Introduction
In spite of the progress made in the treatment and
prevention of human cancer types, gastric carcinoma (as a type of
malignancy that develops from the inner layer of the stomach
tissue) remains the third most common cause of cancer-associated
mortality worldwide (1,2). The wide application of chemotherapy,
radiation treatment and other targeted treatments has substantially
improved the survival of patients with gastric carcinoma during
last several decades. However, surgical resection remains the only
radical treatment for patients with gastric carcinoma who are at an
early stage of the cancer (3). At
present, there is no radical treatment for patients with gastric
carcinoma at advance stages. However, as the majority of patients
with gastric carcinoma are diagnosed at advance stages,
particularly in developing countries, including China (4), survival of those patients remains
poor.
Long non-coding RNAs (lncRNAs) are a group of
non-coding RNA composed of >200 nucleotides (5). It has been reported that lncRNAs are
involved in nearly every aspect of almost all critical
physiological processes and pathological changes in the human body
(6). Neighboring enhancer of FOXA2
(NEF) is a newly discovered lncRNA known to possess functionality
only in the metastasis of hepatocellular carcinoma (7). In the present study, the involvement
of lncRNA-NEF in gastric carcinoma was examined and the clinical
potentials discussed.
Patients and methods
Subjects
A total of 98 patients with gastric carcinoma who
were diagnosed by pathological examination and treated at the
Chengdu Fifth People's Hospital (Sichuan, China) between January
2011 and January 2013 were selected as the patient group. The
patient group included 60 men and 38 women, and the age ranged from
22 to 71 years, with a mean age of 45.7±8.2 years. Patients with
other severe diseases, other gastric diseases or mental disorders
were not included. Staging of primary tumor types were performed
according to the following criteria: Tis, carcinoma in situ,
13 cases; T1, tumor invades muscularis mucosae, lamina propria, or
submucosa, 15 cases; T2, tumor invades muscularis propria, 19
cases; T3, tumor penetrates subserosal connective tissue but no
invasion of visceral peritoneum or adjacent structures was found,
20 cases; T4, tumor invades serosa (visceral peritoneum) or its
adjacent structures, 31 cases. At the same time, 32 healthy people
were included to serve as the control group. Control group included
20 men and 12 women, and the age ranged from 21 to 71 years, with a
mean age of 45.9 ±8.8 years. No notable differences in age and sex
were identified between the patient group and control group. The
present study was ethically approved by Ethics Committee of Chengdu
Fifth People's Hospital. All patients provided written informed
consent.
Specimen collections
Tumor tissues and adjacent healthy tissues within 5
cm of the tumor were collected from 45 patients during surgical
operation. The tissues were confirmed to be tumor or healthy
tissues by pathological examinations. Blood (about 15 ml) was also
extracted from elbow vein of all participants. Blood was maintained
at room temperature for 2 h, followed by centrifugation at 1,000 ×
g at room temperature for 20 min to collect serum. All specimens
were stored in liquid nitrogen prior to use.
Cell lines and cell culture
Human gastric cell line SNU-1 (Asian) and Hs 746T
(Caucasian) were purchased from American Type Culture Collection
(Manassas, VA, USA). Cells were cultured with ATCC-formulated
RPMI-1640 Medium containing 10% fetal bovine serum at 37°C in a 5%
CO2 incubator. Cells were harvested during the
logarithmic growth phase for subsequent experiments.
Construction of lncRNA-NEF silencing
and overexpression cell lines
LncRNA-NEF small interfering RNA (siRNA;
5′-GGAGCUGUUUGGGCAAUAATT-3′) and non-specific silencing control
(negative control) were provided by Shanghai GenePharma Co., Ltd.
(Shanghai, China). NEF cDNA was inserted into a pIRSE2-EGFP vector
(Clontech Laboratories, Inc., Mountainview, CA, USA) to establish a
NEF expression vector. Cells of two cell lines (SNU-1 and Hs 746T)
were cultured overnight to reach 80–90% confluence, and
Lipofectamine 2000 reagent (cat no. 11668-019; Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) was then used to
transfect 10 nM vector or 50 nM siRNA (negative control) into
5×105 cells in each well of a 6-well plate at 37°C in a
5% CO2 incubator. Cells were incubated with transfection
reagents, and vectors or siRNAs for 6 h, followed by washing with
fresh cell culture medium to avoid cytotoxicity.
Cell proliferation assay
Cells of both SNU-1 and Hs 746T cell lines were
collected during the logarithmic growth phase to produce cell
suspensions (4×104 cell/ml). Then, a 100 µl cell
suspension containing 4×103 cells was added into each
well of a 96-well plate. Cells were cultured in an incubator (37°C
and 5% CO2) and Cell Counting Kit-8 (CCK-8) solution (10
µl) was added into each well and incubated for 24, 48, 72 and 96 h.
Following incubation at 37°C for an additional 4 h, the optical
density value at 450 nm was measured using the Fisherbrand™
accuSkan™ GO UV/Vis Microplate Spectrophotometer (Thermo Fisher
Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Tumor tissues and adjacent healthy tissues were
ground in liquid nitrogen, followed by the addition of TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) to extract
total RNA. In vitro cultured cells of both SNU-1 and Hs 746T
cell lines were directly mixed with TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) at room temperature to extract
total RNA. Reverse transcription was then performed to synthesize
cDNA, followed by the preparation of PCR reactions using
SYBR®-Green Real-Time PCR Master Mixes (Thermo Fisher
Scientific, Inc.) and the following primers: lncRNA-NEF forward,
5′-CTGCCGTCTTAAACCAACCC-3′ and reverse, 5′-GCCCAAACAGCTCCTCAATT-3′;
human β-actin forward, 5′-GACCTCTATGCCAACACAGT-3′ and reverse,
5′-AGTACTTGCGCTCAGGAGGA-3′. PCR reaction conditions were as
follows: 95°C for 40 sec, followed by 40 cycles of 15 sec at 95°C
and 30 sec at 55°C. Cq values were produced using the
2−ΔΔCq method (8).
Relative expression levels of lncRNA-NEF were normalized to the
endogenous control (β-actin).
Western blot analysis
Total protein extraction was performed using RIPA
solution (Thermo Fisher Scientific, Inc.) on ice with 30 min
incubation, and protein quantification was performed using the BCA
method. Subsequently, 10% SDS-PAGE gel electrophoresis was
performed using 30 µg protein per lane, followed by gel transfer to
polyvinylidene fluoride (PVDF) membranes. PVDF membranes were
incubated with 5% skimmed milk for 1.5 h at room temperature,
followed by incubation with rabbit anti-human primary antibodies
against Runx1 (1:2,000; cat no. ab15309; Abcam, Cambridge, UK) and
GAPDH (1:1,000; cat no. ab8245; Abcam) overnight at 4°C.
Subsequently, membranes were incubated with anti-rabbit
immunoglobulin G horseradish peroxidase-conjugated secondary
antibody (1:1,000; cat no. MBS435036; MyBioSource, Inc., San Diego,
CA, USA) at room temperature for 1.5 h, followed by signal
development using ECL reagents (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Signals were scanned using the MYECL™ Imager
(Thermo Fisher Scientific, Inc.), and the relative expression level
of Runx1 was normalized to the endogenous control GAPDH using Image
J version 1.46 software (National Institutes of Health, Bethesda,
MD, USA).
Statistical analysis
SPSS19.0 (SPSS, Inc., Chicago, IL, USA) was used in
the present study for all statistical analyses. Measurement data
were represented as mean ± standard deviation. Comparisons of
measurement data between two groups and among multiple groups were
performed using a paired Student's t-test and one-way analysis of
variance followed by a least-significant-difference post-hoc test,
respectively. Continuous data were compared using a χ2
test. Receiver operating characteristic (ROC) curve analysis was
performed to evaluate the diagnostic values of serum circulating
lncRNA-NEF for gastric carcinoma. According to the median relative
serum level of circulating lncRNA-NEF (5.02), all patients were
divided into a high expression group and a low expression group.
Survival data were collected during a 5 year follow-up and the
Kaplan-Meier method was used to plot survival curves. Survival
curves were compared using a log rank t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of lncRNA-NEF in cancer
tissues and adjacent healthy tissues
The differential expression of a gene in tumor
tissues and healthy tissues indicate its involvement in the tumor.
In the present study, tumor tissues and adjacent healthy tissues
were collected from 45 patients with gastric carcinoma, and the
expression levels of lncRNA-NEF in those tissues were measured
using RT-qPCR. Among those 45 patients, 41 were revealed to exhibit
a significantly higher expression level of lncRNA-NEF in the
adjacent healthy tissues compared with the cancer tissues,
accounting for 91.1% of all patients (P<0.05; Fig. 1). In contrast, a significantly
lower expression level of lncRNA-NEF in adjacent healthy tissues
compared with cancer tissues was only identified in 2 cases,
accounting for 4.4% of all patients (P<0.05). No significant
difference was identified in the remaining 2 cases, accounting for
4.4% (Fig. 1). The present data
indicates that the downregulation of lncRNA-NEF is likely to be
involved in gastric carcinoma.
Expression of lncRNA-NEF in patients
with gastric carcinoma at different stages of a primary tumor
To further confirm the involvement of lncRNA-NEF in
gastric carcinoma, serum levels of lncRNA-NEF in patients with
gastric carcinoma and healthy controls were also measured using
RT-qPCR. Results revealed that levels of circulating lncRNA-NEF in
the serum were significantly higher in the control group compared
with patients with different stages of gastric carcinoma except
stage Tis (P<0.05; Fig. 2). In
addition, the level of circulating lncRNA-NEF significantly
decreased gradually with the increase of primary tumor stage
(P<0.05; Fig. 2).
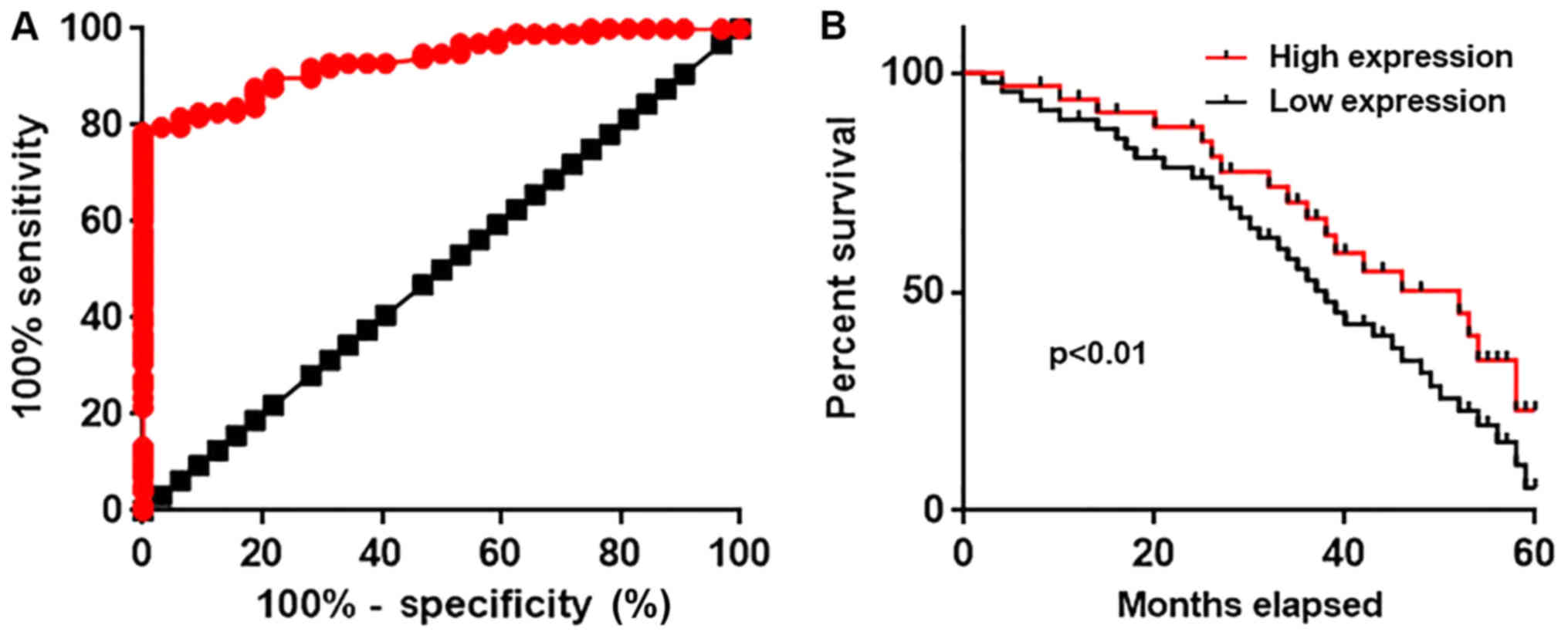
Diagnostic and prognostic value of
serum circulating lncRNA-NEF for gastric carcinoma
Differential expression of a gene may possess
diagnostic potential for disease diagnosis. Therefore, receiver
operating characteristic curve analysis was performed to evaluate
the diagnostic values of serum circulating lncRNA-NEF for gastric
carcinoma. The area under the curve was 0.9349 with 95% confidence
intervals of 0.8958 to 0.9741 (P<0.0001, compared with the line
of identity; Fig. 3A). According
to the median serum level of circulating lncRNA-NEF (5.02), all
patients were divided into a high expression group and a low
expression group. Survival data were collected during a 5 year
follow-up and the Kaplan-Meier method was used to plot survival
curves. Survival curves were compared using a log rank t-test.
Comparison of survival curves revealed that the overall survival
rate of patients with a high serum level of ncRNA-NEF was
significantly higher compared with that of patients with a low
serum level of lncRNA-NEF (P<0.01; Fig. 3B). The present data suggests that
serum circulating lncRNA-NEF has substantial diagnostic and
prognostic value for gastric carcinoma.
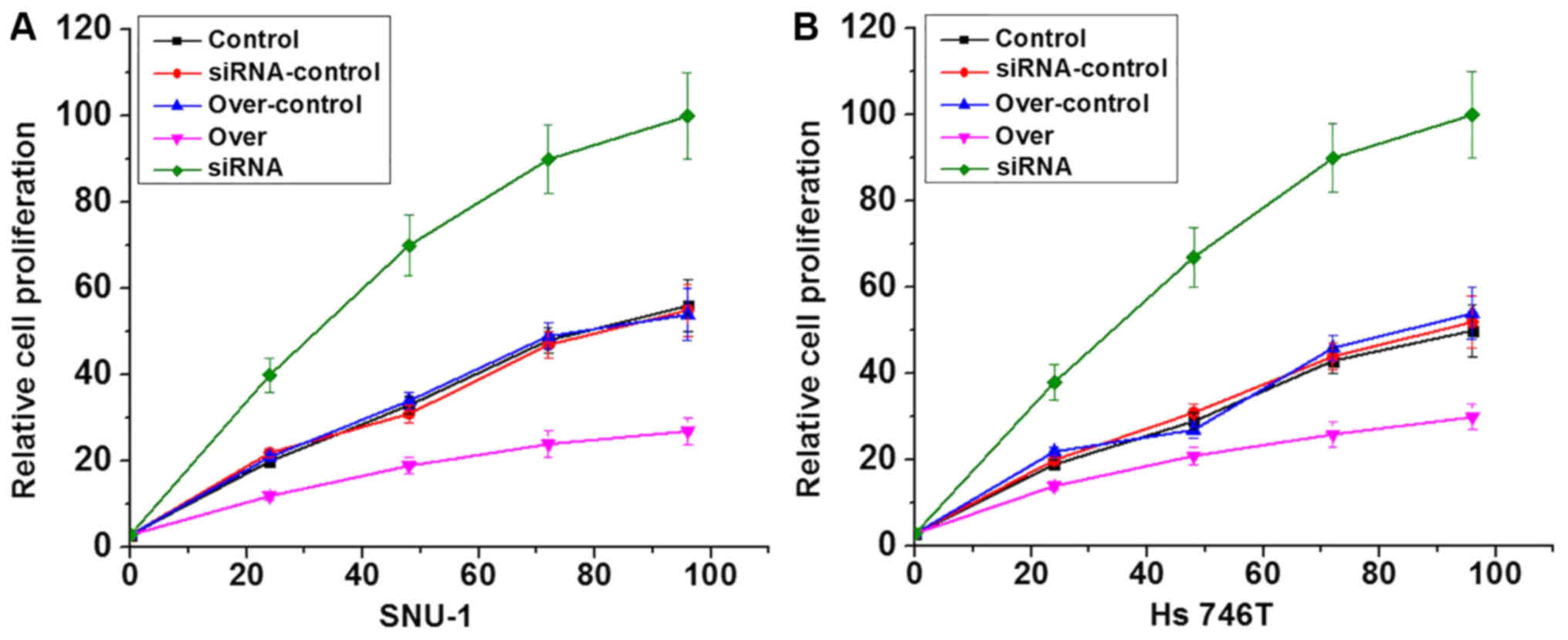
Effects of lncRNA-NEF overexpression
and knockdown on gastric carcinoma cell proliferation
LncRNA-NEF overexpression and siRNA silencing
gastric carcinoma cell lines were constructed and confirmed by
measuring the expression level of lncRNA-NEF through RT-qPCR (data
not shown). Effects of lncRNA-NEF overexpression on cell
proliferation were investigated using a CCK-8 assay. As presented
in Fig. 4, lncRNA-NEF
overexpression clearly inhibited the proliferation of the cells of
two gastric carcinoma cell lines compared with the control cells,
whilst siRNA silencing notably promoted proliferation compared with
the control cells, confirming the inhibitory effects of lncRNA-NEF
on gastric carcinoma cell proliferation.
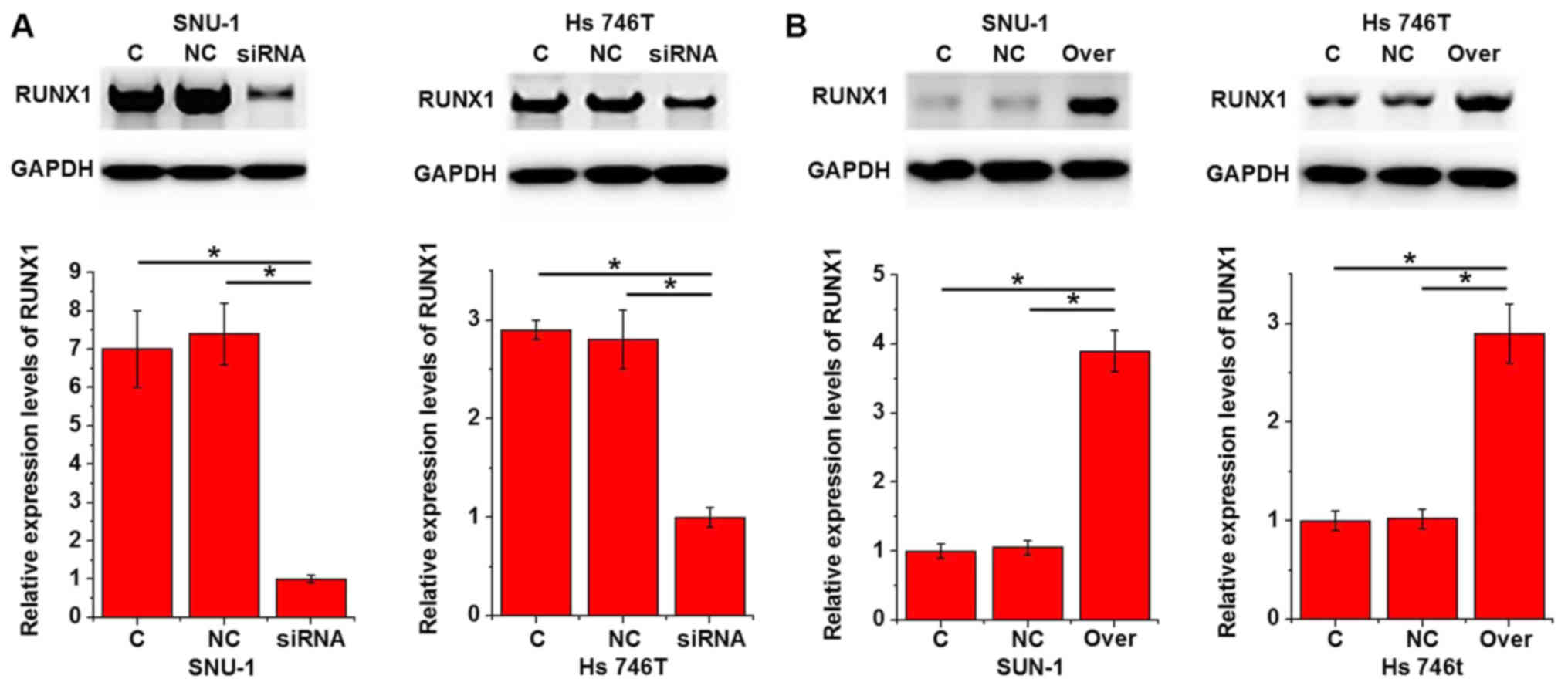
Effects of lncRNA-NEF knockdown and
overexpression on Runx1 expression
Runx1 is a key player in the regulation of tumor
growth in different types of cancers including gastric carcinoma
(9). Therefore, the effects of
lncRNA-NEF knockdown and overexpression on Runx1 expression were
investigated using western blot analysis. As presented in Fig. 5A, lncRNA-NEF knockdown
significantly inhibited the expression levels of Runx1 in two
gastric carcinoma cell lines compared with the control cells
(P<0.05). In contrast, lncRNA-NEF overexpression significantly
promoted the expression levels of Runx1 in the two gastric
carcinoma cell lines compared with the controls (P<0.05;
Fig. 5B).
Discussion
The onset, development and progression of gastric
carcinoma are accompanied with changes in the expression patterns
of a large set of lncRNAs. In a previous study, Cao et al
(10) identified 88 differentially
expressed lncRNAs in gastric carcinoma. LncRNA H19 was revealed to
be upregulated in cancer tissues compared with adjacent healthy
tissues around the tumor in the majority of patients with gastric
carcinoma, which supports its function as an oncogenic gene in the
pathogenesis of this disease (11). In contrast, maternally expressed 3
(which functions as a tumor suppressor gene) is downregulated in
gastric carcinoma (12). In
addition to altered expression patterns during the development and
progression of gastric carcinoma, a number of lncRNAs, including
urothelial cancer associated 1, also demonstrated an altered
expression pattern during the application of chemotherapy, which in
turn affects treatment outcomes (13). LncRNA-NEF is a newly discovered
lncRNA with downregulated expression in hepatocellular carcinoma
(7). In the present study,
significantly reduced expression levels of LncRNA-NEF were
identified in the majority of patients with gastric carcinoma
compared with healthy controls. In addition, reduced serum levels
of circulating lncRNA-NEF were also observed in patients with
gastric carcinoma compared with healthy controls, and serum
lncRNA-NEF levels gradually decreased with the increase of primary
tumor stages. These data suggest that the downregulation of
lncRNA-NEF is involved in the pathogenesis of gastric
carcinoma.
In spite of the progression made in the development
of treatment strategies, treatment of gastric carcinoma remains
challenging due to the low early diagnosis rate (14). Therefore, identifying how to
improve the diagnosis of gastric carcinoma at an early stage is a
major task for treatment. Circulating lncRNAs have been proven to
be sensitive biomarkers for the identification of certain
pathological processes including the development and progression of
gastric carcinoma. For instance, the abnormally upregulated
expression of lncRNA H19 has been proven to effectively distinguish
patients with gastric carcinoma from healthy controls (15). In the present study, lncRNA-NEF was
proven to be a sensitive diagnostic marker for gastric carcinoma.
Accurate prediction of prognosis is also critical for the survival
of patients with malignancies (16,17).
In the present study, high levels of serum circulating lncRNA-NEF
were proven to be associated with poorer survival conditions of
patients with gastric carcinoma. These data suggest that serum
circulating lncRNA-NEF may serve as a promising diagnostic and
prognostic biomarker for gastric carcinoma. However, it is worth
mentioning that lncRNA-NEF is a newly discovered lncRNA with
unknown expression patterns in other diseases. Therefore, multiple
markers should be combined to improve the diagnosis.
A previous study has demonstrated that lncRNA-NEF is
involved in the metastasis, but not growth, of hepatocellular
carcinoma (7). In the present
study, lncRNA-NEF was revealed to exert inhibitory effects on the
proliferation of gastric carcinoma cells, indicating the different
pathogenesis of those two types of malignancies. Runx1 is a
transcription factor that participates in different human
malignancies by regulating cell proliferation (18,19).
LncRNA H19, as a oncogenic lncRNA, promotes the proliferation of
gastric carcinoma cells by inhibiting the expression of Runx1
(9,20). In the present study, NEF
overexpression promoted, and NEF siRNA silencing inhibited, Runx1
expression in SNU-1 (Asian) and Hs 746T (Caucasian) cell lines,
each originating from different ethnic backgrounds. These data
suggest that lncRNA-NEF inhibited the proliferation of gastric
carcinoma cells by inhibiting the expression of Runx1, and the
function of lncRNA-NEF was unlikely to be affected by different
ethnic backgrounds, which has been proven to influence factors in
other types of malignancies (21).
In conclusion, NEF was significantly downregulated
in gastric carcinoma. Serum NEF may serve as a sensitive diagnostic
and prognostic marker for gastric carcinoma. NEF overexpression
promoted, and NEF siRNA silencing inhibited, gastric carcinoma cell
proliferation. Additionally, NEF overexpression promoted, and NEF
siRNA silencing inhibited, Runx1 expression. Therefore, it was
concluded that lncRNA NEF participates in the regulation of cancer
cell proliferation by regulating Runx1 expression. However, the
present study remains challenged by the small sample size. Further
studies with a bigger sample size are required in order to further
confirm the conclusions.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XW and MW conceived and designed the study. XW, XJ,
LZ, ZW and HH performed the experiments. MW wrote the paper. XW,
XJ, LZ, ZW and HH reviewed and edited the manuscript. All authors
read and approved the manuscript and agree to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Institutional Review Board of Chengdu Fifth People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Roder DM: The epidemiology of gastric
cancer. Gastric Cancer. 5:5–11. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Plummer M, Franceschi S and Muñoz N:
Epidemiology of gastric cancer. IARC Sci Publ. 157:311–326.
2004.
|
|
3
|
Orditura M, Galizia G, Sforza V,
Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J,
Savastano B, Mabilia A, et al: Treatment of gastric cancer. World J
Gastroenterol. 20:1635–1649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Perkel JM: ‘Visiting noncodarnia’.
Biotechniques. 54:303–304. 2013. View Article : Google Scholar
|
|
6
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Gen. 12:861–874. 2011. View Article : Google Scholar
|
|
7
|
Liang WC, Ren JL, Wong CW, Chan SO, Waye
MM, Fu WM and Zhang JF: LncRNA-NEF antagonized epithelial to
mesenchymal transition and cancer metastasis via cis-regulating
FOXA2 and inactivating Wnt/β-catenin signaling. Oncogene.
37:1445–1456. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu G, Xiang T, Wu QF and Wang WX: Long
noncoding RNA H19-derived miR-675 enhances proliferation and
invasion via RUNX1 in gastric cancer cells. Oncol Res. 23:99–107.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cao WJ, Wu HL, He BS, Zhang YS and Zhang
ZY: Analysis of long non-coding RNA expression profiles in gastric
cancer. World J Gastroenterol. 19:3658–3664. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun M, Xia R, Jin F, Xu T, Liu Z, De W and
Liu X: Downregulated long noncoding RNA MEG3 is associated with
poor prognosis and promotes cell proliferation in gastric cancer.
Tumor Bio. 35:1065–1073. 2014. View Article : Google Scholar
|
|
13
|
Shang C, Guo Y, Zhang J and Huang B:
Silence of long noncoding RNA UCA1 inhibits malignant proliferation
and chemotherapy resistance to adriamycin in gastric cancer. Cancer
Chemother Pharm. 77:1061–1067. 2016. View Article : Google Scholar
|
|
14
|
Smyth EC, Verheij M, Allum W, Cunningham
D, Cervantes A and Arnold D; ESMO Guidelines Committee, : Gastric
cancer: ESMO clinical practice guidelines for diagnosis, treatment
and follow-up. Annal Onco. 27 Suppl 5:v38–v49. 2016. View Article : Google Scholar
|
|
15
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Reports.
5:115162015. View Article : Google Scholar
|
|
16
|
Rugge M, Fassan M and Graham DY:
Epidemiology of gastric cancer. Gastric Cancer. Strong V: Springer;
Cham: pp. 23–32. 2015, View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cristescu R, Lee J, Nebozhyn M, Kim KM,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Keita M, Bachvarova M, Morin C, Plante M,
Gregoire J, Renaud MC, Sebastianelli A, Trinh XB and Bachvarov D:
The RUNX1 transcription factor is expressed in serous epithelial
ovarian carcinoma and contributes to cell proliferation, migration
and invasion. Cell Cyc. 12:972–986. 2013. View Article : Google Scholar
|
|
19
|
Hong D, Andrew J, Fritz KF, Fitzgerald MP,
Stein JL, Lian J and Stein G: Runx1 possesses anti-tumor activity
and inhibits stemness in breast cancer cells. Cancer Res. 77 Suppl
13:S5534–S5542. 2017. View Article : Google Scholar
|
|
20
|
Zhuang M, Gao W, Xu J, Wang P and Shu Y:
The long non-coding RNA H19-derived miR-675 modulates human gastric
cancer cell proliferation by targeting tumor suppressor RUNX1.
Biochem Biophys Res Commun. 448:315–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hoffmann TJ, Van Den Eeden SK, Sakoda LC,
Jorgenson E, Habel LA, Graff RE, Passarelli MN, Cario CL, Emami NC,
Chao CR, et al: A large multiethnic genome-wide association study
of prostate cancer identifies novel risk variants and substantial
ethnic differences. Cancer Discov. 5:878–891. 2015. View Article : Google Scholar : PubMed/NCBI
|