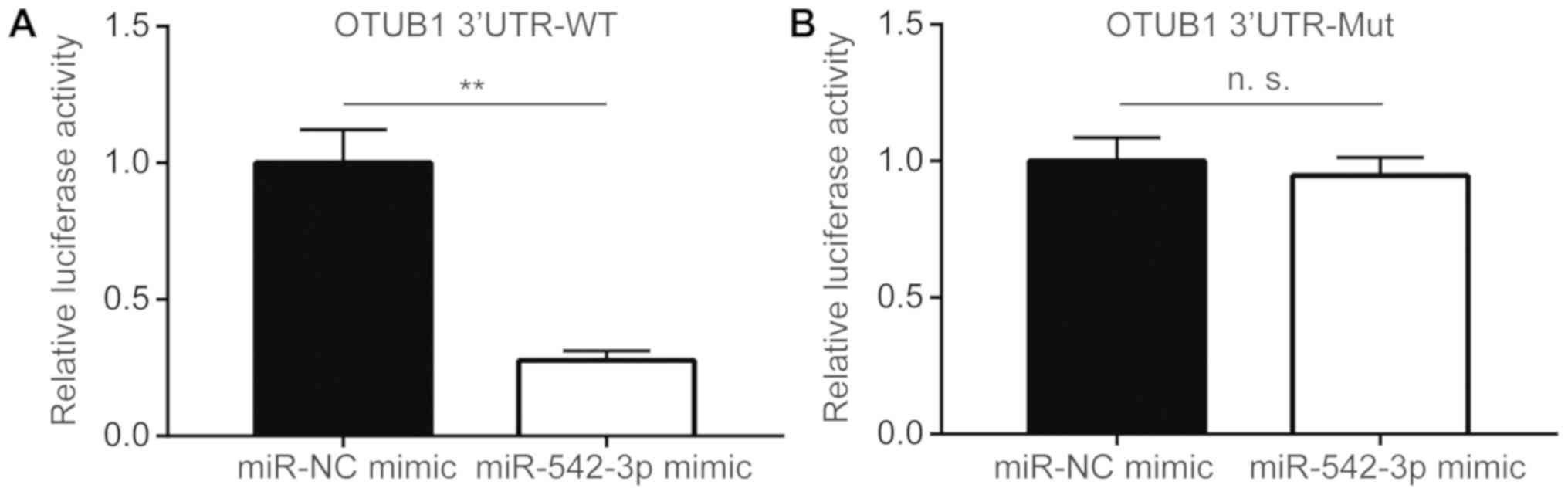
OTUB1 is a target gene of miR-542-3p
in esophageal cancer cells
To confirm whether miR-542-3p directly regulated
OTUB1 expression, dual-luciferase reporter assay was
performed in KYSE150 cells. Transfection of miR-542-3p mimic
significantly reduced luciferase activity of OTUB1 3′UTR-WT
compared with cells transfected with miR-NC mimic (Fig. 3A), whereas overexpression of
miR-542-3p did not alter the luciferase activity of OTUB1 3′UTR-Mut
(Fig. 3B). These data suggested
that miR-542-3p may bind directly to the putative binding site in
3′UTR of OTUB1 to repress its expression.
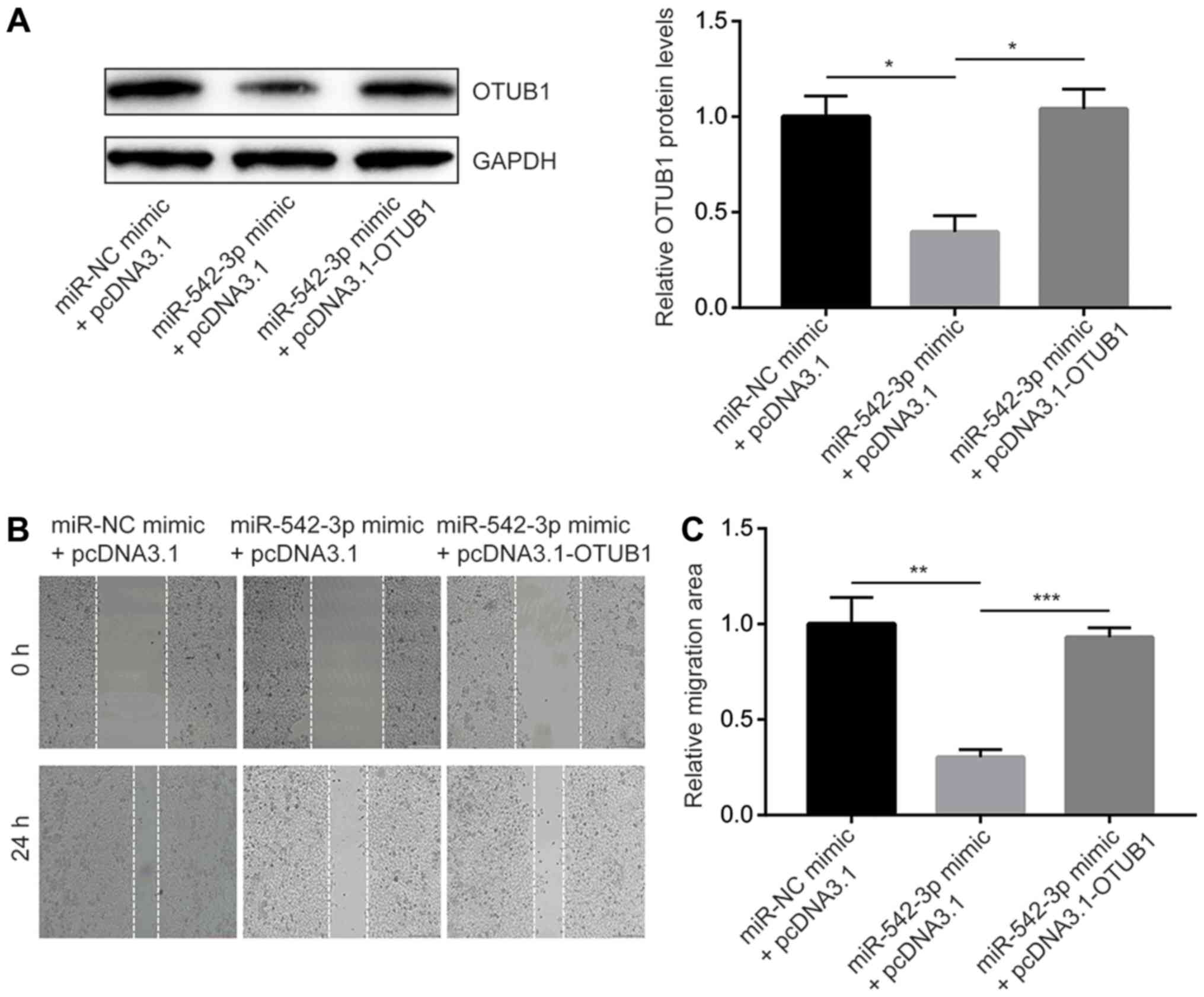
miR-542-3p inhibits esophageal cancer
cell migration and invasion through regulation of OTUB1
To study the biological function of miR-542-3p in
esophageal cancer cells, wound healing and cell invasion assays
were performed to detect cell migratory and invasive ability of
cells transfected with the miR-542-3p mimic either with or without
overexpression of OTUB1.
Transfection of miR-542-3p mimic reduced OTUB1
protein expression whereas co-transfection of miR-542-3p mimic and
recombinant OTUB1 reversed the downregulation of OTUB1 in
KYSE150 cells (Fig. 4A). In the
wound healing assays, overexpression of miR-542-3p significantly
inhibited cell migration towards the wound areas, which indicated
that the migration ability of cells was reduced (Fig. 4B and C). In addition,
overexpression of OTUB1 reversed migration inhibition
induced by miR-542-3p mimic (Fig. 4B
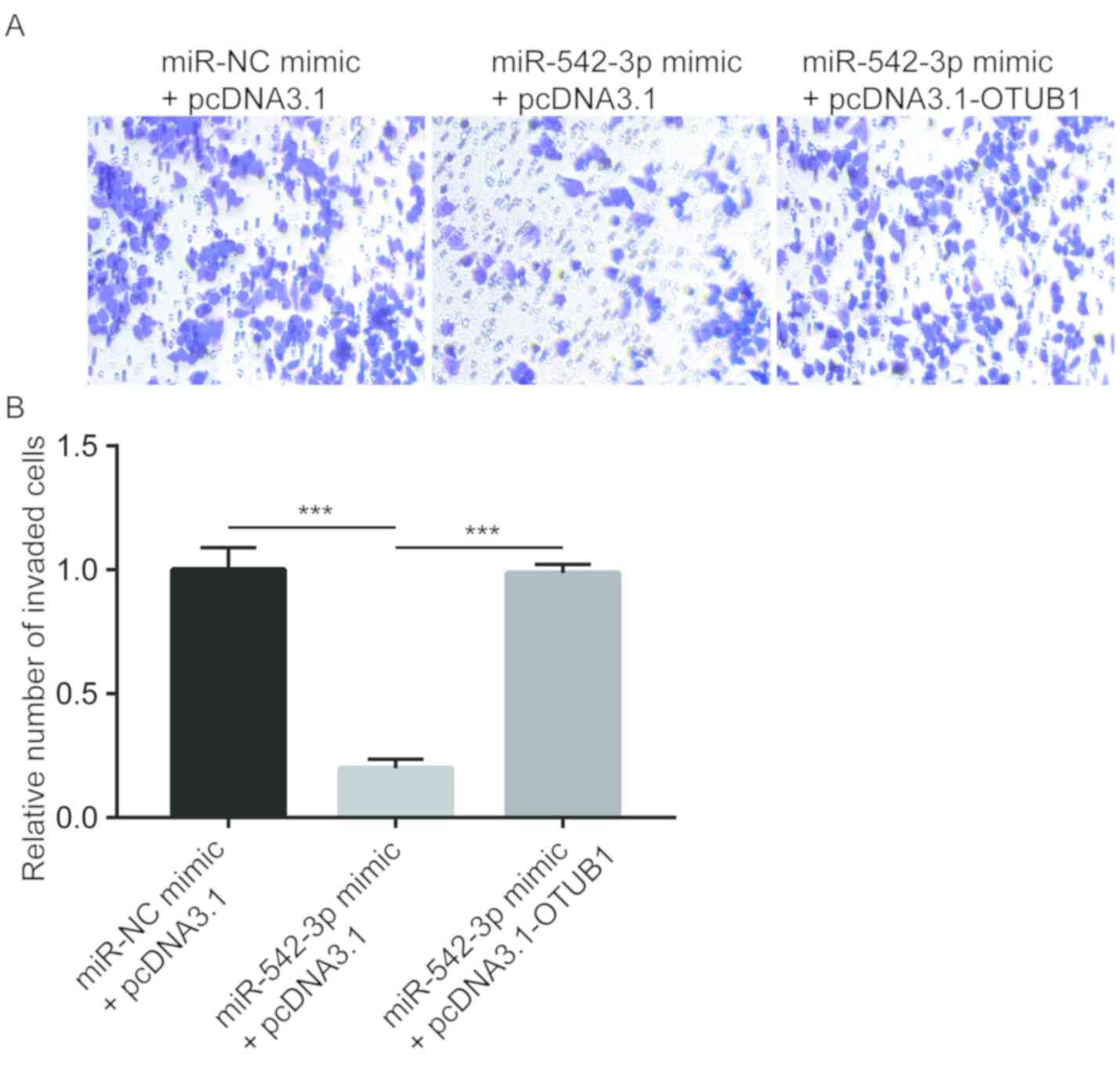
and C). Similarly, overexpression of miR-542-3p significantly
inhibited the invasive ability of the cells, which was reversed by
overexpression of OTUB1 (Fig.
5). The results demonstrated that miR-542-3p may inhibit the
migratory and invasive abilities of esophageal cancer cells through
repression of OTUB1.
Discussion
miRNAs regulate a number of oncogenes and tumor
suppressors in cells, which can lead to the initiation and
progression of cancer (21,22).
Several miRNAs have been identified as key regulators of esophageal
cancer development and accurate predictors of clinical outcome for
patients with esophageal cancer (23–25).
Decreased expression of miR-542-3p has been observed in certain
types of cancers, including hepatocellular carcinoma, osteosarcoma,
colorectal cancer and melanoma (13,26–29).
In the present study, miR-542-3p expression and function were
investigated in esophageal cancer. RT-qPCR revealed that miR-542-3p
expression was decreased in tumor tissues compared with adjacent
normal tissues from patients with esophageal cancer. In KYSE150
cells, overexpression of miR-542-3p markedly reduced cell migratory
and invasive abilities. Thus, consistent with its tumor suppressor
role in other cancer types, miR-542-3p may also function as a tumor
suppressor in esophageal cancer.
OTUB1 is a cysteine protease that removes ubiquitin
from modified proteins to stabilize target proteins (30). OTUB1 is involved in the regulation
of several proteins that are pivotal for the progression of cancer,
such as estrogen receptor, p53 and forkhead box M1 (17,30,31).
Recently, OTUB1 was demonstrated to promote the metastasis of
esophageal cancer by stabilizing Snail (18). miR-542-3p has been demonstrated to
bind to 3′UTR of OTUB1 mRNA to downregulate OTUB1 in
colorectal cancer cells (13).
Consistent with the previous study, an increase in OTUB1
mRNA expression was observed in tumor tissues compared with
adjacent normal tissues from patients with esophageal cancer.
Additionally, OTUB1 expression levels were inversely
correlated with miR-542-3p expression levels in tumor tissues, and
in KYSE150 cells, overexpression of miR-542-3p decreased
OTUB1 expression.
Snail is a member of the Snail
superfamily, which functions in cell survival and cell
differentiation of cancer cells (32). Furthermore, the development and
metastasis of cancer were blocked by Snail suppression
(33). Recently, Snail
silencing was found to inhibit cell migration of esophageal cells
(34). Notably, in KYSE150 cells,
overexpression of miR-542-3p decreased Snail expression.
Finally, cell migration and invasion inhibition
induced by miR-542-3p overexpression was partially attenuated by
co-transfection of recombinant OTUB1 in KYSE150 cells. These
results demonstrated that miR-542-3p may regulate OTUB1 to
inhibit cell metastasis of esophageal cancer, which were consistent
with a previous study about the role of OTUB1 in esophageal
cancer (18). Future studies are
needed to determine whether the expression of miR-542-3p may be
used as a biomarker to predict distant metastasis and overall
survival of patients with esophageal cancer.
The results of the present study indicated a
potential tumor suppressor role for miR-542-3p in esophageal
cancer. Overexpression of miR-542-3p inhibited migration and
invasion of esophageal cancer cells. Therefore, upregulation of
miR-542-3p may be a potential treatment approach for patients with
esophageal cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and or/analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JuS and WY designed the study. JuS, JiS and YD
acquired and interpreted the data. YD and JiS collected clinical
samples. WY prepared the manuscript and supervised the study.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Sheyang People's Hospital (Sheyang, China). All
patients signed written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rustgi AK and El-Serag HB: Esophageal
carcinoma. N Engl J Med. 371:2499–2509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics for Hispanics/Latinos, 2012. CA Cancer J Clin.
62:283–298. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Deng HY, Alai G, Luo J, Li G, Zhuo ZG and
Lin YD: Cancerous esophageal stenosis before treatment was
significantly correlated to poor prognosis of patients with
esophageal cancer: A meta-analysis. J Thorac Dis. 10:4212–4219.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu J, Chen Q, Liu P, Jia W, Chen Z and Xu
Z: Integration of mRNA and miRNA analysis reveals the molecular
mechanism underlying salt and alkali stress Tolerance in tobacco.
Int J Mol Sci. 20:E23912019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ye Y, Li SL and Wang SY: Construction and
analysis of mRNA, miRNA, lncRNA, and TF regulatory networks reveal
the key genes associated with prostate cancer. PLoS One.
13:e01980552018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Liang Y and Lu Q: MicroRNA
epigenetic alterations: Predicting biomarkers and therapeutic
targets in human diseases. Clin Genet. 74:307–315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feber A, Xi L, Luketich JD, Pennathur A,
Landreneau RJ, Wu M, Swanson SJ, Godfrey TE and Litle VR: MicroRNA
expression profiles of esophageal cancer. J Thorac Cardiovasc Surg.
135:255–260; discussion 260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan L, Yuan P, Yuan H, Wang Z, Run Z,
Chen G, Zhao P and Xu B: miR-542-3p inhibits colorectal cancer cell
proliferation, migration and invasion by targeting OTUB1. Am J
Cancer Res. 7:159–172. 2017.PubMed/NCBI
|
|
14
|
Althoff K, Lindner S, Odersky A, Mestdagh
P, Beckers A, Karczewski S, Molenaar JJ, Bohrer A, Knauer S,
Speleman F, et al: miR-542-3p exerts tumor suppressive functions in
neuroblastoma by downregulating Survivin. Int J Cancer.
136:1308–1320. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Karunarathna U, Kongsema M, Zona S, Gong
C, Cabrera E, Gomes AR, Man EP, Khongkow P, Tsang JW, Khoo US, et
al: OTUB1 inhibits the ubiquitination and degradation of FOXM1 in
breast cancer and epirubicin resistance. Oncogene. 35:1433–1444.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baietti MF, Simicek M, Abbasi Asbagh L,
Radaelli E, Lievens S, Crowther J, Steklov M, Aushev VN, Martínez
García D, Tavernier J and Sablina AA: OTUB1 triggers lung cancer
development by inhibiting RAS monoubiquitination. EMBO Mol Med.
8:288–303. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Zhou X, Xu M, Weng W, Zhang Q,
Yang Y, Wei P and Du X: OTUB1-catalyzed deubiquitination of FOXM1
facilitates tumor progression and predicts a poor prognosis in
ovarian cancer. Oncotarget. 7:36681–36697. 2016.PubMed/NCBI
|
|
18
|
Zhou H, Liu Y, Zhu R, Ding F, Cao X, Lin D
and Liu Z: OTUB1 promotes esophageal squamous cell carcinoma
metastasis through modulating Snail stability. Oncogene.
37:3356–3368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Agarwal V, Bell GW, Nam J and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. Aug 4–2015.(Epub ahead of print). doi: 10.7554/eLife.05005.
View Article : Google Scholar
|
|
21
|
Lee K and Ferguson LR: MicroRNA biomarkers
predicting risk, initiation and progression of colorectal cancer.
World J Gastroenterol. 22:7389–7401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paliouras AR, Monteverde T and Garofalo M:
Oncogene-induced regulation of microRNA expression: Implications
for cancer initiation, progression and therapy. Cancer Lett.
421:152–160. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fang Y, Fang D and Hu J: MicroRNA and its
roles in esophageal cancer. Med Sci Monit. 18:RA22–RA30. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Wainscott C and Xi Y: MicroRNA
provides insight into understanding esophageal cancer. Thorac
Cancer. 2:134–142. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qi B, Liu SG, Qin XG, Yao WJ, Lu JG, Guo
L, Wang TY, Li HC and Zhao BS: Overregulation of microRNA-212 in
the poor prognosis of esophageal cancer patients. Genet Mol Res.
13:7800–7807. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu W, Dang S, Feng Q, Liang J, Wang Y and
Fan N: MicroRNA-542-3p inhibits the growth of hepatocellular
carcinoma cells by targeting FZD7/Wnt signaling pathway. Biochem
Biophys Res Commun. 482:100–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu Y, You J, Li F, Wang F and Wang Y:
MicroRNA-542-3p suppresses tumor cell proliferation via targeting
Smad2 inhuman osteosarcoma. Oncol Lett. 15:6895–6902.
2018.PubMed/NCBI
|
|
28
|
Rang Z, Yang G, Wang YW and Cui F:
miR-542-3p suppresses invasion and metastasis by targeting the
proto-oncogene serine/threonine protein kinase, PIM1, in melanoma.
Biochem Biophys Res Commun. 474:315–320. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wiener R, DiBello AT, Lombardi PM, Guzzo
CM, Zhang X, Matunis MJ and Wolberger C: E2 ubiquitin-conjugating
enzymes regulate the deubiquitinating activity of OTUB1. Nat Struct
Mol Biol. 20:1033–1039. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun XX, Challagundla KB and Dai MS:
Positive regulation of p53 stability and activity by the
deubiquitinating enzyme Otubain 1. EMBO J. 31:576–592. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stanisić V, Malovannaya A, Qin J, Lonard
DM and O'Malley BW: OTU Domain-containing ubiquitin
aldehyde-binding protein 1 (OTUB1) deubiquitinates estrogen
receptor (ER) alpha and affects ERalpha transcriptional activity. J
Biol Chem. 284:16135–16145. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mansoori B, Sandoghchian Shotorbani S and
Baradaran B: RNA interference and its role in cancer therapy. Adv
Pharm Bull. 4:313–321. 2014.PubMed/NCBI
|
|
34
|
Hemmatzadeh M, Mohammadi H, Babaie F,
Yousefi M, Ebrazeh M, Mansoori B, Shanehbandi D and Baradaran B:
Snail-1 silencing by siRNA inhibits migration of TE-8 esophageal
cancer cells through downregulation of metastasis-related genes.
Adv Pharm Bull. 8:437–445. 2018. View Article : Google Scholar : PubMed/NCBI
|