Introduction
Cervical cancer is a preventable disease, and
reductions in its incidence and mortality have been achieved
(1); however, it remains the
second common malignancy in women worldwide (2). Research into the molecular mechanisms
associated with the pathogenesis of cervical cancer is important
for its treatment and prevention (3).
microRNAs (miRNAs) are a class of short, non-coding
RNAs ~22 nucleotides long that regulate gene expression by binding
to the 3′ untranslated regions (3′-UTRs) of target mRNAs in a
sequence-specific manner, resulting in translational repression
and/or gene silencing (4,5). A number of studies have demonstrated
that miRNAs, functioning as either onco-miRNAs or tumor
suppressors, perform important roles during cancer progression
(6–8). At present, >2,500 miRNAs have been
identified in humans (miRBase database version 20.0) (9). However, few studies have investigated
the association between miRNA and the tumorigenesis of cervical
cancer (10). miRNA (miR)-491-5p,
which is a mature form of miR-491, functions as a tumor suppressor
gene in vitro, as it induces the apoptosis and inhibits the
proliferation of ovarian (11),
colorectal (12), pancreatic
(13) and breast (14) cancer cells. In addition, miR-491-5p
inhibits the invasion of glioma (15), breast (16) and oral squamous (17) cancer cells. However, the role of
miR-491 in cervical cancer cells remains unknown.
Jumonji domain containing 2A (JMJD2A), a member of
the JmjC domain-containing family of JMJD2 proteins
(JMJD2A-JMJD2D), recognizes di- and tri-methylated histone H3
lysine 9 (H3K9) and H3K36, and trimethylated H1.4K26 as substrates
(18). This leads to the promotion
of an open chromatin state and contributes to transcriptional
activation and the regulation of cancer-associated genes, including
those involved in the cell cycle, cell proliferation, apoptosis,
invasion and metastasis (19).
JMJD2A is involved in several types of cancer, including ductal
carcinoma, lung, breast, ovarian, bladder and colon cancer, renal
adenocarcinoma, and head and neck squamous cell carcinoma (20–24).
It has been demonstrated that miR-491-5p exerts inhibitory effects
on breast cancer cell growth by directly targeting the 3′UTR of
JMJD2B mRNA and blocking the estrogen receptor (ER)α-mediated
signaling pathway (25).
Additionally, a previous study indicated that JMJD2A may contribute
to breast tumor formation by stimulating ERα activity (26). These results suggest that JMJD2A
may exhibit its oncogenic function by regulating the expression of
miR-491-5p.
Thus, the aim of the present study was to
investigate the function of JMJD2A in human cervical cancer and
determine whether its role is dependent on miR-491-5p. The
expression of JMJD2A in human cervical cancer cell lines was
evaluated to determine whether it is an oncogenic protein.
Additionally, the association of JMJD2A levels with overall and
disease-free survival rates and the potential of JMJD2A as an
independent prognostic factor for adverse outcomes were
investigated. Furthermore, the effects of JMJD2A on cervical cancer
cell growth and apoptosis were evaluated.
Materials and methods
Human specimens
A total of 38 primary cervical epithelial carcinoma
tissues were collected from patients (n=38; mean age of 50±0.7
years old). Normal cervical tissues were collected from patients
who underwent hysterectomy as a result of benign gynecological
diseases (n=20; mean age of 46±0.9 years old). Specimens were
obtained from patients admitted to the Banan People's Hospital of
Chongqing (Chongqing, China) between November 2014 and July 2016.
Informed consent was obtained, and the present study was approved
by the Ethics Committee of the Banan People's Hospital of
Chongqing. Tissue samples were immediately snap-frozen in liquid
nitrogen and stored at −80°C prior to use. The mean JMJD2A level of
the cervical cancer tissues was evaluated using western blotting as
later detailed. Tissues exhibiting lower JMJD2A expression compared
with the mean were classified as the JMJD2A low group, while
tissues exhibiting higher JMJD2A expression compared with the mean
were classified as the JMJD2A high group. The mean miR-491-5p level
of the cervical cancer tissues was evaluated using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) as
later detailed. Tissues in which miR-491-5p levels were lower than
the mean level were classified as the miR-491-5p low group, while
tissues with higher miR-491-5p levels compared with the mean level
were classified as the miR-491-5p high group.
Cell culture and oligonucleotide
transfection
The human cervical cancer cell lines HeLa, CaSki,
C-4-I, SiHa and C-33 A were obtained from the Cell Bank of
Shanghai, Institute of Biochemistry and Cell Biology, Chinese
Academy of Sciences (Shanghai, China). Human cervical cancer cells
were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco,
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.)
under an atmosphere of 5% CO2 at 37°C. Normal human
cervical cells were grown in a 100-mm plastic dish and cultured
under the aforementioned conditions. All procedures involving
clinical specimens were approved by the Banan People's Hospital of
Chongqing.
The locked-nucleic-acid-modified oligonucleotide
(LNA) of miR-491-5p (LNA-miR-491-5p) and negative control (LNA-NC)
were synthesized and transfected into cervical cancer cells, as
previously described (27).
Cell viability assay
The viability of cervical cancer cells was
determined using an MTS kit (CellTiter 96 AQ; Promega Corporation,
Madison, WI, USA). Briefly, cells (5×103 cells/well)
were seeded in 96-well plates. After 12 h, a fresh mixture of MTS
and phenazine methosulfate was added and the cells were incubated
for 2–4 h at 37°C. An MR7000 microplate reader (Dynatech
International, LLC, Melville, NY, USA) was used to measure the
absorbance at 490 nm.
Colony formation assay
Cells were trypsinized and plated on 6-well plates
and cultured for 2 weeks under an atmosphere of 5% CO2
at 37°C. Following fixation with 4% paraformaldehyde for 30 min at
room temperature, the colonies were stained with 1% crystal violet
for 30 sec at room temperature. The number of colonies, defined as
>50 cells/colony, were counted from nine random visual fields
using an inverted microscope (magnification, ×200; IX83; Olympus
Corporation, Tokyo, Japan).
Apoptosis analysis
Apoptosis was detected using an Annexin V-FITC/PI
double staining kit (Beyotime Institute of Biotechnology, Jiangsu,
China) according to the manufacturer's protocol. Cells were
harvested and stained with Annexin V-FITC and PI. Cell samples were
analyzed using a flow cytometer (FACScan; BD Biosciences, Franklin
Lakes, NJ, USA). Data were analysed using FlowJo software version
10.0.5 for Microsoft (Tree Star, Inc., Ashland, OR, USA).
Vector construction and
transfection
Adenovirus-based JMJD2A overexpression (Ad-JMJD2A)
was performed as described previously (28). Briefly, cells (2×104)
were infected with Ad-JMJD2A (1×1010 PFU/ml; 1 µl) and
Ad-negative control (Ad-NC; 1 µl) using LipoFiter™ reagent (Hanbio,
Shanghai, China). HEK293 cells were used to generate the
third-generation adenovirus. For deletion of JMJD2A, short hairpin
(sh) RNA (sh-JMJD2A) sequences were adopted (Ambion; Thermo Fisher
Scientific, Inc.). In addition, cells (2×105) were
transfected with sh-JMJD2A (6 µl) or sh-negative control (sh-NC; 6
µl) using HiPerFect Transfection Reagent (6 µl; Qiagen GmbH,
Hilden, Germany). The cells were harvested 48 h post-transfection
and the RNA was then extracted for RT-qPCR analysis to determine
the overexpression or knockdown efficiency.
RNA isolation and RT-qPCR
Post-transfection, total RNA (tRNA) was extracted
from human cervical cancer cells using TRIzol® (Thermo
Fisher Scientific, Inc.), following the manufacturer's protocol.
Following spectrophotometric measurement (260 nm) of tRNA
concentration using an UV spectrophotometer and subsequent analysis
of the quality of the tRNA using 2% agarose gels stained with
ethidium bromide at room temperature for 30 min, 3 mg tRNA was
subjected to reverse transcription (RT) using M-MLV reverse
transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) at 25°C
for 10 min, 37°C for 100 min and 90°C for 5 sec and 4°C for 5 min.
RT-qPCR was performed using the miScript SYBR-Green PCR kit (Qiagen
GmbH) on an ABI 7900 Real-time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The sequences of PCR primers used
were as follows: JMJD2A forward, 5′-ATCCCAGTGCTAGGATAATGACC-3′ and
reverse, 5′-ACTCTTTTGGAGGAACCCTTG-3′; miR-491-5p forward,
5′-ACACTCCAGCTGGGAGTGGGGAACCCTTC-3′ and reverse,
5′-TGGTGTCGTGGAGTCG-3′; GAPDH forward, 5′-TGACGCTGGGGCTGGCATTG-3′
and reverse, 5′-GCTCTTGCTGGGGCTGGTGG-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The thermocycling conditions used were as follows: 95°C for 15 min;
followed by 40 cycles of 94°C for 15 sec, 55°C for 30 sec and 70°C
for 30 sec. All assays were performed in triplicate. Quantitative
analysis of the relative expression of RNA was calculated using the
2−ΔΔCq method (29).
The mRNA expression values of JMJD2A and miR-491-5p were normalized
to the internal controls GAPDH and U6, respectively.
Western blot analysis
Following transfection, protein was isolated from
human cervical cancer cells and cervical cancer tissues using a
radioimmunoprecipitation assay lysis buffer kit (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at 4°C for 30 min. Protein
concentrations were quantified using a Bio-Rad protein assay
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Proteins (20
µg/lane) were separated by 10–12% SDS-PAGE. The separated proteins
were then transferred onto a PVDF membrane (EMD Millipore,
Billerica, MA, USA) at 250 mA for 2 h. The membranes were blocked
with 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) for 1 h at room temperature and then incubated with
primary antibody at 4°C overnight: Anti-JMJD2A (cat. no. ab105953;
1:1,000), anti-Bcl-2 (cat. no. ab32124; 1:1,000), anti-Bax (cat.
no. ab32503; 1:1,000), anti-p21 (cat. no. ab109520; 1:1,000),
anti-caspase-3 (cat. no. ab4051; 1:500), active-caspase-3 (cat. no.
ab13847; 1:500), and anti-GAPDH (cat. no. ab9483; 1:1,000; all
purchased from Abcam, Cambridge, MA, USA). Following 5 washes with
TBST, the membranes were incubated with horseradish
peroxidase-conjugated goat anti-rabbit immunoglobulin G (cat. no.
A50-106P; 1:1,000; Origene Technologies, Inc, Beijing, China) for 1
h at room temperature. Each band was visualized using an enhanced
chemiluminescence kit (EMD Millipore). Quantitiative analysis was
performed using Alpha View Analysis Tools (AlphaViewSA software
version 3.2.2; ProteinSimple, San Jose, CA, USA).
Mouse xenograft experiment
Mouse xenograft experiments were performed as
described previously (30). A
total of 40 female athymic nude (nu/nu) mice (3–4 weeks old;
weighing 14–16 g) were purchased from Shanghai SLAC Laboratory
Animal Co., Ltd. (Shanghai, China). The mice were bred and
maintained under specific pathogen-free conditions. Animals were
housed under a controlled temperature (25±1°C) and humidity (50%),
with a 12/12 h light/dark cycle and free access to food and water.
HeLa, c-4-1 or SiHa cells (5×106) transfected with
Ad-JMJD2A, Sh-JMJD2A or the corresponding controls (Ad-NC and
Sh-NC) were inoculated subcutaneously into the left and right
flanks of the mice (n=10/group). The mice were sacrificed and the
tumors were weighed 3 weeks after inoculation. The weight of the
animals at the time of sacrifice was 12–18 g and there was no
significant change in the weight of each animal over the
experimental period. Tumor volumes were calculated using the
following formula: π/6 × a2 × b, where a is the short
axis and b is the long axis. The largest subcutaneous tumor
detected in the present study had a diameter of 1.9 cm. No mouse
exhibited multiple subcutaneous tumors. Notably, discomfort was
considered a humane endpoints and euthanasia by isoflurane
inhalation was applied to immediately sacrifice those animals
exhibiting such symptoms. The protocol was approved by the Ethics
Committee of Banan People's Hospital of Chongqing.
Statistical analysis
Statistical analysis was performed using SPSS 13.0
(SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ±
standard error of the mean. One-way analysis of variance followed
by Dunnett's post hoc test was used to analyze differences among
multiple groups. The association between JMJD2A levels and
miR-491-5p was tested using Spearman rank correlation. Survival
rate analysis was performed using the Kaplan-Meier method, and
differences between the groups were tested using the log-rank
test.
Results
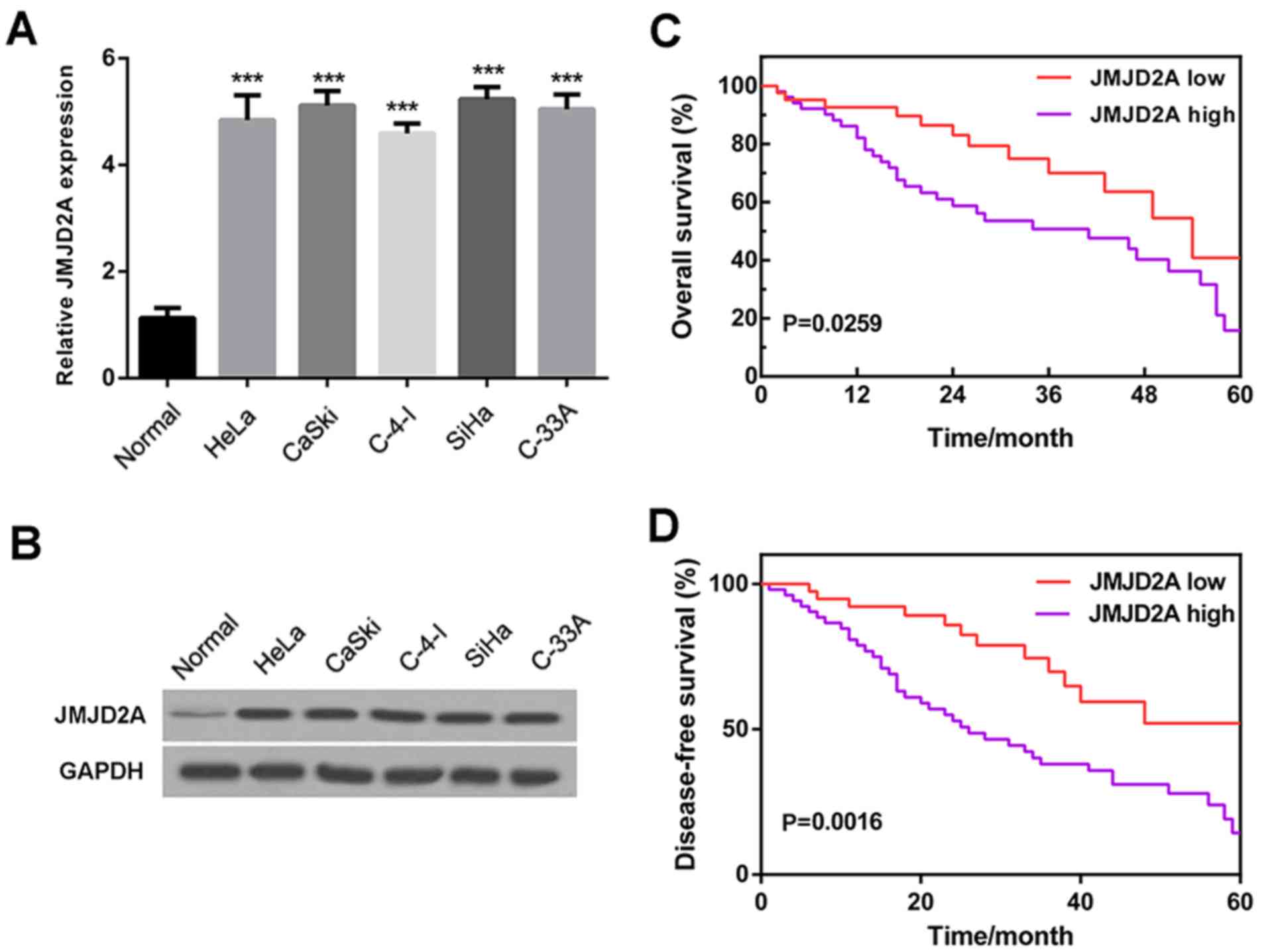
JMJD2A is highly expressed in cervical
cancer cells and strongly associated with survival rate
To investigate the role of JMJD2A in cervical
cancer, the expression of JMJD2A in five cervical cancer cell lines
(HeLa, CaSki, C-4-I, SiHa and C-33 A) was examined. The reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
results (Fig. 1A) and western blot
analysis (Fig. 1B) revealed that
JMJD2A mRNA and protein levels were clearly higher in cervical
cancer cell lines compared with normal cervical cells, suggesting
that JMJD2A is highly expressed in human cervical cancer.
To further investigate the association of JMJD2A
with cervical cancer, the 60-month overall survival rates and
disease-free survival rates of patients with low (lower than the
mean level) and high (higher than the mean level) JMJD2A expression
levels were analyzed. The results demonstrated that a low JMJD2A
level was associated with high overall and disease-free survival
rates, while a high JMJD2A level was associated with poor overall
and disease-free survival rates (Fig.
1C and D). These observations indicate that JMJD2A has an
association with survival rate and may potentially serve as an
independent prognostic factor.
JMJD2A overexpression induces the
growth of cervical cancer cells
To explore whether JMJD2A participates in the growth
and apoptosis of cervical cancer cells, JMJD2A was knocked down or
overexpressed in the human cervical cancer cell lines HeLa, C-4-I,
and SiHa. The knockdown and overexpression efficiencies were
confirmed by RT-qPCR (Fig. 2A).
Next, the cell viability at different time-points after
transfection (0, 24, 48 and 72 h) was determined. The results
demonstrated that JMJD2A knockdown (sh-JMJD2A) significantly
reduced the number of surviving cells, and the inhibition of cell
growth appeared to be time-dependent. In addition, JMJD2A knockdown
significantly decreased the number of colonies in vitro. In
the mouse xenograft model, tumor weights for mice implanted with
the sh-JMJD2A HeLa, C-4-I and SiHa cells were significantly reduced
compared with those in the respective sh-NC group (Fig. 2B-F). Conversely, JMJD2A
overexpression (Ad-JMJD2A) increased the proliferation and colony
formation of cervical cancer cells in vitro, and increased
the weight of the xenograft tumors compared with those in the
respective control group (Ad-NC). These findings indicate that
JMJD2A regulates cervical cancer growth in vitro.
 | Figure 2.JMJD2A overexpression induces growth
and inhibits apoptosis of cervical cancer cells. (A) The knockdown
and overexpression efficiencies of Sh-JMJD2A and Ad-JMJD2A were
confirmed by reverse transcription-quantitative polymerase chain
reaction. JMJD2A overexpression increased the number of (B) HeLa,
(C) C-4-I and (D) SiHa cells, and MJD2A knockdown demonstrated the
opposite effect. The cell numbers were evaluated at 0, 24, 48 and
72 h post-transfection. (E) JMJD2A overexpression increased the
colony number of HeLa, C-4-I and SiHa cells, and JMJD2A knockdown
demonstrated the opposite effect. The number of colonies was
counted at 2 weeks after transfection. (F) JMJD2A overexpression
increased the weights of HeLa, C-4-I and SiHa cell-derived tumors,
and JMJD2A overexpression demonstrated the opposite effect. Tumor
weight was evaluated 3 weeks after the inoculation of the cervical
cancer cells into female athymic nude (nu/nu) mice. (G) JMJD2A
overexpression inhibited cellular apoptosis in HeLa, C-4-I, and
SiHa cells, and JMJD2A overexpression demonstrated the opposite
effect. Data are presented as the mean ± standard error of the
mean. *P<0.05 and ***P<0.001 vs. Ad-NC; #P<0.05,
##P<0.01 and ###P<0.001 vs. sh-NC. JMJD2A, jumonji domain
containing 2A; sh, short hairpin; Ad, adenovirus; NC, negative
control. |
JMJD2A overexpression inhibits
apoptosis of cervical cancer cells
Flow cytometric analysis (Fig. 2G) revealed that JMJD2A knockdown
induced cellular apoptosis in HeLa, C-4-I and SiHa cells, whereas
JMJD2A overexpression significantly attenuated cellular apoptosis
in these cell lines, resulting in a markedly low apoptotic rate.
The results of western blot analysis (Fig. 3A) demonstrated that JMJD2A
knockdown upregulated the expression of pro-apoptotic proteins
(Bax, p21 and active caspase-3) and downregulated the level of the
anti-apoptotic protein Bcl-2. However, it exhibited no clear effect
on the expression of the apoptosis-related protein pro
caspase-3.
 | Figure 3.JMJD2A knockdown induces miR-491-5p
expression in cervical cancer cells. (A) Western blotting
demonstrates that JMJD2A knockdown induced the expression of
pro-apoptotic proteins (active caspase-3, Bax and p21) and
inhibited the expression of the anti-apoptotic protein Bcl-2 in
HeLa cells. (B) miR-491-5p mRNA levels are downregulated in HeLa,
CaSki, C-4-I, SiHa and C-33 A human cervical cancer cell lines.
***P<0.001 vs. normal. (C) The miR-491-5p and JMJD2A levels were
negatively correlated in the human cervical cancer specimens
(n=38). (D) JMJD2A knockdown significantly increased the level of
miR-491-5p, and JMJD2A overexpression significantly decreased the
level of miR-491-5p in cervical cancer cell lines. ***P<0.001
vs. Ad-NC; ###P<0.001 vs. sh-NC. (E) Kaplan-Meier survival rate
analysis demonstrates that the overall survival rate is poor for
cervical cancer patients with low miR-491-5p expression. The mean
miR-491-5p level of the cervical cancer tissues was evaluated using
reverse transcription-quantitative polymerase chain reaction. The
cases whose miR-491-5p level was lower than the mean miR-491-5p
level were enrolled to the miR-491-5p low group, while the others
were enrolled in the miR-491-5p high group. (F) miR-491-5p levels
following the transfection of LNA-miR-491-5p and LNA-NC into
sh-JMJD2A and sh-NC cells. ***P<0.001 vs. LNA-NC + Sh-NC;
###P<0.001 vs. LNA-miR-491-5p + Sh-JMJD2A. JMJD2A, jumonji
domain containing 2A; sh, short hairpin; Ad, adenovirus; NC,
negative control; Bax, Bcl-2-like protein 4; Bcl-2, B-cell
lymphoma. |
JMJD2A knockdown induces miR-491-5p
expression in cervical cancer cells
miR-491-5p has been reported to act as a tumor
suppressor and is downregulated in a number of cancer cell lines
(31–33). The results of the present study
demonstrated that miR-491-5p was downregulated in the human
cervical cancer lines HeLa, CaSki, C-4-I, SiHa and C-33 A (Fig. 3B). To explore the correlation
between JMJD2A and miR-491-5p levels in the cervical cancer
tissues, linear regression analysis was performed. The data
demonstrated that the miR-491-5p and JMJD2A levels were
significantly and negatively correlated (Fig. 3C). In addition, when JMJD2A was
knocked down or overexpressed in cervical cancer cells, significant
inductive and suppressive effects on miR-491-5p expression were
observed, respectively (Fig. 3D).
The miR-491-5p mRNA levels of the cervical cancer tissues were used
to divide the patients into miR-491-5p low and miR-491-5p high
expression groups. Kaplan-Meier curves were plotted for analysis of
the overall survival rates (Fig.
3E) and indicated that a low miR-491-5p level predicted a poor
overall survival rate. Together, these results suggest that
miR-491-5p expression is downregulated in cervical cancer and a low
miR-491-5p expression level is predictive of a poor survival
rate.
miR-491-5p knockdown reverses the
effects of sh-JMJD2A on cervical cancer growth
In order to examine the potential regulative
mechanism, LNA-miR-491-5p and LNA-NC were respectively transfected
into sh-JMJD2A and sh-NC cells. The miR-491-5p levels of the
transfected were validated using RT-qPCR (Fig. 3F).
The cell number at the 0, 24, 48 and 72 h
time-points, the colony number and apoptotic cell rates were
detected in the transfected HeLa, C-4-I and SiHa cell lines. The
results (Fig. 4) demonstrated that
following the knockdown of miR-491-5p (sh-NC + LNA-miR-491-5p), the
cell and colony numbers were significantly higher, and the
apoptotic rate was significantly lower than those in the control
(sh-NC + LNA-NC). In addition, JMJD2A knockdown (sh-JMJD2A +
LNA-NC) significantly decreased the cell and colony numbers, and
induced cell apoptosis in comparison with the control (sh-NC +
LNA-NC). In the cells with knockdown of miR-491-5p and JMJD2A
(sh-JMJD2A + LNA-miR-491-5p), the effects of JMJD2A knockdown were
significantly attenuated, as evidenced by the increase of cell and
colony numbers and the reduction of cell apoptotic rate compared
with those in the sh-JMJD2A + LNA-NC group, and the results were
comparable to those in the control group (sh-NC + LNA-NC).
Discussion
Although the functions of JMJD2A in various types of
cancer have been demonstrated previously, its role in human
cervical cancer remains unclear. The present study identified that
JMJD2A is overexpressed in human cervical cancer, which is
consistent with a previous study reporting that JMJD2A is
overexpressed in human breast, lung, head and neck, uterine,
endometrial and ovarian cancer, and stomach and renal
adenocarcinoma (34). In addition,
log-rank analysis in the present study indicated that high JMJD2A
expression predicts poor overall and disease-free survival rates.
These results suggest that JMJD2A is overexpressed in human
cervical cancer and may be an independent prognostic factor.
The potential mechanisms of JMJD2A in various types
of cancer have been investigated in previous studies. In one study,
the overexpression of JMJD2A was detected in human breast cancer
cells, and it was observed that JMJD2A forms a complex with
estrogen receptor (ER)α in vivo, and the downregulation of
JMJD2A decreased the expression of cyclin D1, a prominent ERα
target gene and cell cycle regulator (26). Another study demonstrated that
JMJD2A promotes cellular transformation by blocking cellular
senescence through transcriptional repression of the tumor
suppressor CHD5 (23).
Furthermore, JMJD2A has been reported to be involved in human
carcinogenesis through regulation of the G1/S transition in human
bladder and lung cancers (22).
Another study suggested that the JMJD2A level correlates with the
level of the pro-apoptotic microRNA miR-34a in gastric cancer
tissues and JMJD2A represses the expression of miR-34a by
decreasing its promoter activity (35).
The potential mechanism underlying the roles of
JMJD2A in human cervical cancer was investigated in the present
study. JMJD2A was found to regulate cervical cancer cell growth and
apoptosis; more specifically, JMJD2A deficiency induced cervical
cancer cell apoptosis by upregulating pro-apoptotic proteins (Bax,
p21 and active caspase-3) and downregulating the anti-apoptotic
protein Bcl-2. In addition, miR-491-5p, which is reported to act as
tumor suppressor in a number of different cancers, was demonstrated
to be negatively correlated with the expression of JMJD2A in
cervical cancer. The present study also revealed that JMJD2A
knockdown significantly upregulated the expression of miR-491-5p.
The significant inhibition of cell growth and increased apoptosis
of cervical cancer cells induced by sh-JMJD2A were markedly
reversed by the suppression of miR-491-5p. Together, these results
indicate that JMJD2A participates in the proliferation and
apoptosis of human cervical cancer cells and its role is partly
mediated via the regulation of miR-491-5p. However, the mechanism
underlying the effects on the regulation of miR-491-5p expression
requires further exploration.
The mechanism by which miR-491-5p acts in tumor
suppression was not fully investigated in the present study.
Previous studies found that miR-491-5p suppressed the growth of
oral squamous cell carcinoma, pancreatic cancer and glioblastoma by
targeting GIT1, EGFR and CDK6 genes (14,17,36).
Another study indicated that miR-491-5p targeting TP53 and
Bcl-XL induced cell apoptosis in SW1990 pancreatic
cancer cells through a mitochondria-mediated pathway (13). However, the genes that miR-491-5p
targets in human cervical cancer have not yet been identified.
In summary, JMJD2A was identified as an oncogenic
protein in human cervical cancer. Its effects on cell and colony
numbers, tumor weight and apoptosis may be mediated by the
downregulation of miR-491-5p, which serves a role as an
anti-onco-miRNA in cervical cancer. Therefore, JMJD2A could serve
as an independent prognostic factor and potential target for
intervention in cervical cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL performed the experiments; YW designed the study
and reviewed the manuscript; ZX analysed and interpreted the data;
HH acquired the data, and drafted and edited the manuscript.
Ethics approval and consent to
participate
Patient specimens were obtained with informed
consent and the present study was approved by the Ethics Committee
of the Banan People's Hospital of Chongqing.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Comprehensive cervical cancer control, . A
guide to essential practice. World Health Organization. (Geneva,
Switzerland). 2006.
|
|
3
|
Burd EM: Human papillomavirus and cervical
cancer. Clin Microbiol Rev. 16:1–17. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Calin GA and Croce CM: MicroRNA-cancer
connection: The beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu T, Liu K, Wu Y, Fan J, Chen ZJ, Li C,
Yang Q and Wang Z: MicroRNA-9 inhibits the proliferation of oral
squamous cell carcinoma cells by suppressing expression of CXCR4
via the Wnt/β-catenin signaling pathway. Oncogene. 33:5017–5027.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cho WC, Chow AS and Au JS: Restoration of
tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung
adenocarcinoma patients with epidermal growth factor receptor
mutation. Eur J Cancer. 45:2197–2206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tang T, Wong HK, Gu W, Yu MY, To KF, Wang
CC, Wong YF, Cheung TH, Chung TK and Choy KW: MicroRNA-182 plays an
onco-miRNA role in cervical cancer. Gynecol Oncol. 129:199–208.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Denoyelle C, Lambert B, Meryet-Figuière M,
Vigneron N, Brotin E, Lecerf C, Abeilard E, Giffard F, Louis MH,
Gauduchon P, et al: miR-491-5p-induced apoptosis in ovarian
carcinoma depends on the direct inhibition of both BCL-XL and EGFR
leading to BIM activation. Cell Death Dis. 5:e14452014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nakano H, Miyazawa T, Kinoshita K, Yamada
Y and Yoshida T: Functional screening identifies a microRNA,
miR-491 that induces apoptosis by targeting Bcl-X(L) in colorectal
cancer cells. Int J Cancer. 127:1072–1080. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo R, Wang Y, Shi WY, Liu B, Hou SQ and
Liu L: MicroRNA miR-491-5p targeting both TP53 and Bcl-XL induces
cell apoptosis in SW1990 pancreatic cancer cells through
mitochondria mediated pathway. Molecules. 17:14733–14747. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Leivonen SK, Sahlberg KK, Mäkelä R, Due
EU, Kallioniemi O, Børresen-Dale AL and Perälä M: High-throughput
screens identify microRNAs essential for HER2 positive breast
cancer cell growth. Mol Oncol. 8:93–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yan W, Zhang W, Sun L, Liu Y, You G, Wang
Y, Kang C, You Y and Jiang T: Identification of MMP-9 specific
microRNA expression profile as potential targets of anti-invasion
therapy in glioblastoma multiforme. Brain Res. 1411:108–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rutnam ZJ and Yang BB: The non-coding 3′
UTR of CD44 induces metastasis by regulating extracellular matrix
functions. J Cell Sci. 125:2075–2085. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang WC, Chan SH, Jang TH, Chang JW, Ko
YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, et al:
miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral
squamous cell carcinoma invasion and metastasis. Cancer Res.
74:751–764. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Berry WL and Janknecht R: KDM4/JMJD2
histone demethylases: Epigenetic regulators in cancer cells. Cancer
Res. 73:2936–2942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qi H, Jing Z, Xiaolin W, Changwu X,
Xiaorong H, Jian Y, Jing C and Hong J: Histone demethylase JMJD2A
inhibition attenuates neointimal hyperplasia in the carotid
arteries of balloon-injured diabetic rats via transcriptional
silencing: Inflammatory gene expression in vascular smooth muscle
cells. Cell Physiol Biochem. 37:719–734. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li BX, Li J, Luo CL, Zhang MC, Li H, Li
LL, Xu HF, Shen YW, Xue AM and Zhao ZQ: Expression of JMJD2A in
infiltrating duct carcinoma was markedly higher than fibroadenoma,
and associated with expression of ARHI, p53 and ER in infiltrating
duct carcinoma. Indian J Exp Biol. 51:208–217. 2013.PubMed/NCBI
|
|
21
|
Li BX, Zhang MC, Luo CL, Yang P, Li H, Xu
HM, Xu HF, Shen YW, Xue AM and Zhao ZQ: Effects of RNA
interference-mediated gene silencing of JMJD2A on human breast
cancer cell line MDA-MB-231 in vitro. J Exp Clin Cancer Res.
30:902011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kogure M, Takawa M, Cho HS, Toyokawa G,
Hayashi K, Tsunoda T, Kobayashi T, Daigo Y, Sugiyama M, Atomi Y, et
al: Deregulation of the histone demethylase JMJD2A is involved in
human carcinogenesis through regulation of the G(1)/S transition.
Cancer Lett. 336:76–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mallette FA and Richard S: JMJD2A promotes
cellular transformation by blocking cellular senescence through
transcriptional repression of the tumor suppressor CHD5. Cell Rep.
2:1233–1243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kauffman EC, Robinson BD, Downes MJ,
Powell LG, Lee MM, Scherr DS, Gudas LJ and Mongan NP: Role of
androgen receptor and associated lysine-demethylase coregulators,
LSD1 and JMJD2A, in localized and advanced human bladder cancer.
Mol Carcinog. 50:931–944. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hui Z, Yiling C, Wenting Y, XuQun H,
ChuanYi Z and Hui L: miR-491-5p functions as a tumor suppressor by
targeting JMJD2B in ERα-positive breast cancer. FEBS Lett.
589:812–821. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Berry WL, Shin S, Lightfoot SA and
Janknecht R: Oncogenic features of the JMJD2A histone demethylase
in breast cancer. Int J Oncol. 41:1701–1706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rajwanshi VK, Håkansson AE, Sørensen MD,
Pitsch S, Singh SK, Kumar R, Nielsen P and Wengel J: The eight
stereoisomers of LNA (Locked Nucleic Acid): A remarkable family of
strong RNA binding molecules we acknowledge the Danish Natural
Science Research Council, the Danish Technical Research Council,
and Exiqon A/S for financial support. Ms Britta M. Dahl is thanked
for oligonucleotide synthesis, Dr. Carl E. Olsen for MALDI-MS
analysis, and Ms. Karen Jørgensen for recording CD spectra. Angew
Chem Int Ed Engl. 39:1656–1659. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shinoura N, Muramatsu Y, Nishimura M,
Yoshida Y, Saito A, Yokoyama T, Furukawa T, Horii A, Hashimoto M,
Asai A, et al: Adenovirus-mediated transfer of p33ING1 with p53
drastically augments apoptosis in gliomas. Cancer Res.
59:5521–5528. 1999.PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y,
Yao H, Liu X, Ke Y, Si J and Zhou T: MiR-375 frequently
downregulated in gastric cancer inhibits cell proliferation by
targeting JAK2. Cell Res. 20:784–793. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gong F, Ren P, Zhang Y, Jiang J and Zhang
H: MicroRNAs-491-5p suppresses cell proliferation and invasion by
inhibiting IGF2BP1 in non-small cell lung cancer. Am J Transl Res.
8:485–495. 2016.PubMed/NCBI
|
|
32
|
Xu Y, Hou R, Lu Q, Zhang Y, Chen L, Zheng
Y and Hu B: MiR-491-5p negatively regulates cell proliferation and
motility by targeting PDGFRA in prostate cancer. Am J Cancer Res.
7:2545–2553. 2017.PubMed/NCBI
|
|
33
|
Sun R, Liu Z, Tong D, Yang Y, Guo B, Wang
X, Zhao L and Huang C: miR-491-5p, mediated by Foxi1, functions as
a tumor suppressor by targeting Wnt3a/β-catenin signaling in the
development of gastric cancer. Cell Death Dis. 8:e27142017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Black JC, Manning AL, Van Rechem C, Kim J,
Ladd B, Cho J, Pineda CM, Murphy N, Daniels DL, Montagna C, et al:
KDM4A lysine demethylase induces site-specific copy gain and
rereplication of regions amplified in tumors. Cell. 154:541–555.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu CE, Liu YC, Zhang HD and Huang GJ:
JMJD2A predicts prognosis and regulates cell growth in human
gastric cancer. Biochem Biophys Res Commun. 449:1–7. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li X, Liu Y, Granberg KJ, Wang Q, Moore
LM, Ji P, Gumin J, Sulman EP, Calin GA, Haapasalo H, et al: Two
mature products of MIR-491 coordinate to suppress key cancer
hallmarks in glioblastoma. Oncogene. 34:1619–1628. 2015. View Article : Google Scholar : PubMed/NCBI
|