Introduction
Allergic asthma is a chronic airway disorder
characterized by symptoms that include wheeze, cough, shortness of
breath and chest tightness (1,2). An
estimated 300,000,000 individuals worldwide suffer from asthma
which imposes a substantial economic burden on individuals and
society (3). Cumulative evidence
from human and animal model systems suggest that asthma is
associated with polarized T-helper (Th)2 responses to inhaled
allergens (4,5). Th2 cells secrete cytokines, including
interleukin (IL)-4, IL-5 and IL-13, which induce isotype switching
to immunoglobulin E (IgE) by B lymphocytes. The cross-linking of
allergen-specific IgE bound to the receptor on the surface of mast
cells and basophils results in the release of histamine and
leukotriene, and leads to bronchoconstriction and vasodilation. The
earlier contact between an allergen and innate immune cells,
including dendritic cells (DCs), natural killer (NK) cells and
bronchial epithelial cells, is thought to direct the subsequent
antigen-specific T-cell response (6,7).
Inhaled allergens activate the Toll-like receptor (TLR) class of
pattern recognition receptors involved in innate immunity and
induce the synthesis of innate cytokines for initiating the
development of Th2 adaptive responses (8,9).
NK cells are critical in innate immune responses and
make up 10% of resident lymphocytes in the lungs of humans and mice
(10). Evidence has emerged that
NK cells are important in various lung diseases. NK cells are best
noted for their ability to produce interferon (IFN)-γ, however,
they also produce a number of other cytokines, including tumor
necrosis factor (TNF)-α, granulocyte-macrophage colony-stimulating
factor, IL-5, IL-13 and C-C motif chemokine ligand 5/RANTES
(11). Human NK cells can be
divided into four subsets based on the relative expression of
markers CD16 and CD56. The CD16+ CD56− NK
subset, the majority of which is in the peripheral blood, is potent
cytotoxic cells, and the CD16− CD56+ NK
subset is found mostly in the lymphoid organs and produces high
levels of cytokines (12,13). In mice, CD11b and CD27 reflect
distinct population and functional specialization in human NK
cells. The CD11b− CD27+ NK subset, which is
typically observed in the bone marrow, is a potent producer of
cytokines and is less cytotoxic than the CD11b+
CD27− NK subset that resides primarily in the spleen,
peripheral blood and lungs (14,15).
The present study examined the dynamic behavior of
NK cells and their subsets during the development of asthma using
an ovalbumin (OVA)-induced mouse model. The percentage of NK cells
in the lung was decreased following OVA sensitization and
challenge. However, the NK cells showed a high level of activation
and secreted more Th2 cytokines than Th1 cytokines. In addition, an
increased proportion of CD11b− NK cells was observed in
the process of OVA induction. CD11b− CD27− NK
cells were the primary NK subset producing Th2 cytokines. Together,
these findings provide novel insights for understanding the
mechanism of asthma.
Materials and methods
Mice
Female BALB/c mice (6–8 weeks old; 18–20 g; n=6 each
group) were purchased from Guangdong Medical Laboratory Animal
Center, Foshan (Guangdong, China). The mice were housed in a 12 h
light/dark cycle at 22°C under specific pathogen-free conditions
and allowed access to food and water ad libitum. The animal
experiments were performed according to the Institutional
Guidelines for the Animal Care and Use Committee of Shenzhen
University (Shenzhen, China).
OVA-induced allergic asthma model
The mice were sensitized with an intraperitoneal
(i.p.) injection of 10 µg OVA (Grade V; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) emulsified in aluminium hydroxide gel
(InvivoGen, San Diego, CA, USA) on days 0 and 7. On days 14–17 the
animals were challenged with 6% OVA aerosol. The aerosol
administrations were performed for 25 min using an ultrasonic
nebulizer (Leyi Industry Co., Ltd., Shanghai, China). Animals in
the control group received an i.p injection of PBS emulsified in
100 µl of aluminium hydroxide gel and were challenged with PBS
aerosols. All mice were sacrificed 24 h following the final OVA
challenge for subsequent assays (Fig.
1).
Bronchoalveolar lavage fluid (BALF)
collection
Tracheotomy was performed, and a cannula was
inserted into the trachea. PBS (0.6 ml ×2) was instilled into the
left lungs following ligation of the right lungs, and BALF was
collected. The cells in the BALF were centrifuged at 300 × g at 4°C
for 10 min in a cytocentrifuge and then stained with Wright-Giemsa
(Quick Wright-Giemsa Stain, Jiancheng Bioengineering Institute,
Nanjing, China). The cells were identified as macrophages,
eosinophils, neutrophils and lymphocytes under magnification, ×200
using a Leica DM LB microscope (Leica Microsystems, Inc., Buffalo
Grove, IL, USA).
Blood collection
Blood samples were obtained from the retro-orbital
venous plexus. The blood was stored at room temperature for 1 h,
then centrifuged at 1,500 × g at room temperature for 15 min. The
serum was collected for IgE and OVA-specific IgE assays.
Lung histology
The right lung tissues were fixed in 4%
paraformaldehyde, paraffinized, cut into 4-µm sections, and then
stained with hematoxylin and eosin for examining cell infiltration
and with periodic acid-schiff for the detection of mucin in goblet
cells (mucus-secreting cells) by light microscopy.
ELISA
The quantitation of cytokines IL-4, IL-5 and IL-13
in the BALF and the serum levels of IgE were measured by sandwich
ELISA using paired antibodies (BD Biosciences, San Jose, CA, USA).
The serum levels of OVA-IgE were measured using a mouse
OVA-specific IgE ELISA kit (Cusabio Biotech, Wuhan, China). The
sensitivity of detection was 4 pg/ml for IL-4, IL-5 and IL-13, and
5 ng/ml for IgE.
Lungs tissue preparation
The lungs from the mice, perfused with PBS
containing 0.5 mM EDTA, were immediately excised and cut into
pieces. As collagenase digestion downregulates the density of CD27
on lymphocytes, the lung pieces were ground with a stirring rod
accompanied with the addition of complete RPMI-1640 (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) on a 200-mesh stainless
steel gauze, as described previously (16). The cells were filtrated,
centrifuged at 300 × g at 4°C for 5 min and then resuspended in
PBS.
Flow cytometric analysis
The cells isolated from the lungs were resuspended
in PBS and the viable cells were counted using a solution of 0.4%
Trypan blue in PBS. The cells were seeded at a density of
2×105 cells/ml in complete RPMI-1640 containing 1X Cell
Stimulation Cocktail plus protein transport inhibitors (Invitrogen;
Thermo Fisher Scientific, Inc.) and incubated at 37°C for 16 h.
Following stimulation, the cells were washed in flow cytometry
buffer (eBioscience) and were then incubated at 4°C for 30 min with
the optimal concentration of PE/Cy7-conjugated anti-CD3 mAb (cat.
no. 100219; 1:1,000), PerCP/Cy5.5-conjugated anti-CD11b mAb (cat.
no. 101227; 1:1,000), APC-conjugated anti-CD27 mAb (cat. no.
124211; 1:1,000; all Biolegend, Inc., San Diego, CA, USA),
FITC-conjugated anti-CD49b (cat. no. 11-5971-81; 1:1,000) and
PE-conjugated anti-CD69 mAb (cat. no. 12-0691-82; 1:1,000; both
eBioscience; Thermo Fisher Scientific, Inc.). To stain for
intracellular IFN-γ, IL-5 and IL-13, the cells were fixed and
permeabilized with Cytofix/Cytoperm (BD Biosciences) and then
stained with anti-mouse IFN-γ (cat. no. 12-7311-82; 1:1,000), IL-5
(cat. no. 12-7052-82; 1:1,000) and IL-13 mAb (cat. no. 12-7133-82;
1:1,000; all eBioscience; Thermo Fisher Scientific, Inc.). The
cells were washed twice and then detected by flow cytometry
(FACSCalibur; BD Biosciences) and analyzed using FlowJo 7.6
software (FlowJo LLC, Ashland, OR, USA).
Statistical analysis
Data are shown as the mean ± standard error of the
mean. Statistical significance of differences was determined with
Student's unpaired two-tailed t-test. The software package GraphPad
Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) was used for
data analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Successful establishment of a mouse
allergic asthma model with OVA
Analysis of the inflammatory cells in the BALF
samples revealed that the proportion of eosinophils was
significantly increased following OVA challenge (Fig. 2A, green arrow), which constituted
almost 50% of the total BALF cells (Fig. 2B). In addition, all mice challenged
with OVA had increased total cell numbers and various inflammatory
cell numbers in BALF in comparison with the PBS-challenged mice
(Fig. 2B). On histological
observation, the basement membrane of the bronchus specimens of the
mice challenged with OVA became thickened compared with those of
the control mice. Additionally, an increase in inflammatory cells
around the airways was detected by repeated OVA challenge compared
with the PBS control (Fig. 2C). By
contrast, the OVA-challenged mice, but not the PBS-challenged mice,
developed marked goblet cell hyperplasia and mucus hypersecretion
in the bronchi (Fig. 2D). Th2
cytokines have been considered to be the main orchestrator of the
allergic airway inflammation underlying asthma (17). The results of the present study
showed that OVA inhalation in sensitized mice caused marked
increases in the levels of IL-4, IL-5 and IL-13 into the BALF
compared with those in the PBS aerosol control (Fig. 2E). A crucial component of allergic
immune responses is the production of IgE (18). Therefore, the levels of anti-IgE
and OVA-specific IgE (OVA-IgE) were measured in serum of the
OVA-challenged mice and PBS controls. The results demonstrated that
the levels of anti-IgE and anti-OVA IgE were increased in the lungs
of the OVA-challenged mice (Fig.
2F). Together, these data suggest that the mice sensitized and
challenged with OVA were characterized by pathological asthma.
Therefore, these mice were suitable for use as a model mice to
investigate asthma.
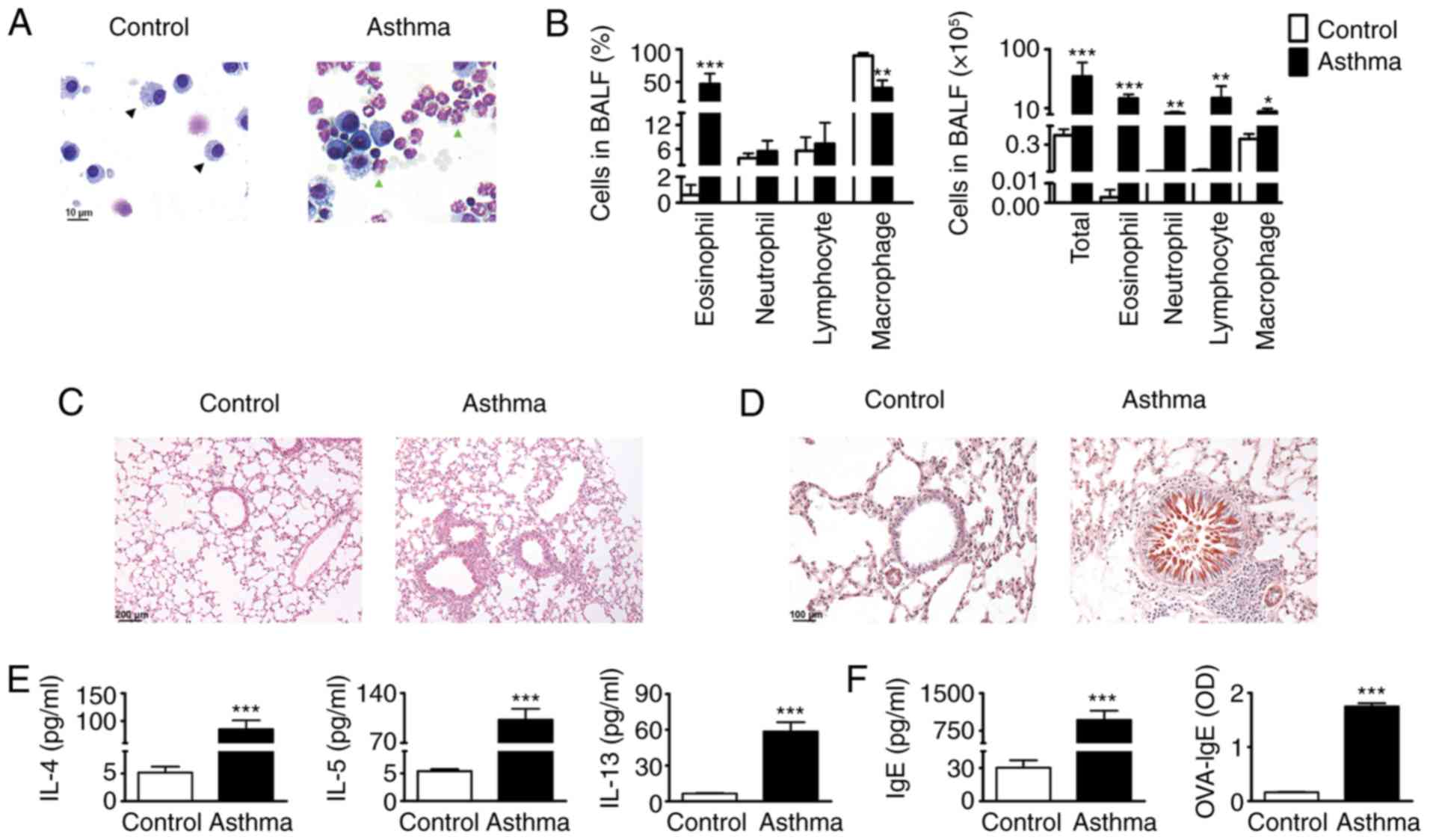 | Figure 2.Successful establishment of an asthma
model. (A) Wright-Giemsa staining shows the macrophages (black
arrow) and eosinophils (green arrow) under microscopy
(magnification, ×400). (B) Statistical analysis of the numbers and
proportions of eosinophils, neutrophils, macrophages and
lymphocytes in BALF from sensitized mice 24 h following final last
saline aerosol or OVA aerosol challenge. (C) Lung sections were
stained with hematoxylin and eosin and (D) periodic acid-schiff,
and examined under microscopy (magnification, ×200). (E) Levels of
IL-4, IL-5 and IL-13 in BALF and (F) IgE and OVA-IgE in serum were
analyzed using ELISA (n=6). *P<0.05, **P<0.01 and
***P<0.001, vs. control group. OVA, ovalbumin; BALF,
bronchoalveolar lavage fluid; IgE, immunoglobin E; IL,
interleukin. |
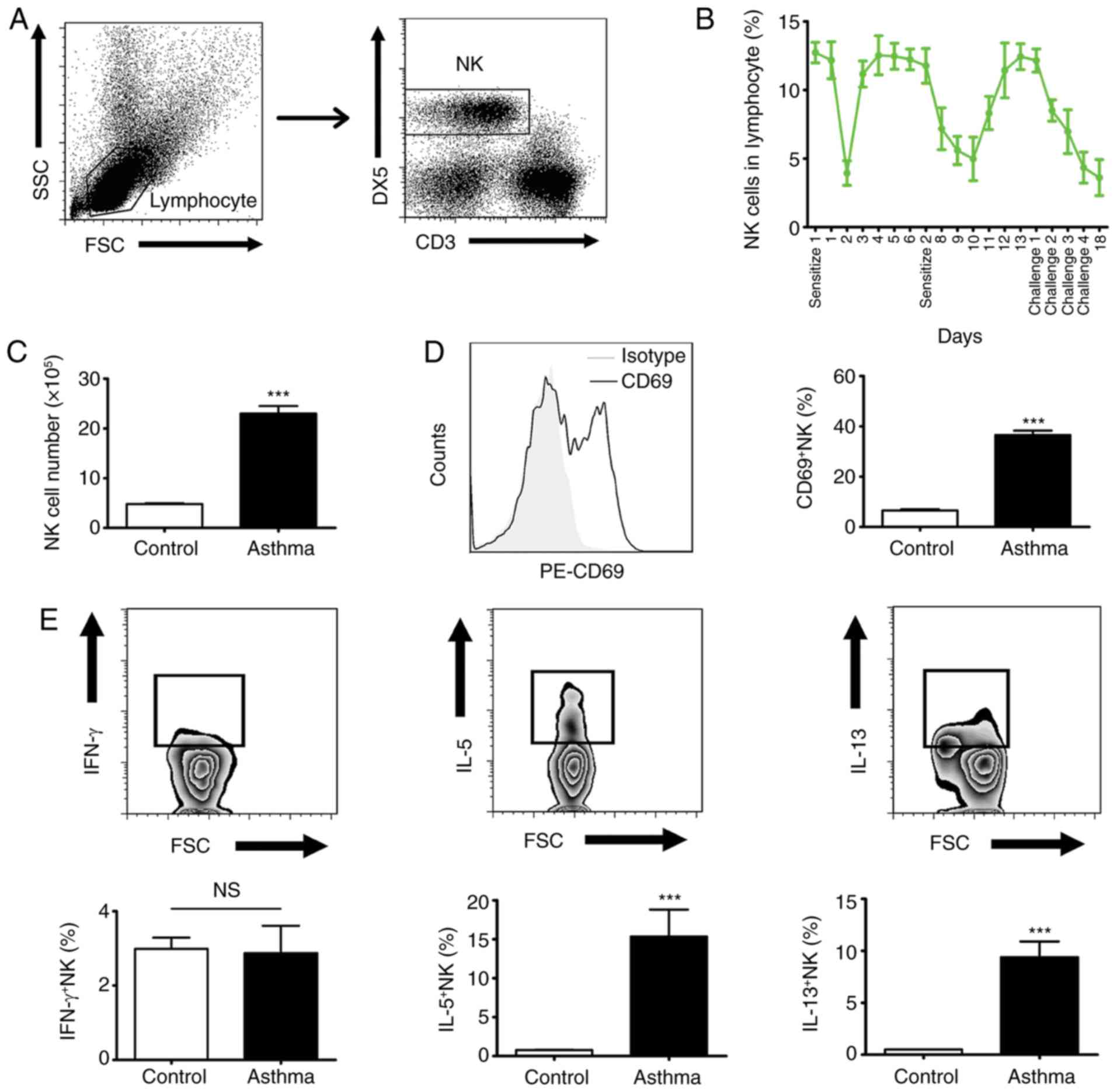
Dynamic changes of NK cells in the
development of asthma
NK cells reside in large numbers in the lungs, and
are crucial in host survival by killing infected cells and/or
producing cytokines (19). In the
present study, the dynamic changes of NK cells were investigated
during the development of asthma in the established OVA-induced
mouse model. All the lymphocytes obtained from lungs were gated
according to forward scatter (FSC) and side scatter (SSC), and NK
cells (CD3− DX5+) within the lymphocytes were
gated for the following analysis (Fig.
3A). The results showed that the proportion of NK cells in the
lymphocytes was markedly decreased following OVA sensitization and
challenge (Fig. 3B). Despite this,
increased numbers of NK cells were observed in the asthma group
compared with the control group (Fig.
3C). In addition, the activity of the NK cells was
significantly enhanced following OVA treatment in the asthma group
(Fig. 3D). The results showed that
higher levels of IL-5 and IL-13, rather than IFN-γ, were produced
by the NK cells following OVA aerosol challenge (Fig. 3E). These data suggest that,
although NK cells are not the crucial type of lymphocyte involved
in asthma, they are likely to be involved in the development of
asthma by secreting Th2 cytokines.
CD11b− CD27− NK cells are the
primary NK subset which produce Th2 cytokines in asthma. Murine NK
cells can be divided into four subsets according to the surface
density of CD11b and CD27, the most immature subset is
CD11b− CD27−, followed by immature
CD11b− CD27+, mature CD11b+
CD27+ and the most mature CD11b+
CD27− subset (14). The
present study aimed to determine whether the lung NK cell phenotype
was dynamically regulated with the development of asthma. The
results showed that the percentages of the CD11b−
CD27− NK and CD11b− CD27+ NK
subsets gradually increased during the development of asthma
(Fig. 4A). By contrast, the
percentages of the CD11b+ CD27+ NK and
CD11b+ CD27− NK subsets reduced with OVA
sensitization and challenge (Fig.
4A). Subsequently, the cytokine secretion capacity of these NK
subsets following OVA challenge was analyzed. Previous studies have
suggested that immature NK cells are potent producers of cytokines
and mature NK cells exhibit potent cytotoxic capabilities (15). Consistent with these findings, the
results of the present study showed that CD11b− NK
subsets, rather than CD11b+ NK subsets, produced more
Th2 cytokines IL-5 and IL-13 following OVA challenge (Fig. 4B). Furthermore, the flow cytometry
results showed that CD11b− CD27− NK cells
were the primary NK subset that produces Th2 cytokines following
OVA challenge (Fig. 4C). These
findings collectively indicated that CD11b−
CD27− NK cells may be the primary NK subset involved in
the development of asthma.
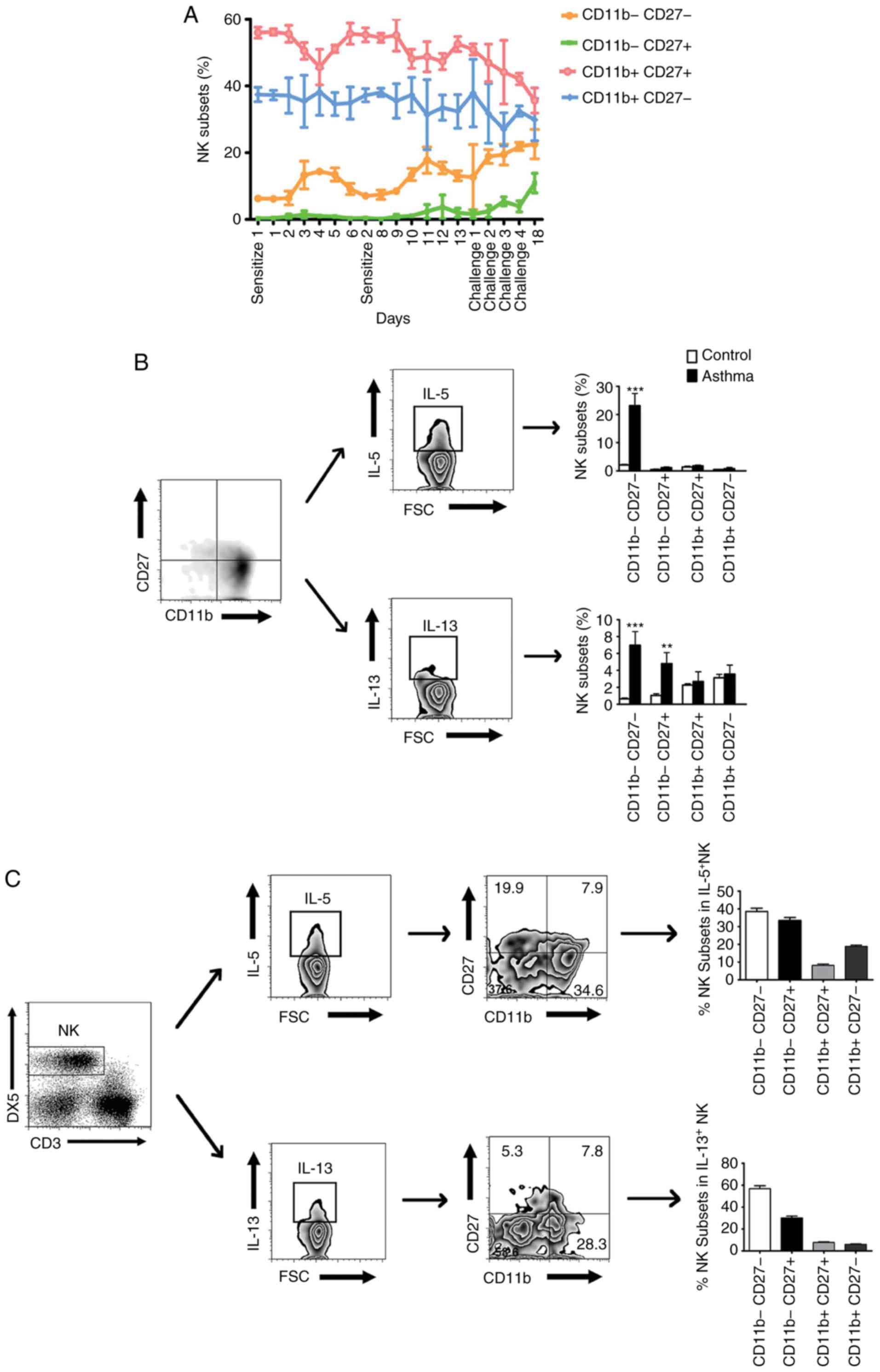 | Figure 4.Dynamic changes of lung NK subsets in
the development of asthma. (A) Percentages of the CD11b−
CD27−, CD11b− CD27+,
CD11b+ CD27+ and CD11b+
CD27− subsets of NK cells obtained from the lung of mice
24 h following sacrifice. (B) Representative FACS histograms
showing the expression of IL-5 and IL-13 in CD11b−
CD27−, CD11b− CD27+,
CD11b+ CD27+ and CD11b+
CD27− subsets of NK cells, respectively. (C) Percentages
of the IL-5+ and IL-13+ NK cell subsets in
total IL-5+ and IL-13+ NK cells, respectively
(n=6). **P<0.01 and ***P<0.001, vs. control group. NK,
natural killer; FSC, forward scatter; IL, interleukin. |
Discussion
Allergic asthma has been considered to be driven by
Th2 cytokines including IL-4, IL-5 and IL-13, which are important
in orchestrating and amplifying allergic inflammation in asthma
(4,20). Th2 cells are considered the primary
source of these cytokines (21).
IL-4 promotes Th2 cell differentiation from uncommitted Th0 cells
and IgE production by B cells (22,23).
IL-5 is known to drive eosinophil maturation and differentiation,
goblet cell hyperplasia and mucus secretion (24). IL-13 has several actions similar to
those of IL-4 by inducing IgE secretion, and has been implicated in
airway remodeling, including mucus hypersecretion and goblet cell
hyperplasia (25). Although
evidence has clearly indicated the significance of Th2 cells in
asthma, cumulative studies have revealed that innate lymphoid cells
also have substantial involvement in asthma. Bronchial epithelial
cells, which are the first line of defense against inhaled
pathogens and particles, are important for allergen uptake and the
initiation of allergic inflammation by releasing Th2-promoting
cytokines, including IL-25, thymic stromal lymphopoietin and IL-33
(26). In addition, airway
dendritic cells (DCs) are responsible for the initiation and
maintenance of adaptive Th2 responses in asthma as they could
recognize antigens through pattern recognition receptors such as
TLRs and NLRs (27). The high
frequency and number of NK cells in the lungs indicate the
prominent role for these cells in the local immune system of the
airways. In addition, the early response of NK cells to infection
or antigens and subsequent interaction with other cells, including
DCs, makes these cells potentially ideal candidates for influencing
T cell responses. However, so far, the role of NK cells in asthma
remains to be fully elucidated. Clinical investigations have shown
that NK cells in the peripheral blood of patients with asthma were
higher in number and exhibited higher cytotoxicity compared with
healthy controls (28). With
regard to animal models, several studies have indicated that NK
cells may contribute to the initiation and development of allergic
airway inflammation. Korsgren et al (29) found that the depletion of NK cells
prior to OVA sensitization led to decreased production of Th2
cytokines and systemic IgE antibodies. However, anti-NK1.1 antibody
may also knock out NK T cells, which have been demonstrated to be
required for allergen-induced airway inflammation. Subsequently,
Ple et al (30) showed that
eosinophilic airway inflammation was reduced when NK cells were
depleted following OVA challenge using anti-asialo GM1 antibodies.
A later study by Mathias et al (31) observed that the depletion of NK
cells using anti-Ly49 mAbs led to a decrease in airway
inflammation, Th2 cytokine secretion and OVA-specific antibody
production. Although the use of these antibodies did not influence
NK T cells, GM1 and Ly49 are also expressed on T cell subsets.
Together with experiments in mice, a requirement for NK cells in
the development of asthma was revealed with these experiment
methods. However, the mechanism of NK cells in asthma remains to be
fully elucidated.
NK cells have a variety of biological effects,
including exocytosis of cytotoxic granules and synthesis of
cytokines (10). Although first
identified by their cytotoxic activity against virally infected
cells and tumors, NK cells also have a potent cytokine secretion
capacity. Previous data have shown that NK cell cytokine production
may be governed in part by the milieu during inflammation (32). As a general rule, NK cells secrete
a large amount of IFN-γ in response to IL-12 and IL-18 stimulation
at an early stage of infection (33). However, in vitro experiments
have revealed that NK cells in the spleen and liver also produce
the IL-13 cytokine following co-stimulation with IL-18 and IL-12
(34). McDermott et al
(35) demonstrated that NK cells
secreted high levels of IL-13, which acted on the intestinal
epithelial and led to the disruption of intestinal architecture in
a mouse model of nematode infection. In addition, it has been
observed that the NK cells from atopic patients with asthma
released higher levels of IL-5 and IL-13 compared with healthy
individuals (36). In the present
study, it was found that NK cells secreted high levels of IL-5 and
IL-13 in an OVA-induced mouse model of asthma. In addition, the
percentage of lung NK cells in lymphocytes declined following OVA
sensitization and challenge. These results support the previous
conclusion that Th2 cells are the foremost cell types involved in
asthma (37,38). However, increased numbers and
enhanced activity of NK cells were detected following OVA aerosol
challenge in the experiments, which were consistent with the
phenomenon observed clinically. Together, the data obtained in the
present study and previous reports indicate that NK cells may be
involved the development of asthma by producing Th2 cytokines.
It has been suggested that CD11b−
CD27−, CD11b− CD27+,
CD11b+ CD27+, and CD11b+
CD27− are discrete stages of NK cell in vivo
maturation. The mature NK cells (CD11b+) make up the
majority of NK cells circulating in peripheral blood and in
non-lymphoid tissues, including the spleen and lung (12). These NK subsets have potent
cytotoxic function and low cytokine production upon activation
(39,40). By contrast, immature NK cells
(CD11b−) are most abundant within the bone marrow and
lymph nodes and are efficient producers of cytokines (41,42).
Consistent with previous evidence, the results of the present study
showed that the majority of lung NK cells within normal mice were
CD11b+ NK subsets, constituting ~90% of the NK cells.
These CD11b+ NK subsets gradually decreased following
OVA induction whereas the immature CD11b− NK subsets
increased, revealing that the changes in circumstance during the
development of asthma have an impact on the NK subsets.
Furthermore, the CD11b− NK subsets secreted more Th2
cytokines (IL-5 and IL-13) following OVA challenge, whereas the
cytokine-producing capacity of the CD11b+ NK subsets had
no notable changes. These data demonstrated that immature NK cell
subsets have an increased ability to secrete cytokines than mature
NK cell subsets, as reported previously. Of note, the flow
cytometry results showed that the CD11b−
CD27− NK cells are the primary NK subset that secrete
Th2 cytokines, suggesting that the CD11b−
CD27− NK cells may be the primary NK subset involved in
the development of asthma.
In conclusion, the dynamic behavior of lung NK cells
and their subsets during the development of asthma were analyzed in
the present study. The results suggest that NK cells may be
involved in the pathogenesis of asthma by secreting Th2
cytokines.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81273305), the Basic
Research Program of Science and Technology Plan of Shenzhen (grant
no. JC201505280578A) and the Special Program of Construction
National Innovative City of Shenzhen (grant no. 301201003010).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZC and LW designed the project and performed the
experiments. ZC wrote the manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Department of Immunology, School of Medicine, Shenzhen
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Jayasinghe H, Kopsaftis Z and Carson K:
Asthma bronchiale and exercise-induced bronchoconstriction.
Respiration. 89:505–512. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reddel HK and Levy ML; Global Initiative
for Asthma Scientific Committee and Dissemination and
Implementation Committee, : The GINA asthma strategy report: What's
new for primary care? NPJ Prim Care Respir Med. 25:150502015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Becker AB and Abrams EM: Asthma
guidelines: The global initiative for asthma in relation to
national guidelines. Curr Opin Allergy Clin Immunol. 17:99–103.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fajt ML and Wenzel SE: Asthma phenotypes
and the use of biologic medications in asthma and allergic disease:
The next steps toward personalized care. J Allergy Clin Immunol.
135:299–311. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wenzel SE: Asthma phenotypes: The
evolution from clinical to molecular approaches. Nat Med.
18:716–725. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu S, Kim HY, Chang YJ, DeKruyff RH and
Umetsu DT: Innate lymphoid cells and asthma. J Allergy Clin
Immunol. 133:943–951. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
DeKruyff RH, Yu S, Kim HY and Umetsu DT:
Innate immunity in the lung regulates the development of asthma.
Immunol Rev. 260:235–248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim HY, DeKruyff RH and Umetsu DT: The
many paths to asthma: Phenotype shaped by innate and adaptive
immunity. Nat Immunol. 11:577–584. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McKenzie AN: Type-2 innate lymphoid cells
in asthma and allergy. Ann Am Thorac Soc. 11 (Suppl 5):S263–S270.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Campbell KS and Hasegawa J: Natural killer
cell biology: An update and future directions. J Allergy Clin
Immunol. 132:536–544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Varchetta S, Oliviero B, Mavilio D and
Mondelli MU: Different combinations of cytokines and activating
receptor stimuli are required for human natural killer cell
functional diversity. Cytokine. 62:58–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ndhlovu LC, Lopez-Vergès S, Barbour JD,
Jones RB, Jha AR, Long BR, Schoeffler EC, Fujita T, Nixon DF and
Lanier LL: Tim-3 marks human natural killer cell maturation and
suppresses cell-mediated cytotoxicity. Blood. 119:3734–3743. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Messaoudene M, Fregni G,
Fourmentraux-Neves E, Chanal J, Maubec E, Mazouz-Dorval S,
Couturaud B, Girod A, Sastre-Garau X, Albert S, et al: Mature
cytotoxic CD56(bright)/CD16(+) natural killer cells can infiltrate
lymph nodes adjacent to metastatic melanoma. Cancer Res. 74:81–92.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chiossone L, Chaix J, Fuseri N, Roth C,
Vivier E and Walzer T: Maturation of mouse NK cells is a 4-stage
developmental program. Blood. 113:5488–5496. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Holmes ML, Huntington ND, Thong RP, Brady
J, Hayakawa Y, Andoniou CE, Fleming P, Shi W, Smyth GK,
Degli-Esposti MA, et al: Peripheral natural killer cell maturation
depends on the transcription factor Aiolos. EMBO J. 33:2721–2734.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Z, Chen X, Xu Y, Xiong P, Fang M, Tan
Z, Gong F and Zheng F: Collagenase digestion down-regulates the
density of CD27 on lymphocytes. J Immunol Methods. 413:57–61. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Allan RS, Zueva E, Cammas F, Schreiber HA,
Masson V, Belz GT, Roche D, Maison C, Quivy JP, Almouzni G and
Amigorena S: An epigenetic silencing pathway controlling T helper 2
cell lineage commitment. Nature. 487:249–253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gray LEK and Sly PD: Update in asthma
2017. Am J Respir Crit Care Med. 197:1108–1115. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Poli A, Michel T, Patil N and Zimmer J:
Revisiting the functional impact of NK cells. Trends Immunol.
39:460–472. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peters SP: Asthma phenotypes: Nonallergic
(intrinsic) asthma. J Allergy Clin Immunol Pract. 2:650–652. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schatz M and Rosenwasser L: The allergic
asthma phenotype. J Allergy Clin Immunol Pract. 2:645–649. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Steinke JW and Borish L: Th2 cytokines and
asthma. Interleukin-4: Its role in the pathogenesis of asthma and
targeting it for asthma treatment with interleukin-4 receptor
antagonists. Respir Res. 2:66–70. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Munitz A, Brandt EB, Mingler M, Finkelman
FD and Rothenberg ME: Distinct roles for IL-13 and IL-4 via IL-13
receptor alpha1 and the type II IL-4 receptor in asthma
pathogenesis. Proc Natl Acad Sci USA. 105:7240–7245. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rosenwasser LJ and Rothenberg ME: IL-5
pathway inhibition in the treatment of asthma and Churg-Strauss
syndrome. J Allergy Clin Immunol. 125:1245–1246. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ingram JL and Kraft M: IL-13 in asthma and
allergic disease: Asthma phenotypes and targeted therapies. J
Allergy Clin Immunol. 130:829–844. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lambrecht BN and Hammad H: Asthma and
coagulation. N Engl J Med. 369:1964–1966. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van Helden MJ and Lambrecht BN: Dendritic
cells in asthma. Curr Opin Immunol. 25:745–754. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Deniz G, van de Veen W and Akdis M:
Natural killer cells in patients with allergic diseases. J Allergy
Clin Immunol. 132:527–535. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Korsgren M, Persson CG, Sundler F, Bjerke
T, Hansson T, Chambers BJ, Hong S, Van Kaer L, Ljunggren HG and
Korsgren O: Natural killer cells determine development of
allergen-induced eosinophilic airway inflammation in mice. J Exp
Med. 189:553–562. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ple C, Barrier M, Amniai L, Marquillies P,
Bertout J, Tsicopoulos A, Walzer T, Lassalle P and Duez C: Natural
killer cells accumulate in lung-draining lymph nodes and regulate
airway eosinophilia in a murine model of asthma. Scand J Immunol.
72:118–127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mathias CB, Guernsey LA, Zammit D, Brammer
C, Wu CA, Thrall RS and Aguila HL: Pro-inflammatory role of natural
killer cells in the development of allergic airway disease. Clin
Exp Allergy. 44:589–601. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Perricone R, Perricone C, De Carolis C and
Shoenfeld Y: NK cells in autoimmunity: A two-edg'd weapon of the
immune system. Autoimmun Rev. 7:384–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Haeberlein S, Sebald H, Bogdan C and
Schleicher U: IL-18, but not IL-15, contributes to the
IL-12-dependent induction of NK-cell effector functions by
Leishmania infantum in vivo. Eur J Immunol. 40:1708–1717. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hoshino T, Winkler-Pickett RT, Mason AT,
Ortaldo JR and Young HA: IL-13 production by NK cells:
IL-13-producing NK and T cells are present in vivo in the absence
of IFN-gamma. J Immunol. 162:51–59. 1999.PubMed/NCBI
|
|
35
|
McDermott JR, Humphreys NE, Forman SP,
Donaldson DD and Grencis RK: Intraepithelial NK cell-derived IL-13
induces intestinal pathology associated with nematode infection. J
Immunol. 175:3207–3213. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Aktas E, Akdis M, Bilgic S, Disch R, Falk
CS, Blaser K, Akdis C and Deniz G: Different natural killer (NK)
receptor expression and immunoglobulin E (IgE) regulation by NK1
and NK2 cells. Clin Exp Immunol. 140:301–309. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fahy JV: Type 2 inflammation in
asthma-present in most, absent in many. Nat Rev Immunol. 15:57–65.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Holgate ST: Innate and adaptive immune
responses in asthma. Nat Med. 18:673–683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sceneay J, Chow MT, Chen A, Halse HM, Wong
CS, Andrews DM, Sloan EK, Parker BS, Bowtell DD, Smyth MJ and
Möller A: Primary tumor hypoxia recruits CD11b+/Ly6Cmed/Ly6G+
immune suppressor cells and compromises NK cell cytotoxicity in the
premetastatic niche. Cancer Res. 72:3906–3911. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yanagisawa K, Exley MA, Jiang X, Ohkochi
N, Taniguchi M and Seino K: Hyporesponsiveness to natural killer
T-cell ligand alpha-galactosylceramide in cancer-bearing state
mediated by CD11b+ Gr-1+ cells producing nitric oxide. Cancer Res.
66:11441–11446. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Clinthorne JF, Beli E, Duriancik DM and
Gardner EM: NK cell maturation and function in C57BL/6 mice are
altered by caloric restriction. J Immunol. 190:712–722. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mao Y, Sarhan D, Steven A, Seliger B,
Kiessling R and Lundqvist A: Inhibition of tumor-derived
prostaglandin-e2 blocks the induction of myeloid-derived suppressor
cells and recovers natural killer cell activity. Clin Cancer Res.
20:4096–4106. 2014. View Article : Google Scholar : PubMed/NCBI
|