Introduction
Glioblastoma is the most common and aggressive
primary brain tumor AND originates from the glial cells in adults
(1,2). Glioblastoma is characterized by the
appearance of vascular proliferation, aggressive invasion and
necrosis around human normal brain tissues (3). A previously study identified that
glioblastoma accounts for ~75% of all malignant tumors associated
with the brain (4). According to
characteristics of pathologic evaluation and infiltrative growth,
different malignant grades result in diverse glioblastoma shapes
(5). Despite the progression of
treatment from solely surgical intervention to radiotherapy,
chemotherapy or targeted treatments, these current treatment
options are not effective and the overall survival for most
patients with GBM remains poor (6,7),
with a median overall survival following surgical resection of
12–14 months (8). Glioblastoma
patients usually have a poor prognosis with a 5 year survival rate
of <5% (9). Therefore, it is
necessary to identify an effective molecular biomarker that can
predict the development and progression and could be developed into
a novel therapeutic approach for GBM.
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNAs, which are >200 nucleotides in length and
participate in multiple biological processes, including cell
differentiation and transcriptional regulation (10,11).
LncRNAs exhibit special profiles in various cancers, regulating
disease progression and serving as a predictor of patient outcomes.
Previous studies identified that lncRNAs function in various
aspects of cell biology and can potentially contribute to tumor
development, including in GBM (12–14).
These studies revealed the importance of lncRNAs and suggest a
novel potential therapeutic strategy for the treatment of GBM.
Although the molecular mechanism and biological function of
lncRNA-mediated tumor progression remains largely unknown, previous
studies have suggested that lncRNAs can function as competitive
endogenous RNAs (ceRNAs) that can sequester microRNAs (miRNAs)
(15), which are endogenously
expressed non-coding RNAs of ~22 nucleotides in length that
participate in tumor progression (16). Another study confirmed the
existence of a widespread interaction network of competitive
endogenous RNAs (ceRNAs), in which lncRNAs may exert functions by
targeting miRNAs and regulating their function role (17). The lncRNA Unigene56159 is located
on chromosome 3 and has been reported to be upregulated in
hepatocellular carcinoma cells and associated with poor patient
prognosis (18).
miRNAs are highly conserved among species, and play
important roles in a variety of biological and pathological
processes. Dysregulation of miRNAs in glioma has also been
reported, and certain miRNAs have been functionally involved in
glioma. Previous studies have demonstrated that miR-194-5p may
exerts as a tumor-suppressor gene and is down-regulated in many
tumors, including glioma (19–23).
However, the molecular mechanism of miR-194-5p deregulation and how
such deregulation contributes to glioma tumorigenesis remains
unclear.
Thus, the present study aimed to investigate the
interaction between Unigene56159 and miRNA (miR)-194-5p in GBM
progression. It demonstrated that Unigene56159 overexpressed in GBM
tissues and cell lines and that Unigene56159 may negatively
regulate miR-194-5p levels and promote proliferation and invasion,
which may provide insight into a potential novel treatment option
for GBM.
Materials and methods
Cell lines and clinical tissues
Human GBM cell lines (U251, T98G, LN229, SHG44 and
A172) were purchased from The Cell Bank of Type Culture Collection
of the Chinese Academy of Sciences. Normal human astrocyte cells
(NHA) cells were obtained from the American Type Culture
Collection. Cells were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.), supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and incubated under humidified conditions at 37°C
and 5% CO2.
Human GBM samples and adjacent normal brain samples
were collected from 50 patients undergoing surgical resection at
Tianjin Medical University General Hospital (Tianjin, China)
between June 2013 and June 2017. These glioma samples were from 33
males and 17 females with age ranging from 23–75 years (median, 49
years). All GBM samples were examined by two senior pathologists.
Written informed consent was obtained from all patients prior to
enrollment in the study; the study was approved by The
Institutional Review Board of Tianjin Medical University General
Hospital.
Data acquisition and Gene Ontology
(GO) term enrichment analysis with Unigene56159 expression
The edgeR software package (Bioconductor) in R
Studio 3.5.1 (https://www.rstudio.com/) was used to analyze the
aberrantly expressed lncRNAs in normalized gene expression profile
data from The Cancer Genome Atlas (TCGA) GBM database (24,25).
RNA sequencing data of GBM tissues and normal brain tissues were
collected from the TCGA database (http://cancergenome.nih.gov), and 162 GBM cases were
detected in all. For the normalized gene expression profile data,
the edge R package of R software was used to analyze significantly
aberrantly expressed lncRNAs at the level: moderately to GBM
samples vs. normal samples. A log fold change >2 and
false-discovery rate P<0.05 was selected as significantly cutoff
values. Significantly enriched gene sets were investigated. The
clinical data were obtain from Gene Expression Profiling
Interactive Analysis (GEPIA) dataset (http://gepia.cancer-pku.cn/). GO term enrichment
analysis was identified using the Database for Annotation,
Visualization and Integrated Discovery (DAVID) version 6.8
(https://david.ncifcrf.gov/).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from clinical tissues and
GBM cell lines using TRIzol® reagent according to the
manufacturer's instructions (Invitrogen; Thermo Fisher Scientific,
Inc.). A NanoDrop spectrophotometer was used to determine the
concentration of extracted RNA. RT-qPCR was performed in triplicate
on an ABI 7500 HT fast real-time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. qPCR was performed using the SYBR® Premix Ex
Taq™ II kit (Takara Biotechnology Co., Ltd.), according to the
manufacturer's protocol. Primers were: Unigene56159 forward,
5′-GTGAAAAGAAACATTCGAGTGT-3′, and reverse,
5′-TGAAGTAAGCAGGAAAGGGGGA-3′; miR-194-5p forward,
5′-AGTGTGACGTGACATCCGT-3′, and reverse, 5′-GCAGCTCAGTAACAGTCCGC-3′;
PCNA forward, 5′-TTTGGTGCAGCTCACCCTG-3′, and reverse.
5′-CGCGTTATCTTCGGCCCTTA-3′; MMP-2 forward,
5′-CAGGACATTGTCTTTGATGGCATCGC-3′, and reverse,
5′-TGAAGAAGTAGCTATGACCACCGCC-3′; MMP-9 forward,
5′-ATCCCCCACCTTTACCA-3′, and reverse 5′-TCAGAACCGACCCTACAA-3′; U6
forward, 5′-TGTGGGCATCAATGATTTGG-3′ and reverse,
5′-ACACCATGTATCCGGGTCAAT-3′; GAPDH forward
5′-CCATGTTCGTCATGGTGTG-3′ and reverse, 5′-GGTGCTAAGCAGTTGGTGGTG-3′.
The cycling conditions were: 95°C for 10 min, then 40 cycles at
95°C for 15 sec, and 60°C for 60 sec. U6 was used as a control to
normalize the miR-194-5p expression. Relative expression levels
were calculated using the 2−ΔΔCq method and normalized
to the internal reference gene (26).
Cell transfection
Unigene56159 small interfering RNA (siRNA) and the
negative control (si-NC), miR-194-5p mimic or inhibitor, and their
respective negative control (miR-NC; 20 µM)were obtained from
Shanghai GenePharma Co., Ltd. Sequences were: Unigenge56159 siRNA
forward, 5′-GGAGUGAGAUGUCAAAUAACA-3′, and reverse,
5′-UUAUUAGACAUCACACUCCAU-3′; si-NC forward,
5′-UUCUACGAAUGUGUCACCUTT-3′, and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′; miR-194-5p mimics forward,
5′-UGUAACAGCAACUCCAUGUGGA-3′, and reverse,
5′-CACAUGGAGUUGCUGUUACAUU-3′; miR-194-5p inhibitor forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACUUCGGAGAATT-3′; miR-NC forward,
5′-CAGUACUUUUGUGUAGUACAA-3′ and reverse,
5′-UUAACUAAUAUUUCAUCCAUA-3′. The Lipofectamine® 2000 kit
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for
transfection according the manufacturer's protocol at 37°C for 4 h.
Then the supernatant was removed and fresh medium was added. The
sample was collected for experiment at 24 h after transfection.
Dual-luciferase reporter assay
The TargetScan database (www.targetscan.org) and the starBase database
(http://starbase.sysu.edu.cn/) were used
to investigate target miRNAs interacting with Unigene56159 through
complementary sequences. From the statistically relevant microRNAs,
the top 5 in terms of their prediction score were selected,
including miR-194-5p, miR-124-3p, miR-130a-3p, miR-148a-3p and
miR-543: miR-194-5p achieved the highest score in two databases.
The human Unigene56159 Luc-reporter (Genepharm, Inc.) was
transfected into the ligation site of the Unigene56159
3′-untranslated region (UTR) PCR product. U251 cells were cultured
in 6-well plates at 3×105 cells/wells and co-transfected
with pmirGLO-Unigene56159-3′UTR-wild-type (WT) or
pmirGLO-Unigene56159-3′UTR-mutant (MUT), and miR-194-5p or miR-NC
mimics. Then cells was incubated in a 37°C, 5% CO2
humidified atmosphere for 4 h and then supernatant was removed. At
48 h post-transfection, luciferase activity was detected using the
Luciferase Assay system (Promega Corporation), and normalized to
Renilla luciferase activity.
Cell proliferation and colony
formation assays
Transfected cells were collected 24 h
post-transfection and cultured at a density of 2×103
cells/well in 96-well plates. Proliferation assays were performed
using a Cell Counting Kit-8 (Beyotime Institute of Biotechnology)
according to manufacturer's protocols, and measured at an
absorbance of 450 nm at 0, 24, 48 and 72 h (Infinite F50, Tecan
Group, Ltd.).
U251 and T98G Cells (~200) were seeded into 6-well
plates and cultured in 10% FBS at 37°C for 12 days to allow for
colony formation. Subsequently, cells were fixed with 4%
polyoxymethylene for 10 min at room temperature before being
stained with 10% Giemsa (30 min) (Sigma-Aldrich; Merck KGaA). The
number of colonies (>50 cells) was calculated under light
microscope (Nikon Corp., Tokyo, Japan).
Cell invasion assays
U251 and T98G cells (5×104) were seeded
in the upper chambers of Transwell plates precoated with
Matrigel® (Corning Life Sciences) in serum-free DMEM
(Gibco; Thermo Fisher Scientific, Inc.). DMEM supplemented with 20%
FBS was added to the lower chambers and the cells were incubated at
37°C in a 5% CO2 humidified atmosphere for 24 h.
Following incubation, non-invasive cells in the upper chamber were
removed using a cotton swab. The invasive cells in the lower
chamber were fixed using 4% paraformaldehyde for 10 min and stained
with hematoxylin and eosin for 5 min at room temperature. Stained
cells were manually counted under a light microscope (Nikon
Corporation; magnification ×100).
Western blotting
Total protein was extracted from patient tissue
samples (approximately 250 mg/case) and cell lines (U251 and T98G)
using RIPA buffer (Pierce; Thermo Fisher Scientific, Inc.). Protein
concentrations were determined using the BCA protein assay kit
(Bio-Rad Laboratories, Inc.), and 40 µg protein was separated by
10% SDS-PAGE. Separated proteins were transferred onto a PVDF
membrane (EMD Millipore; Merck KGaA) and blocked for 1 h at room
temperature with TBS containing 5% non-fat milk (w/v). The
membranes were incubated overnight at 4°C with the following
primary antibodies: rabbit anti-proliferating cell nuclear antigen
PCNA (Rabbit polyclonal antibody, cat. no. 10205-2-AP, 1:500; Wuhan
Sanying Biotechnology), rabbit MMP-2 (Rabbit polyclonal antibody,
cat. no 10373-2-AP, 1:500; Wuhan Sanying Biotechnology), rabbit
anti-MMP-9 (Rabbit polyclonal antibody, cat. no 10375-2-AP, 1:500;
Wuhan Sanying Biotechnology) and mouse anti-GAPDH (Mouse Monoclonal
Antibody, cat. no. sc-47724, 1:1,000; Santa Cruz Biotechnology,
Inc.), as the loading control. Images of the western blots were
captured using a ChemiDoc™ MP Imaging System (Bio-Rad Laboratories,
Inc.).
Immunofluorescence staining
U251 and T98G cell (1×105) were culture
on cell slides (glass slides stained with 0.1% poly-L-Lysine
overnight)and then fixed with 4% paraformaldehyde for 20 min and
incubated with 0.1% Triton X-100 for 10 min at room temperature.
Slides were subsequently washed with PBS twice for 5 min and
incubated with 5% BSA for 1 h at room temperature (CAS
Number:9048-46-8, Sigma Aldrich; Merck KGaA), then incubated with
primary antibodies against MMP-2 (Rabbit polyclonal antibody, cat.
no. 10373-2-AP, 1:100; Wuhan Sanying Biotechnology) at 4°C
overnight. Following primary antibody incubation, slides were
incubated with fluorescence-labeled rabbit secondary antibody
(Rhodamine TRITC-conjugated Goat Anti-Rabbit IgG, Catalog No.:
SA00007-2, 1:100, Wuhan Sanying Biotechnology) at room temperature
for 1 h. The nuclei were stained with DAPI for 10 min. Slides were
visualized using a fluorescent microscope (magnification ×400).
Statistical analysis
One-way ANOVA with post hoc Tukey's test or
Student's t-test was used to compare between groups. Survival
curves were drawn using the log-rank test with GraphPad Prism 5.0
(GraphPad Software, Inc.). The correlation between Unigene56159
expression and the clinicopathological characteristics of patients
with glioma was analyzed using the χ2 test or Fisher's
exact test. Statistical analysis was performed using SPSS 19.0 (IBM
Corp.) and data was displayed as mean ± SD. Experiments were
independently conducted in triplicate. P<0.05 was considered to
indicate a statistically significant difference.
Results
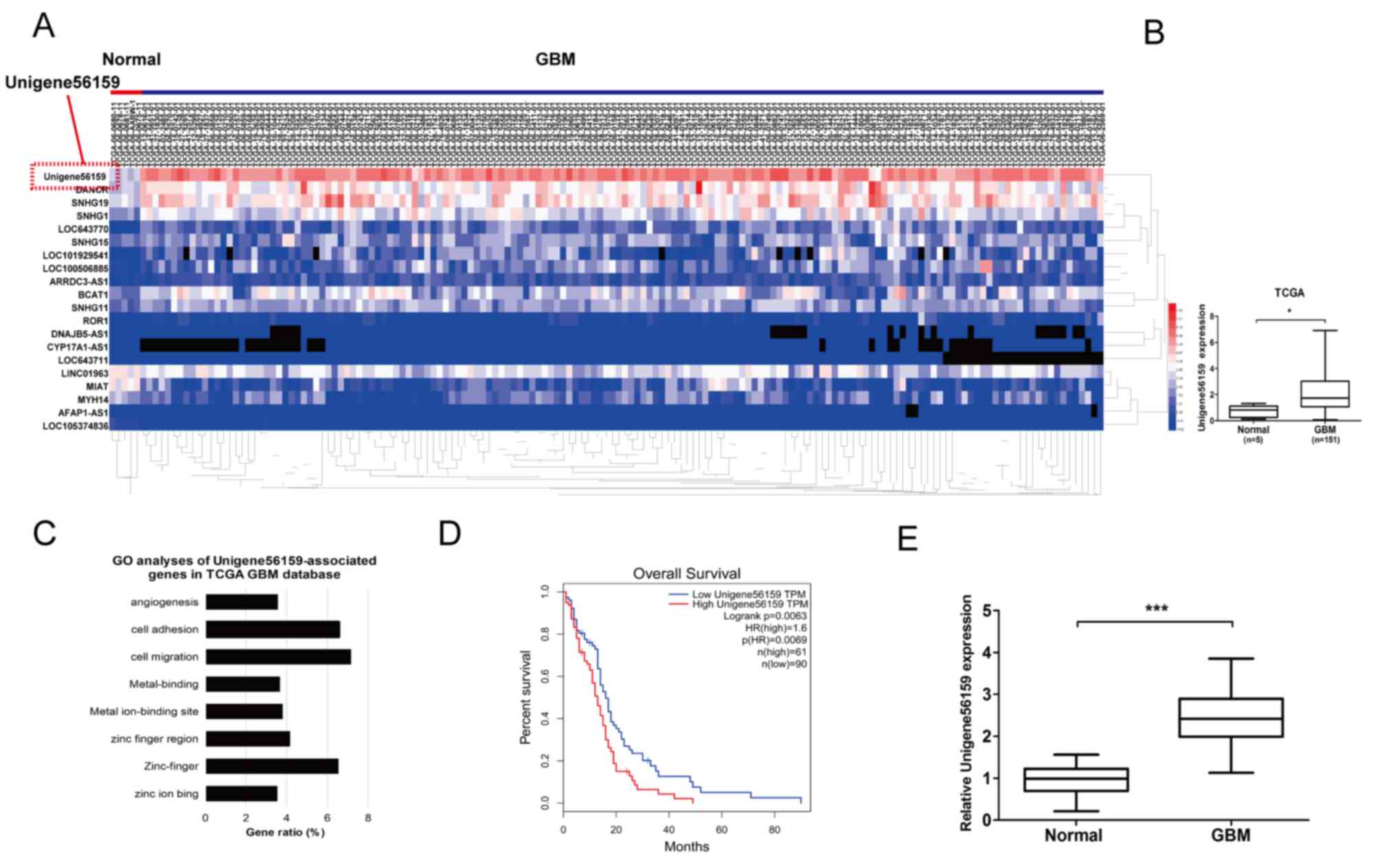
Unigene56159 is upregulated in GBM
tissue and correlates with poor prognosis
A log2-fold change (FC) of >2 and a false
discovery rate (P<0.05) were selected as the cut-off values
based on the Benjamini-Hochberg method (27). The expression of Unigene56159 was
obtained and 151 cases of valid data collected. The top 20
differentially expressed lncRNAs meeting this criteria were
collected, and these lncRNAs were identified according to the level
of log2FC (Fig. 1A). Among the
differentially expressed lncRNAs, Unigene56159 expression levels
were the highest in GBM, demonstrating markedly upregulated
expression levels compared with normal brain tissue (Fig. 1A and B). To determine putative
functions of Unigene56159, the associated gene expression profiles
collected from the GO database were analyzed. The most prominent
biological processes included cell migration, cell adhesion and
zinc-finger activity (Fig. 1C).
Moreover, the clinical data collected from the TCGA database
(GEPIA) revealed that high levels of Unigene56159 in GBM were
associated with a poorer overall survival compared with low
Unigene56159 expression levels according to the median survival
time of patients (http://gepia.cancer-pku.cn/) (Fig. 1D). The expression level of
Unigene56159 was also significantly associated with patient's
Karnofsky performance scale scores from the data of our clinical
sample (n=50; Table I; P=0.018).
Subsequently, RT-qPCR analysis was used to assess Unigene56159
expression levels in GBM and adjacent normal brain tissues for 50
patients from the present study. Unigene56159 expression was
significantly increased in the GBM tissue compared with normal
brain tissue (Fig. 1E). These
findings suggested that Unigene56159 may function as an oncogene
for GBM progression.
 | Table I.Correlation between the expression of
Unigene56159 and the clinicopathological feature in patients'
glioma tissues. |
Table I.
Correlation between the expression of
Unigene56159 and the clinicopathological feature in patients'
glioma tissues.
|
|
| miR-194-5p
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | Cases (n=50) | Low | High | P-value |
|---|
| Age (years) |
|
|
|
|
|
<60 | 26 | 2 | 24 | 0.095 |
|
≥60 | 24 | 6 | 18 |
|
| Sex |
|
|
|
|
|
Male | 33 | 3 | 30 | 0.063 |
|
Female | 17 | 5 | 12 |
|
| Karnofsky
Performance Score |
|
|
|
|
|
<60 | 36 | 3 | 33 | 0.018 |
|
≥60 | 14 | 5 | 9 |
|
| Mean tumor diameter
(cm) |
|
|
|
|
|
<5 | 27 | 5 | 22 | 0.4 |
| ≥5 | 23 | 3 | 20 |
|
| Necrosis on
MRI |
|
|
|
|
|
Yes | 34 | 4 | 30 | 0.234 |
| No | 16 | 4 | 12 |
|
| Seizure |
|
|
|
|
|
Yes | 9 | 3 | 6 | 0.117 |
| No | 41 | 5 | 36 |
|
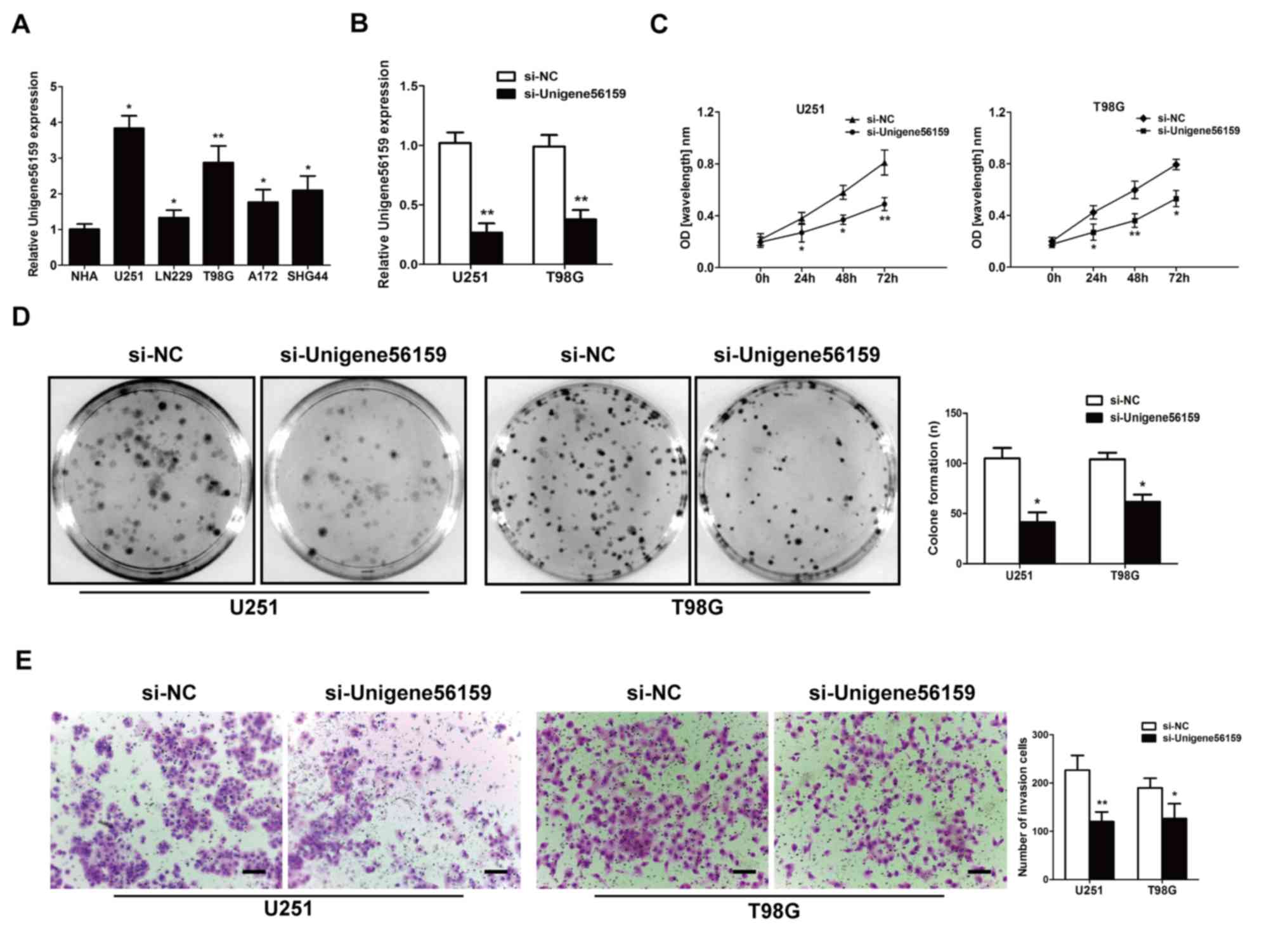
Downregulated Unigene56159 expression
suppresses GBM cell proliferation and invasion
The expression of Unigene56159 was evaluated in GBM
cell lines (U251, LN229, T98G, A172 and SHG44) compared with the
normal astrocyte cell line (NHA) by RT-qPCR assay (P-values=0.0101,
0.0435, 0.0094, 0.02436 and 0.03435; Fig. 2A). It was noted that the
Unigene56159 level were higher in U251 and T98G than in other cell
lines. To identify the effect of Unigene56159 on the proliferative
and invasive ability of GBM, U251 and T98G cells were selected and
transfected with either si-Unigene56159 to knockdown Unigene56159
gene expression or with the si-NC. The transfection efficiency of
si-Unigene56159 was high, exhibiting significantly decreased
expression levels in the si-Unigene56159-transfected cells compared
with the si-NC in both U251 and T98G cell lines (Fig. 2B). In both GBM cell lines, the
knockdown of Unigene56159 resulted in a significant decrease
compared with si-NC group at the similar time points in their
proliferative capacity following 24–72 h (Fig. 2C). Furthermore, Unigene56159
silencing significantly reduced the number of colonies (>50
cells) (Fig. 2D), in addition to
the invasive capacity in both U251 and T98G cell lines compared
with respective si-NC transfected cells (Fig. 2E). These data demonstrated a
suppressive function of both proliferative and invasive processes
following Unigene56159 silencing in GBM cells in vitro.
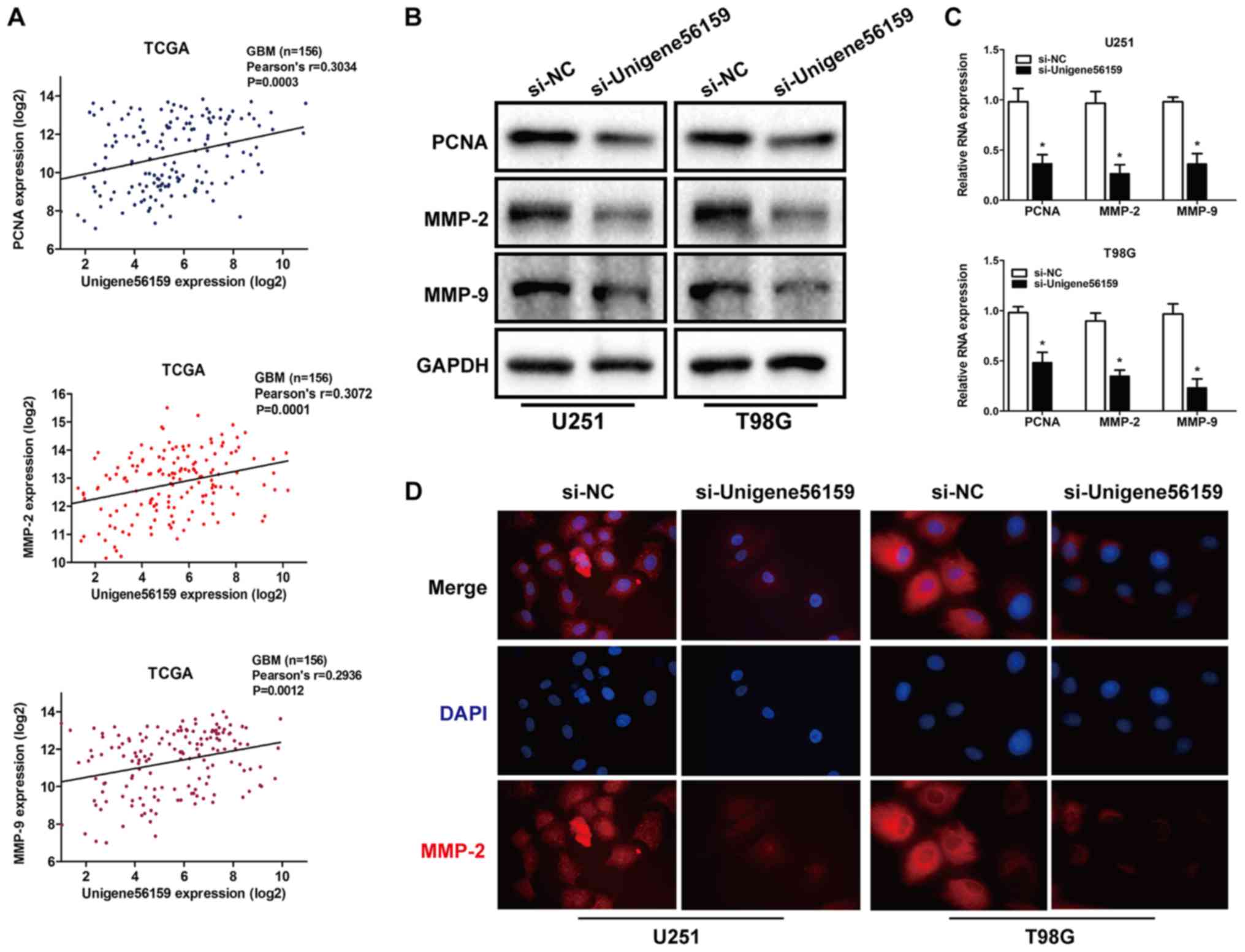
Invasion and proliferation serve a vital role in
tumor progression (28,29). Using data from the TCGA database,
the expression of Unigene56159 was obtained and 151 cases of valid
data collected and a significant positive correlation was
identified between Unigene56159 expression and expression levels of
the tumor proliferation marker; PCNA (r=0.3034; P=0.0003) and
invasion markers MMP-2 (r=0.3072; P=0.0001) and MMP-9 (r=0.2936;
P=0.0012; Fig. 3A).
To further determine whether reduced Unigene56159
expression may affect the proliferative and invasive capacity of
GBM, mRNA and protein expression levels of PCNA, MMP-2 and MMP-9
were assessed using RT-qPCR and western blot analysis,
respectively, and MMP-2 levels were also detected by
immunofluorescence. As shown in Fig.
3B, Unigene56159 knockdown decreased the protein level of
proliferation and invasion markers and inhibited the mRNA
expression of PCNA, MMP-2 and MMP-9 (Fig 3C). Then PCNA, MMP-2 and MMP-9 were
costained in U251 and T98G cells with immunofluorescence staining
assays, the result further showed that unigene56159 silencing
suppressed the MMP-2 expression in glioma cells (Fig. 3D). These results demonstrated that
Unigene56159 silencing significantly decreased the expression
levels of both proliferation- and invasion-related biomarkers.
Taken together, these results suggested that Unigene56159 may be
associated with both proliferation and invasion in GBM.
Correlation between Unigene56159 and
miR-194-5p
The complementary sequence between Unigene56159 and
miR-194-5p was identified using both the TargetScan database and
starBase database. A dual-luciferase reporter assay was
subsequently used to identify the putative miR-194-5p target site
(Fig. 4A). Transfection efficiency
was evaluated by increasing or decreasing miR-194-5p expression
levels in U251 and T98G cell lines (Fig. 4B). Notably, miR-194-5p
significantly decreased the luciferase activity of the
Unigene56159-3′UTR-WT U251 cells, but not the
Unigene56159-3′UTR-MUT U251 cells (Fig. 4C). To determine the role of
Unigene56159 on the expression of miR-194-5p, U251 and T98G cells
were transfected with siRNA-Unigene56159. The expression of
miR-194-5p significantly increased in both U251 and T98G cell lines
upon Unigene56159 knockdown compared with si-NC transfected cells
(Fig. 4D). To further explore
this, miR-194-5p expression levels in the 50 GBM samples were
compared with normal brain samples and it was found that the
expression was significantly decreased in GBM compared with normal
tissue (P<0.001; Fig. 4E). In
addition, the level of Unigene56159 expression was significantly
negatively correlated with miR-194-5p in GBM patient samples
(r=−0.4046; P=0.0036; Fig. 4F).
These data confirmed that miR194-5p represses Unigene56159 in
glioma cells.
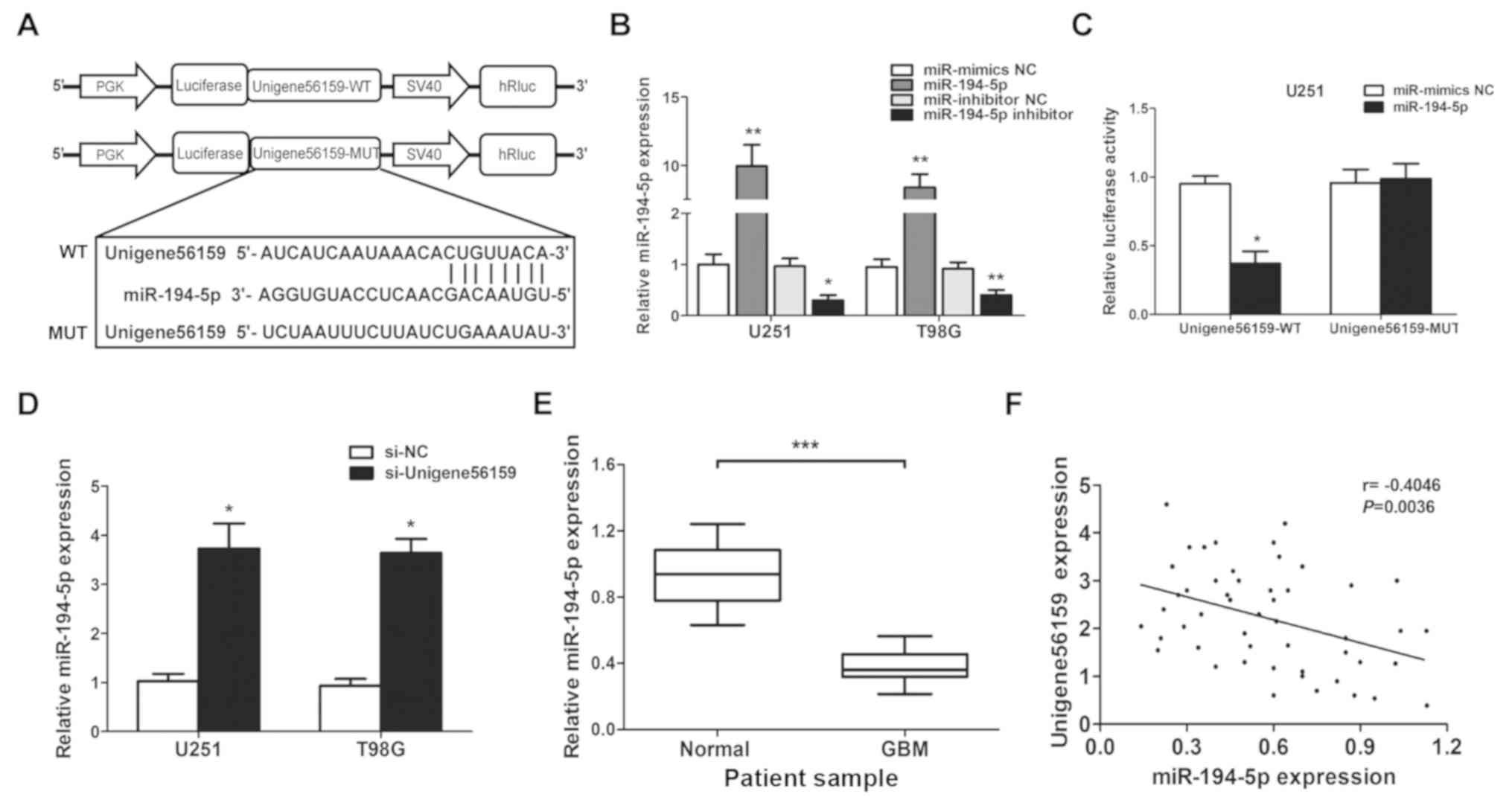 | Figure 4.Long non-coding RNA Unigene56159 is
targeted by miR-194-5p at the 3′UTR. (A) The target site of
miR-194-5p in the 3′UTR region of Unigene56159. (B) miR-194-5p
expression is increased by miR-194-5p mimics compared with
miR-mimics NC (**P<0.01 in U251 and T98G cells), or suppressed
by miR-194-5p inhibitor compared with miR-inhibitor NC (*P<0.05
and **P<0.01), respectively, in U251 and T98G cells. (C) The
relative luciferase activity was detected following co-transfection
of miR-194-5p mimics vs. miR-NC with Unigene56159-WT (*P<0.05)
or Unigene56159-MUT in U251 or T98G cells using the dual-luciferase
reporter assay.. (D) The expression levels of miR-194-5p were
examined by RT-qPCR following transfection with si-Unigene56159 or
the si-NC in GBM cell lines *P<0.05 vs. si-NC group. (E)
Expression levels of miR-194-5p in GBM were measured using RT-qPCR
analysis. ***P<0.001 vs. normal tissues. (F) Pearson's
correlation coefficient analysis between Unigene56159 and
miR-194-5p expression levels. The experiments were repeated three
times.. GBM, glioblastoma multiforme; WT, wild-type; MUT, mutant;
miR, microRNA; NC, negative control; siRNA, small interfering RNA;
UTR, untranslated region; RT-qPCR, reverse
transcription-quantitative PCR. |
miR-194-5p impedes the effect of
Unigene56159 in GBM cells
There were 162 cases of miR-194-5p expression with
survival time in TCGA database. The Kaplan–Meier curve demonstrated
that a high level of miR-194-5p was positively correlated with the
overall survival of patients with glioma from GEPIA database
(Fig. 5A). In GBM cell lines,
miR-194-5p was found to be significantly decreased compared to NHA
cells (Fig. 5B); based on this,
U251 cells were used to conduct rescue experiments. The level of
miR-194-5p increased when transfected with si-Unigene56159 compared
with si-NC. However, after adding the miR-194-5p inhibitor in the
si-Unigene56159 group, the miR-194-5p level decreased compared with
miR-inhibitor NC (Fig. 5C).
Furthermore, a colony formation assay indicated that a miR-194-5p
inhibitor impeded the suppression of proliferative ability
following Unigene56159 knockdown (Fig.
5D). Tumor invasive ability was also decreased following
Unigene56159 knockdown, whereas co-transfection with miR-194-5p
inhibitors impeded these effects in U251 cells (Fig. 5E). Altogether, these data
demonstrated that miR-194-5p may abrogate the malignant behavior of
Unigene56159 in GBM cells.
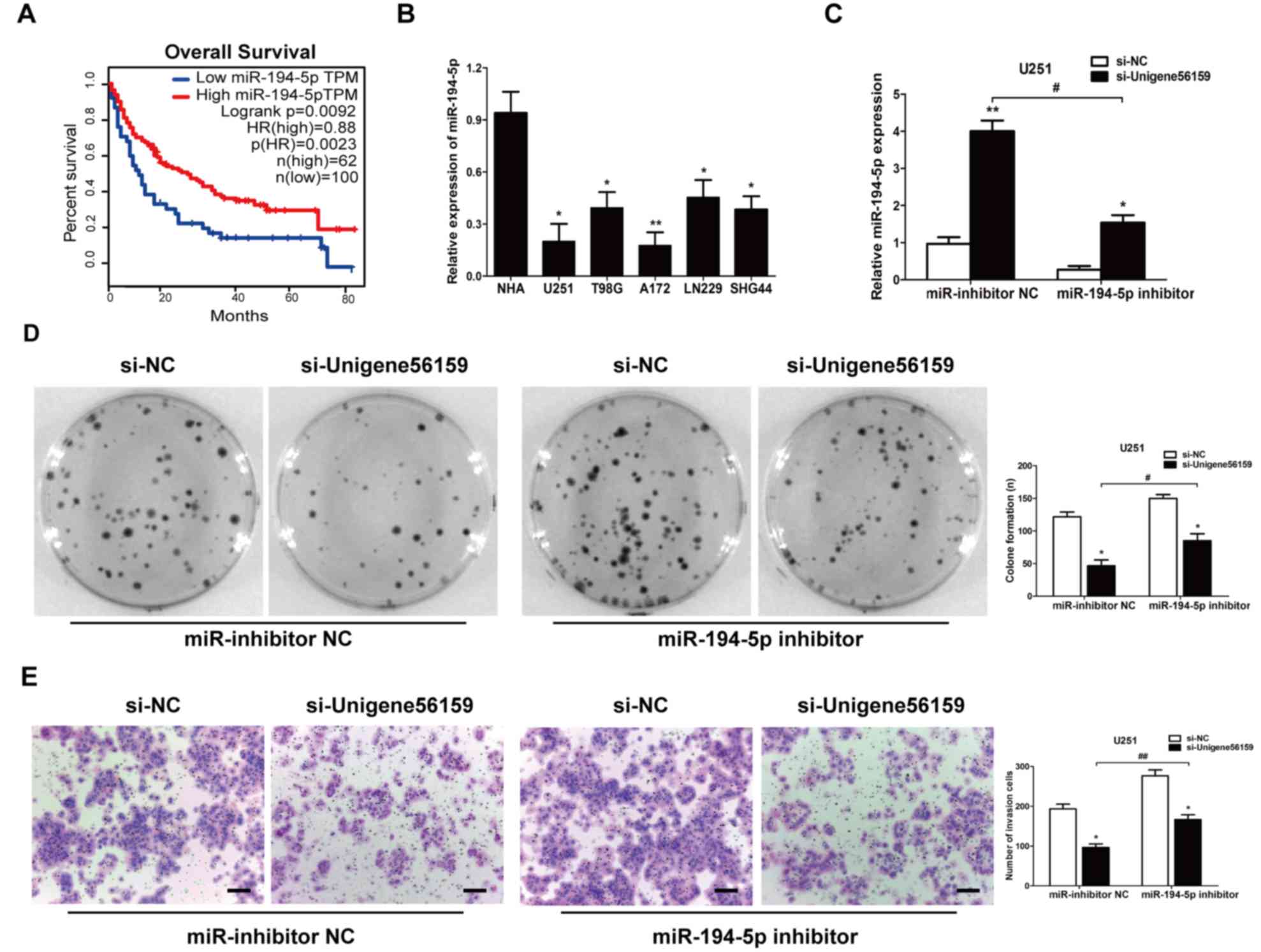 | Figure 5.miR-194-5p inhibits the effect of
long non-coding RNA Unigene56159 in GBM cells. (A) Kaplan-Meier
survival curve analysis of patient data from TCGA database with
high or low levels of miR-194-5p (n=162) (P<0.01). (B)
Expression levels of miR-194-5p in GBM cell lines were examined by
reverse transcription-quantitative PCR. *P<0.05, **P<0.01 vs.
NHA. (C) Gene expression levels of miR-194-5p were identified in
U251 cells co-transfected with (si-Unigene56159) + the miR-194-5p
inhibitor, and with (si-Unigene56159) + miR-inhibitor NC,
*P<0.05 vs. si-NC; and in si-Unigene56159 + miR-194-5p inhibitor
compared with si-Unigene56159+miR-inhibitor NC.
#P<0.05 vs. si-NC. (D) Colony formation assay was
determined in transfected cells. (si-Unigene56159 vs. si-NC) + the
miR-194-5p inhibitor, and with (si-Unigene56159 vs si-NC) +
miR-inhibitor NC; and in si-Unigene56159 + miR-194-5p inhibitor vs.
si-Unigene56159+miR-inhibitor NC. *P<0.05 and
#P<0.05. (E) Invasive ability was investigated by
using Matrigel assays (si-Unigene56159 vs. si-NC) + the miR-194-5p
inhibitor, and with (si-Unigene56159 vs. si-NC) + miR-inhibitor NC;
and in si-Unigene56159 + miR-194-5p inhibitor compared with
si-Unigene56159+miR-inhibitor NC. Scale bar, 500 µm. *P<0.05 and
##P<0.01. The experiment was repeated three times.
NHA, normal human astrocyte; miR, microRNA; NC, negative control;
si, small interfering RNA. |
Discussion
GBM is one of the most prevalent types of malignant
tumor found within the CNS, and the overall survival of patients
with late stage GBM remains poor (1,2).
Therefore, it is essential to investigate novel therapeutic
strategies for patients with GBM. Aberrantly expressed lncRNAs
serve an important role in tumor development within multiple
cancers (30). However, the
underlying mechanism between lncRNAs and GBM is still unclear.
In the present study, high expression levels of
Unigene56159 and low levels of miR-194-5p were found in GBM tissues
and cell lines compared with normal tissues and cells. Following
the suppression of proliferative and invasive capacities of GBM
cell lines after siRNA knockdown, it was suggested that
Unigene56159 may act as an oncogene in GBM. The result confirmed
that miR194-5p repressed Unigene56159 in glioma cells. To further
study the effect of miR-194-5p in GBM transfected with
si-Unigene56159, Unigene56159 was knocked down with siRNA and the
proliferation and invasion ability was decreased. Then the
miR-194-5p inhibitor was added to glioma cells (si-Unigene56159 and
si-NC) and it was found that the suppressive effect was impeded
compared with miR-inhibitor NC group. Together, these results
indicated that gene knockdown of Unigene56159 exerted a suppressive
effect in GBM progression, suggesting a novel therapeutic strategy
for GBM.
lncRNAs act as endogenous miRNA sponges for binding
to miRNAs or participating in the competitive endogenous RNAs
(ceRNA) regulatory network (31).
For example, the lncRNA PVT1 regulates malignant behavior in
xenograft models of breast cancer cells (32), whereas small nucleolar RNA host
gene 5 knockdown restrains the malignant phenotype of gastric
cancer cells by targeting the miR-32/KLF4 axis (33). In addition, the upregulation of
SNHG1 in lung cancer positively correlates with both tumor size and
tumor-node-metastasis stages (34). Lv et al (18) reported that Unigene56159 promotes
epithelial-to-mesenchymal transition processes in hepatocellular
carcinoma cells by regulating miR-140-5p, whilst Lu et al
(35) reported that LINC00673
suppresses the migratory and invasive capacity of non-small cell
lung cancer by sponging miR-150-5p. lncRNAs can function as
competitive endogenous RNAs (ceRNAs) that can sequester miRNAs and
prevent their expression. Then lncRNA nullify their ability to
target protein-coding mRNAs and indirectly affect downstream
biological processes (36,37). Thus, the present study aimed to
investigate whether the lncRNA Unigene56159 could act as a ceRNA
towards miR-194-5p in GBM.
Results from the present study indicated that high
levels of Unigene56159 may correlate with worse overall survival
and that miR194-5p repress the effect of Unigene56159 in glioma
cells. The negative correlation with Unigene56159 was confirmed by
exploring the data in TCGA database. The putative binding site
between Unigene56159 and miR-194-5p was detected by using
luciferase assay. The present study determined that miR-194-5p was
a target of Unigene56159. Unigene56159 silencing can reduce the
proliferation and invasion ability of GBM cell and after adding
with miR-194-5p inhibitor in si-Unigene56159 group, the suppressive
effect of si-Unigene56159 was impeded compared with miR-inhibitor
NC. These results provided evidence for a role of Unigene56159 in
GBM and may improve our understanding of the mechanisms underlying
GBM development. This could provide a promising therapeutic target
for the treatment of GBM.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GJ designed the study, performed experiments,
analyzed the data and wrote the manuscript. HD performed the in
vitro experiments. HD and YD analyzed the data and drafted the
manuscript. XY designed and supervised the study, and edited the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients and the study was approved by the ethics committee of
Tianjin Medical University (Tianjin, China). All procedures were
performed in accordance with national (D.L.n.26, March 4th, 2014)
and international laws and policies (directive 2010/63/EU).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lai NS, Wu DG, Fang XG, Lin YC, Chen SS,
Li ZB and Xu SS: Serum microRNA-210 as a potential noninvasive
biomarker for the diagnosis and prognosis of glioma. Br J Cancer.
112:1241–1246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chow KK, Naik S, Kakarla S, Brawley VS,
Shaffer DR, Yi Z, Rainusso N, Wu MF, Liu H, Kew Y, et al: T cells
redirected to EphA2 for the immunotherapy of glioblastoma. Mol
Ther. 21:629–637. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang K, Kievit FM, Jeon M, Silber JR,
Ellenbogen RG and Zhang M: Nanoparticle-mediated target delivery of
TRAIL as gene therapy for glioblastoma. Adv Healthc Mater.
4:2719–2726. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Delfino KR, Serão NV, Southey BR and
Rodriguez-Zas SL: Therapy-, gender- and race-specific microRNA
markers, target genes and networks related to glioblastoma
recurrence and survival. Cancer Genomics Proteomics. 8:173–183.
2011.PubMed/NCBI
|
|
5
|
Koekkoek JA, Postma TJ, Heimans JJ,
Reijneveld JC and Taphoorn MJ: Antiepileptic drug treatment in the
end-of-life phase of glioma patients: A feasibility study. Support
Care Cancer. 24:1633–1638. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Su HK, Zhao HF, Chen ZP and To SS:
Progress in the application of molecular biomarkers in gliomas.
Biochem Biophys Res Commun. 465:1–4. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Delgado-López PD and Corrales-García EM:
Survival in glioblastoma: A review on the treatment modalities.
Clin Transl Oncol. 18:1062–1071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li C, Jing H, Ma G and Liang P: Allicin
induces apoptosis through activation of both intrinsic and
extrinsic pathways in glioma cells. Mol Med Rep. 17:5976–5981.
2018.PubMed/NCBI
|
|
9
|
Mrugala MM: Advances and challenges in the
treatment of glioblastoma: A clinician's perspective. Discov Med.
83:221–230. 2013.
|
|
10
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research. Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grzmil M, Morin P Jr, Lino MM, Merlo A,
Frank S, Wang Y, Moncayo G and Hemmings BA: MAP kinase-interacting
kinase 1 regulates SMAD2- dependent TGF-β signaling pathway in
human glioblastoma. Cancer Res. 71:2392–2402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang S, Wang W, Liu G, Xie S, Li Q, Li Y
and Lin Z: Long non-coding RNA HOTTIP promotes hypoxia-induced
epithelial-mesenchymal transition of malignant glioma by regulating
the miR-101/ZEB1 axis. Biomed Pharmacother. 95:711–720. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng J, Du T, Song Y, Gao Y, Li F, Wu R,
Chen Y, Li W, Zhou H, Yang Y and Pei Z: Knockdown of long noncoding
RNA CCAT2 inhibits cellular proliferation, invasion, and EMT in
glioma cells. Oncol Res. 6:913–921. 2017. View Article : Google Scholar
|
|
15
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sullivan TB, Robert LC, Teebagy PA, Morgan
SE, Beatty EW, Cicuto BJ, Nowd PK, Rieger-Christ KM and Bryan DJ:
Spatiotemporal microRNA profile in peripheral nerve regeneration:
miR-138 targets vimentin and inhibits schwann cell migration and
proliferation. Neural Regen Res. 13:1253–1262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 2:344–357. 2011. View Article : Google Scholar
|
|
18
|
Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang
Y, Liu M and Tang H: Long non-coding RNA unigene56159 promotes
epithelial-mesenchymal transition by acting as a ceRNA of
miR-140-5p in hepatocellular carcinoma cells. Cancer Lett.
2:166–175. 2016. View Article : Google Scholar
|
|
19
|
Zhang Z, Lei B, Wu H, Zhang X and Zheng N:
Tumor suppressive role of miR-194-5p in glioblastoma multiform. Mol
Med Rep. 6:9317–9322. 2017. View Article : Google Scholar
|
|
20
|
Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu
Q, Tong X, Yang W, Xu Q, Huang D and Tu K: A novel lncRNA
MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by
targeting miR-194-5p/FOXA1 axis. Mol Cancer. 1:282019. View Article : Google Scholar
|
|
21
|
Dell'Aversana C, Giorgio C, D'Amato L,
Lania G, Matarese F, Saeed S, Di Costanzo A, Belsito Petrizzi V,
Ingenito C, Martens JHA, et al: MiR-194-5p/BCLAF1 deregulation in
AML tumorigenesis. Leukemia. 11:2315–2325. 2017. View Article : Google Scholar
|
|
22
|
Zhang Q, Wei T, Shim K, Wright K, Xu K,
Palka-Hamblin HL, Jurkevich A and Khare S: Atypical role of sprouty
in colorectal cancer: Sprouty repression inhibits
epithelial-mesenchymal transition. Oncogene. 24:3151–3162. 2016.
View Article : Google Scholar
|
|
23
|
Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove
R, Xu R and Huang W: MiR-194 is a marker of hepatic epithelial
cells and suppresses metastasis of liver cancer cells in mice.
Hepatology. 6:2148–2157. 2010. View Article : Google Scholar
|
|
24
|
Huang Da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Robinson MD, Mccarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 1:139–140. 2010.
View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate - A practical and powerful approach to
multiple testing. J R Stat Soc. 1:289–300. 1995.
|
|
28
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 3:161–174. 2002. View
Article : Google Scholar
|
|
29
|
Mayer A, Takimoto M, Fritz E, Schellander
G, Kofler K and Ludwig H: The prognostic significance of
proliferating cell nuclear antigen, epidermal growth factor
receptor, and mdr gene expression in colorectal cancer. Cancer.
8:2454–2460. 2015.
|
|
30
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: CeRNA in cancer: Possible functions and clinical
implications. J Med Genet. 10:710–718. 2015. View Article : Google Scholar
|
|
32
|
Tang J, Li Y, Sang Y, Yu B, Lv D, Zhang W
and Feng H: LncRNA PVT1 regulates triple-negative breast cancer
through KLF5/beta-catenin signaling. Oncogene. 34:4723–4734. 2018.
View Article : Google Scholar
|
|
33
|
Sun J, Zhang S, Liu Y, Shan B, Zheng D and
Shi J: The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell
proliferation and migration by targeting KLF4. FASEB J. 31:893–903.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cui Y, Zhang F, Zhu C, Geng L, Tian T and
Liu H: Upregulated lncRNA SNHG1 contributes to progression of
non-small cell lung cancer through inhibition of miR-101-3p and
activation of Wnt/β-catenin signaling pathway. Oncotarget.
8:17785–17794. 2017.PubMed/NCBI
|
|
35
|
Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H,
Kong J, Ding K, Shen HM, Wu H, et al: Long non-coding RNA linc00673
regulated non-small cell lung cancer proliferation, migration,
invasion and epithelial mesenchymal transition by sponging
miR-150-5p. Mol Cancer. 16:1182017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karreth FA, Tay Y, Perna D, Ala U, Tan SM,
Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al:
In vivo identification of tumor- suppressive PTEN ceRNAs in an
oncogenic BRAF-induced mouse model of melanoma. Cell. 147:382–395.
2011. View Article : Google Scholar : PubMed/NCBI
|