Long noncoding RNAs (lncRNAs) are a class of RNA
(>200 nucleotides in length) that cannot synthesize proteins
(1–4). These biomolecules are involved in
post-transcriptional regulation (5–8), and
are abnormally expressed in multiple types of solid tumor and
hematopoietic malignancy; lncRNAs are involved in both
carcinogenesis and tumor suppression (9–13).
The expression levels of several lncRNAs have been
reported in multiple myeloma (MM); their clinical significance,
biological functions and potential molecular mechanisms in the
disease have also been investigated (14–16).
MM is the second most frequent hematological malignancy, and
accounts for ~10% of all such malignancies (17–22).
Immunomodulatory drugs (such as lenalidomide and pomamide),
proteasome inhibitors (such as bortezomib and carfilzomib) and
monoclonal antibodies have significantly increased the survival
rate of patients with MM over the past decade (23–25);
however, the treatment of relapsed and partially refractory
patients remains challenging. The pathogenesis and progression of
MM involve complex and heterogeneous genomic alterations (26–30),
including modifications that are influenced by lncRNAs.
Certain lncRNAs have been documented to serve an
important role in the progression of MM, and can be used as
indicators of patient prognosis. For example, metastasis-associated
lung adenocarcinoma transcript 1 (MALAT1) is overexpressed
in MM tissues and various MM cell lines; upregulation of
MALAT1 is significantly associated with poor prognosis,
including overall survival (OS) and progression-free survival (PFS)
(31–33). Nuclear paraspeckle assembly
transcript 1 (NEAT1) has also been reported to serve a
pivotal role in promoting MM, and its elevated expression is
closely associated with poor prognosis (34,35).
The upregulation of urothelial cancer associated 1 (UCA1)
(36), protein disulfide isomerase
family A member 3 pseudogene 1 (PDIA3P) (37), H19 (38), colon cancer associated transcript 1
(CCAT1) (39) and
colorectal neoplasia differentially expressed (CRNDE)
(40) are closely associated with
poor prognosis in MM; these genes may be used as future indicators
in the clinical prognosis of patients with MM. Despite the large
numbers of lncRNAs, only a small number have been associated with
the prognosis of MM; however, numerous as-yet-undiscovered lncRNAs
may also be associated with the progression of MM and patient
outcome. Additionally, the predictive ability of a single indicator
is limited; a prognostic signature composed of numerous indicators
is required to conduct a comprehensive clinical evaluation of tumor
prognosis. Prognostic models that combine several indicators have
been used in a wide variety of tumors (41–45);
however, a prognostic model for MM comprising lncRNAs is yet to be
reported.
The present study screened gene chips with
expression data from patients with MM and selected prognostic
lncRNAs and mRNAs. The associations between the prognostic lncRNAs
and mRNAs were mapped, and certain indicators were selected to
construct a prognostic model. The MM prognostic model presented in
the current study may provide novel insight and directions for the
clinical treatment of MM in the future.
The microarray gene expression profiling data from
the bone marrow of newly diagnosed patients with MM that had not
been treated was obtained from the Genome Expression Omnibus (GEO)
dataset (46,47) with accession number GSE24080
(48). The data from 559 patients
with MM were included for further survival analysis. To separate
lncRNAs and mRNAs, probes from the Affymetrix HG-U133_Plus_2.0
array were re-annotated. For genes that matched >1 probe, the
expression values of all the measurements were calculated using an
average value of the probes. The lncRNAs were extracted according
to their Refseq database label (Release 93) (49) and Ensembl annotations (Release
version 96) (50).
Event-free survival (EFS) generally provides more
reliable endpoint information for survival analysis (51); thus, it was selected as the
survival analysis endpoint in the present study. Univariate Cox
analysis was conducted to select prognosis-associated mRNAs and
lncRNA using the survival package (version 2.44–1.1) in R (version
3.4.4) (R). P<0.005 was considered to be statistically
significant (52). Kaplan-Meier
plot was generated to observe the survival status between different
survival associated mRNA and lncRNA expression levels.
To further investigate the potential molecular
mechanisms of the top 20 prognosis-associated mRNAs, the biological
processes, which were acquired from gene ontology and Kyoto
Encyclopedia of Genes and Genomes (Release 87.1) (KEGG) pathways
(53–55) were examined based on enrichment
analysis using the Clusterprofiler package (version 3.10.1) in R
(56). Protein-protein interaction
(PPI) networks were developed to explore the associations between
each gene using the GeneMANIA plug-in in Cytoscape version 3.6.1
(57,58).
As the prognostic value of a single indicator is
limited, prognostic signatures were produced that combined multiple
indicator candidates. Multivariate Cox analysis was performed on
the top 20 most significant prognosis-associated mRNAs and lncRNAs
to develop prognostic signatures. The performances of these
prognostic signatures were tested using the survivalROC (version
1.0.3) package in R, which provides time-dependent receiver
operator characteristic (ROC) curve estimation (59,60).
The area under curve (AUC) was calculated at 75 months, as fewer
events occurred after this point.
As lncRNAs cannot be transcribed into proteins,
their functional effects are frequently achieved by targeting
mRNAs. To investigate the associations between lncRNAs and mRNAs,
WGCNA was conducted using the WGCNA package (version 1.63) in R
(61,62). The mRNAs were separated into
modules, and correlations between the prognostic mRNAs and lncRNAs
were calculated. A lncRNA-mRNA axis was identified when an
association coefficient >0.4 was obtained. The potential
regulatory network was constructed using Cytoscape software.
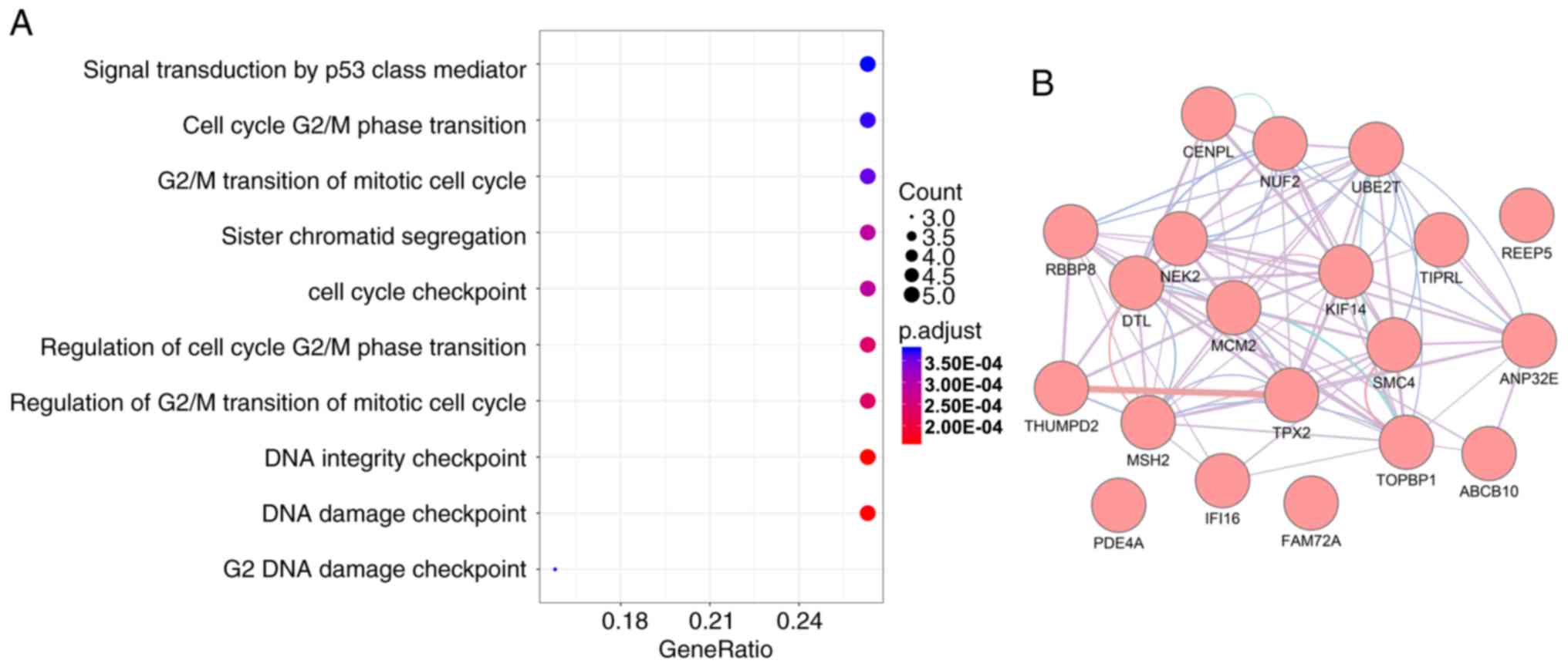
Enrichment analyses for the top 20
prognosis-associated mRNAs were conducted to identify risk pathways
and biological functions associated with these prognostic genes
(Table II). As presented in
Fig. 2A, a number of cell
cycle-associated biological processes were identified, including
signal transduction by a p53-class mediator, cell cycle G2/M
transition and mitotic cell cycle G2/M transition. These categories
are also closely involved in tumor proliferation. The PPI network
revealed that these genes were closely associated with each other
(Fig. 2B). It was also observed
that these prognostic genes were associated with homologous
recombination (Table II).
Collectively, the present results suggested that these genes may
serve an important role in the pathogenesis of MM.
WGCNA was used to separate prognosis-associated
mRNAs into groups, and to explore the associations between lncRNAs
and mRNAs (Fig. 6). The
lncRNA-mRNA network provided novel insight into the regulatory
mechanisms associated with the progression of MM (Fig. 7). A total of 143 mRNAs were
positively or negatively associated with 23 prognosis-related
lncRNAs. NCRNA00201, LOC115110 and RP5-968J1.1
appeared to be the most dominant drivers, as they possessed the
highest number of connected genes.
Currently, there is no precise method to assess the
prognosis of patients with MM. In the present study, the expression
data from a gene chip containing genomic samples from 559 patients
with MM were analyzed, and predictive models were constructed based
on the lncRNA and mRNA expression profiles. Of note, it was
observed that pools of prognostic candidates exhibited greater
predictive power than individual indicators. There may also be a
targeting relationship between the prognosis-associated lncRNAs and
mRNAs. As a previous report has contraindicated the use of a lncRNA
prediction model for the prognosis of MM, the present study
provides novel insight for the clinical diagnosis and treatment of
MM (63).
Previously, two other research groups have analyzed
the gene chip data of GSE24080 to obtain MM prognosis-associated
lncRNAs using different statistical methods. Zhou et al
(64) randomly split the MM cohort
into a training dataset (n=280) and a testing dataset (n=279).
Univariate regression analysis identified 59 lncRNAs that were
associated with the OS of patients. Only four of those lncRNAs
(RP4-803J11.2, RP1-43E13.2, RP11-553L6.5 and ZFY-AS1)
were reported to exhibit a predictive effect following multivariate
regression analysis. These results were inconsistent with a study
by Hu et al (63), which
identified 176 lncRNAs from the GSE24080 and GSE57317 datasets that
appeared to be associated with patient survival. The Hu et
al study employed Kaplan-Meier analysis to determine the
prognostic influence of lncRNAs, identifying 176 lncRNAs, including
RP1-286D6.1, AC008875.2, MTMR9L, AC069360.2 and
AL512791.1, as prospective markers for assessing the
prognosis of patients with MM. Of note, none of the aforementioned
lncRNAs overlapped with the top 20 prognosis-associated lncRNAs
identified in the present study, which conducted survival analysis
using the survival package in R. Among the top 20
prognosis-associated lncRNAs in the present study, a number were
identified as risk factors, including NCRNA00201, AC116904.1,
AC022087.1, C21ORF34, AC004383.4 and RP11-706O15.5. The
remaining 14 lncRNAs may protect against MM. The use of different
statistical tools may partially explain the variations in the
lncRNAs identified in each study. In the previously published
studies that analyzed GSE24080, Hu et al (63) did not conduct ROC analysis to
determine the prognostic values of their lncRNAs, whereas Zhou
et al (64) reported four
lncRNAs (RP4-803J11.2, RP1-43E13.2, RP11-553L6.5 and
ZFY-AS1) together generated an AUC of 0.682 to represent
prognostic performance. In the present study, ROC analysis was
performed using the survivalROC package in R, resulting in an AUC
of 0.739, more favorable than that in Zhou et al (64).
None of the lncRNAs in the presently reported
prognostic model have been previously investigated in MM. At
present, the majority of the top 20 prognosis-associated lncRNAs
have not been reported in any disease. Only three of the lncRNAs
have been previously studied, NCRNA00201, HCG26 and
C21ORF34.
The third previously reported lncRNA that exhibited
potential prognostic value in MM was C21orf34.
C21orf34 has been studied for its role in blood pressure by
the Hypertension Genetic Epidemiology Network; African Americans
and European Americans exhibited associations between blood
pressure and intronic single nucleotide polymorphisms on chromosome
21q21.1 (70). The C21orf34
gene was linked to African American patients, improving
understanding of the pathophysiology of hypertension (70). C21orf34 has also been
studied in malignancy; C21orf34, which is the host gene of
microRNA-125b, was reported to be downregulated in human metastatic
melanoma (71). The role of
C21orf34 in MM is yet to be determined. This study is the
first to identify C21orf34 as an MM prognostic indicator.
Its increased expression levels may predict the improved survival
of patients with MM, suggesting that it may act as a protective
factor against MM. As the prognostic value of C21orf34 was
only determined via gene chip data mining, little is known
regarding the functional role and mechanism of C21orf34 in
MM. Therefore, further investigation is required.
To investigate the potential functional implications
of prognostic markers for the onset and progression of MM, various
bioinformatics computational methods were combined. The most
reliable prognostic biomarkers identified in the present study were
actively involved in cell cycle-associated processes. Sustained and
proliferative signaling has been increasingly acknowledged as a
fundamental trait of cancer cells, so the present findings are not
unexpected (72). Previous studies
reported that cell cycle interference may exert an antitumor
function in MM (73,74). As a result of the complex
mechanisms of tumorigenesis and tumor progression, a single gene is
unlikely to underpin poor prognosis in MM. Therefore, a lncRNA-mRNA
network was proposed to comprehensively explore the molecular
characteristics of MM. The WGCNA results indicated that there may
also be targeting relationships between the prognosis-associated
lncRNAs and mRNAs. These lncRNAs may exert their prognostic effects
by targeting closely associated mRNAs. Investigations of
lncRNA-based regulatory networks are limited, particularly from the
perspective of prognosis. Ronchetti et al (75) previously proposed a network
constructed by lncRNAs and miRNAs. Further research should be
conducted into the functional relationships between mRNAs and
lncRNAs.
Certain shortcomings in the present study should be
stated. A total of 559 cases were included in this study; however,
the reported findings should be confirmed in additional independent
cohorts. Furthermore, the prognostic values of the lncRNAs in this
study were investigated using a gene chip; this single detection
method should be verified by other methods, such as reverse
transcription-quantitative PCR. Additionally, the majority of the
lncRNAs identified in our prognostic model have not been previously
reported. Their specific clinical significance, biological
functions and potential mechanisms of action should be studied in
further experiments. Finally, the molecular associations between
identified lncRNAs and mRNAs in the expression network should be
further investigated. Additional experiments are required to
determine whether the prognosis-associated lncRNAs serve a role in
MM via their corresponding mRNA targets.
In conclusion, the present study constructed a model
that is capable of predicting prognosis in MM and generated a
network with corresponding prognosis-associated mRNAs. Of note, the
clinical significance and function of the majority of the lncRNAs
identified in the present study remain unknown. These results offer
novel perspective for the clinical diagnosis and treatment of MM
and suggest novel directions for investigating the mechanisms
underlying the development of MM.
Not applicable.
The present study was supported in part by the
National Nature Science Foundation of China (grant no. 81560024),
Program of Scientific and Technology Project, Guilin Science
Research and Technology Development (grant no. 2016012706-2),
National Natural Science Foundation of China (grant no. 81460038)
and Guangxi Natural Science Foundation of China (grant no.
2017GXNSFAA198178).
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
YRL, ZPG and ZZY conceived and designed the study,
as well as desiged the figures and tables. FXZ, XTW and ZZY
contributed to the statistical analysis, as well as writing and
correcting the manuscript. All authors read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Xue JY, Huang C, Wang W, Li HB, Sun M and
Xie M: HOXA11-AS: A novel regulator in human cancer proliferation
and metastasis. Onco Targets Ther. 11:4387–4393. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang H, Sun J, Sun Y, Wang C, Gao S, Li W
and Hu JF: Long noncoding RNAs and their epigenetic function in
hematological diseases. Hematol Oncol. 37:15–21. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jia L, Zhang Y, Tian F, Chu Z and Xin H:
Long noncoding RNA colon cancer associated transcript-1 promotes
the proliferation, migration and invasion of cervical cancer. Mol
Med Rep. 16:5587–5591. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang R and Xia T: Long non-coding RNA
XIST regulates PDCD4 expression by interacting with miR-21-5p and
inhibits osteosarcoma cell growth and metastasis. Int J Oncol.
51:1460–1470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Z, Jiang X, Su Z, Li J, Kang P, Li C
and Cui Y: Current insight into a cancer-implicated long noncoding
RNA ZFAS1 and correlative functional mechanisms involved. Pathol
Res Pract. 214:1517–1523. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cai B, Zheng Y, Ma S, Xing Q, Wang X, Yang
B, Yin G and Guan F: BANCR contributes to the growth and invasion
of melanoma by functioning as a competing endogenous RNA to
upregulate Notch2 expression by sponging miR204. Int J Oncol.
51:1941–1951. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ohtsuka M, Ling H, Ivan C, Pichler M,
Matsushita D, Goblirsch M, Stiegelbauer V, Shigeyasu K, Zhang X,
Chen M, et al: H19 noncoding RNA, an independent prognostic factor,
regulates essential Rb-E2F and CDK8-β-catenin signaling in
colorectal cancer. EBioMedicine. 13:113–124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ling ZA, Xiong DD, Meng RM, Cen JM, Zhao
N, Chen G, Li RL and Dang YW: LncRNA NEAT1 promotes deterioration
of hepatocellular carcinoma based on in vitro experiments, data
mining, and RT-qPCR analysis. Cell Physiol Biochem. 48:540–555.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li BL and Wan XP: The role of lncRNAs in
the development of endometrial carcinoma. Oncol Lett. 16:3424–3429.
2018.PubMed/NCBI
|
|
10
|
Zhu Y, Chen P, Gao Y, Ta N, Zhang Y, Cai
J, Zhao Y, Liu S and Zheng J: MEG3 activated by Vitamin D inhibits
colorectal cancer cells proliferation and migration via regulating
clusterin. EBioMedicine. 30:148–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu Q, Yu T, Ou X, Cao D, Xie T and Chen X:
Potential lncRNA diagnostic biomarkers for early gastric cancer.
Mol Med Rep. 16:9545–9552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiong DD, Li ZY, Liang L, He RQ, Ma FC,
Luo DZ, Hu XH and Chen G: The LncRNA NEAT1 accelerates lung
adenocarcinoma deterioration and binds to Mir-193a-3p as a
competitive endogenous RNA. Cell Physiol Biochem. 48:905–918. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun W, Zu Y, Fu X and Deng Y: Knockdown of
lncRNA-XIST enhances the chemosensitivity of NSCLC cells via
suppression of autophagy. Oncol Rep. 38:3347–3354. 2017.PubMed/NCBI
|
|
14
|
Dong H, Jiang S, Fu Y, Luo Y, Gui R and
Liu J: Upregulation of lncRNA NR_046683 serves as a prognostic
biomarker and potential drug target for multiple myeloma. Front
Pharmacol. 10:452019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Butova R, Vychytilova-Faltejskova P,
Souckova A, Sevcikova S and Hajek R: Long non-coding RNAs in
multiple myeloma. Noncoding RNA. 5(pii): E132019.PubMed/NCBI
|
|
16
|
Yu T, Xu Z, Zhang X, Men L and Nie H: Long
intergenic non-protein coding RNA 152 promotes multiple myeloma
progression by negatively regulating microRNA-497. Oncol Rep.
40:3763–3771. 2018.PubMed/NCBI
|
|
17
|
Zhao Y, Xie Z, Lin J and Liu P: MiR-144-3p
inhibits cell proliferation and induces apoptosis in multiple
myeloma by targeting c-Met. Am J Transl Res. 9:2437–2446.
2017.PubMed/NCBI
|
|
18
|
Xia J, Xu H, Zhang X, Allamargot C,
Coleman KL, Nessler R, Frech I, Tricot G and Zhan F: Multiple
myeloma tumor cells are selectively killed by
pharmacologically-dosed ascorbic acid. EBioMedicine. 18:41–49.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao Y, Zhang E, Lv N, Ma L, Yao S, Yan M,
Zi F, Deng G, Liu X, He J, et al: Metformin and FTY720
synergistically induce apoptosis in multiple myeloma cells. Cell
Physiol Biochem. 48:785–800. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Knief J, Reddemann K, Gliemroth J, Brede
S, Bartscht T and Thorns C: ERG expression in multiple myeloma-A
potential diagnostic pitfall. Pathol Res Pract. 213:130–132. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen R, Zhang X, Gao C, Luan C, Wang Y and
Chen B: Treatment and prognostic factors for survival in newly
diagnosed multiple myeloma patients with bortezomib and
dexamethasone regimen: A single Chinese center retrospective study.
Cancer Manag Res. 9:373–380. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jin J, Wang T, Wang Y, Chen S, Li Z, Li X,
Zhang J and Wang J: SRC3 expressed in BMSCs promotes growth and
migration of multiple myeloma cells by regulating the expression of
Cx43. Int J Oncol. 51:1694–1704. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Joao C, Bergantim R, Neves M, Chacim S,
Afonso C, Barradas J, Bernardo M, Coelho H, Esteves G, Fraga C, et
al: Multiple myeloma in elderly patients-a Portuguese multicentric
real-life study. Ann Hematol. 98:1689–1701. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nooka AK, Kaufman JL, Hofmeister CC,
Joseph NS, Heffner TL, Gupta VA, Sullivan HC, Neish AS, Dhodapkar
MV and Lonial S: Daratumumab in multiple myeloma. Cancer.
125:2364–2382. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song X, Wilson KL, Kagan J and Panjabi S:
Cost of peripheral neuropathy in patients receiving treatment for
multiple myeloma: A US administrative claims analysis. Ther Adv
Hematol. 10:20406207198390252019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen WC, Kanate AS, Craig M, Petros WP and
Hazlehurst LA: Emerging combination therapies for the management of
multiple myeloma: The role of elotuzumab. Cancer Manag Res.
9:307–314. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Willenbacher W, Seeber A, Steiner N,
Willenbacher E, Gatalica Z, Swensen J, Kimbrough J and Vranic S:
Towards molecular profiling in multiple myeloma: A literature
review and early indications of its efficacy for informing
treatment strategies. Int J Mol Sci. 19(pii): E20872018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ribatti D and Vacca A: New insights in
Anti-angiogenesis in multiple myeloma. Int J Mol Sci. 19(pii):
E20312018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zeng ZH, Chen JF, Li YX, Zhang R, Xiao LF
and Meng XY: Induction regimens for transplant-eligible patients
with newly diagnosed multiple myeloma: A network meta-analysis of
randomized controlled trials. Cancer Manag Res. 9:287–298. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Abramson HN: The multiple myeloma drug
pipeline-2018: A review of small molecules and their therapeutic
targets. Clin Lymphoma Myeloma Leuk. 18:611–627. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Handa H, Kuroda Y, Kimura K, Masuda Y,
Hattori H, Alkebsi L, Matsumoto M, Kasamatsu T, Kobayashi N, Tahara
KI, et al: Long non-coding RNA MALAT1 is an inducible stress
response gene associated with extramedullary spread and poor
prognosis of multiple myeloma. Br J Haematol. 179:449–460. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu Y, Lin J, Fang H, Fang J, Li C, Chen W,
Liu S, Ondrejka S, Gong Z, Reu F, et al: Targeting the
MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in
multiple myeloma. Leukemia. 32:2250–2262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ronchetti D, Agnelli L, Taiana E, Galletti
S, Manzoni M, Todoerti K, Musto P, Strozzi F and Neri A: Distinct
lncRNA transcriptional fingerprints characterize progressive stages
of multiple myeloma. Oncotarget. 7:14814–14830. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Geng W, Guo X, Zhang L, Ma Y, Wang L, Liu
Z, Ji H and Xiong Y: Resveratrol inhibits proliferation, migration
and invasion of multiple myeloma cells via NEAT1-mediated
Wnt/β-catenin signaling pathway. Biomed Pharmacother. 107:484–494.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu Y and Wang H: LncRNA NEAT1 promotes
dexamethasone resistance in multiple myeloma by targeting
miR-193a/MCL1 pathway. J Biochem Mol Toxicol. 322018.
|
|
36
|
Zhang ZS, Wang J, Zhu BQ and Ge L: Long
noncoding RNA UCA1 promotes multiple myeloma cell growth by
targeting TGF-β. Eur Rev Med Pharmacol Sci. 22:1374–1379.
2018.PubMed/NCBI
|
|
37
|
Yang X, Ye H, He M, Zhou X, Sun N, Guo W,
Lin X, Huang H, Lin Y, Yao R and Wang H: LncRNA PDIA3P interacts
with c-Myc to regulate cell proliferation via induction of pentose
phosphate pathway in multiple myeloma. Biochem Biophys Res Commun.
498:207–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun Y, Pan J, Zhang N, Wei W, Yu S and Ai
L: Knockdown of long non-coding RNA H19 inhibits multiple myeloma
cell growth via NF-κB pathway. Sci Rep. 7:180792017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen L, Hu N, Wang C, Zhao H and Gu Y:
Long non-coding RNA CCAT1 promotes multiple myeloma progression by
acting as a molecular sponge of miR-181a-5p to modulate HOXA1
expression. Cell Cycle. 17:319–329. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meng YB, He X, Huang YF, Wu QN, Zhou YC
and Hao DJ: Long noncoding RNA CRNDE promotes multiple myeloma cell
growth by suppressing miR-451. Oncol Res. 25:1207–1214. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
He RQ, Zhou XG, Yi QY, Deng CW, Gao JM,
Chen G and Wang QY: Prognostic signature of alternative splicing
events in bladder urothelial carcinoma based on spliceseq data from
317 cases. Cell Physiol Biochem. 48:1355–1368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Formicola D, Petrosino G, Lasorsa VA,
Pignataro P, Cimmino F, Vetrella S, Longo L, Tonini GP, Oberthuer
A, Iolascon A, et al: An 18 gene expression-based score classifier
predicts the clinical outcome in stage 4 neuroblastoma. J Transl
Med. 14:1422016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou M, Zhang Z, Zhao H, Bao S and Sun J:
A novel lncRNA-focus expression signature for survival prediction
in endometrial carcinoma. BMC Cancer. 18:392018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim HY, Lee DH, Lee JH, Cho YY, Cho EJ, Yu
SJ, Kim YJ and Yoon JH: Novel biomarker-based model for the
prediction of sorafenib response and overall survival in advanced
hepatocellular carcinoma: A prospective cohort study. BMC Cancer.
18:3072018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liang L, Zeng JH, Qin XG, Chen JQ, Luo DZ
and Chen G: Distinguishable prognostic signatures of left- and
right-sided colon cancer: A study based on sequencing data. Cell
Physiol Biochem. 48:475–490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41((Database Issue)):
D991–D995. 2013.PubMed/NCBI
|
|
48
|
Shi L, Campbell G, Jones WD, Campagne F,
Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, et al: The
microarray quality control (MAQC)-II study of common practices for
the development and validation of microarray-based predictive
models. Nat Biotechnol. 28:827–838. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
O'Leary NA, Wright MW, Brister JR, Ciufo
S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B,
Ako-Adjei D, et al: Reference sequence (RefSeq) database at NCBI:
Current status, taxonomic expansion, and functional annotation.
Nucleic Acids Res. 44:D733–D745. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zerbino DR, Achuthan P, Akanni W, Amode
MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et
al: Ensembl 2018. Nucleic Acids Res. 46:D754–D761. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ioannidis JPA: The proposal to lower P
value thresholds to .005. JAMA. 319:1429–1430. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lin P, He RQ, Ma FC, Liang L, He Y, Yang
H, Dang YW and Chen G: Systematic analysis of survival-associated
alternative splicing signatures in gastrointestinal
pan-adenocarcinomas. EBioMedicine. 34:46–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liu LM, Xiong DD, Lin P, Yang H, Dang YW
and Chen G: DNA topoisomerase 1 and 2A function as oncogenes in
liver cancer and may be direct targets of nitidine chloride. Int J
Oncol. 53:1897–1912. 2018.PubMed/NCBI
|
|
61
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Langfelder P and Horvath S: Fast R
functions for robust correlations and hierarchical clustering. J
Stat Softw. 46(pii): i112012.PubMed/NCBI
|
|
63
|
Hu AX, Huang ZY, Zhang L and Shen J:
Potential prognostic long non-coding RNA identification and their
validation in predicting survival of patients with multiple
myeloma. Tumour Biol. 39:10104283176945632017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhou M, Zhao H, Wang Z, Cheng L, Yang L,
Shi H, Yang H and Sun J: Identification and validation of potential
prognostic lncRNA biomarkers for predicting survival in patients
with multiple myeloma. J Exp Clin Cancer Res. 34:1022015.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Thierry G, Beneteau C, Pichon O, Flori E,
Isidor B, Popelard F, Delrue MA, Duboscq-Bidot L, Thuresson AC, van
Bon BW, et al: Molecular characterization of 1q44 microdeletion in
11 patients reveals three candidate genes for intellectual
disability and seizures. Am J Med Genet A. 158A:1633–1640. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sutaria DS, Jiang J, Azevedo-Pouly ACP,
Lee EJ, Lerner MR, Brackett DJ, Vandesompele J, Mestdagh P and
Schmittgen TD: Expression profiling identifies the noncoding
processed transcript of HNRNPU with proliferative properties in
pancreatic ductal adenocarcinoma. Noncoding RNA. 3(pii):
E242017.PubMed/NCBI
|
|
67
|
He W, Wei D, Cai, Chen S, Li S and Chen W:
Altered long non-coding RNA transcriptomic profiles in ischemic
stroke. Hum Gene Ther. 29:719–732. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu YD, Li Y, Feng SX, Ye DS, Chen X, Zhou
XY and Chen SL: Long noncoding RNAs: Potential regulators involved
in the pathogenesis of polycystic ovary syndrome. Endocrinology.
158:3890–3899. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Low JS, Chin YM, Mushiroda T, Kubo M,
Govindasamy GK, Pua KC, Yap YY, Yap LF, Subramaniam SK, Ong CA, et
al: A genome wide study of copy number variation associated with
nasopharyngeal carcinoma in Malaysian Chinese identifies CNVs at
11q14.3 and 6p21.3 as candidate loci. PLoS One. 11:e01457742016.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Simino J, Shi G, Arnett D, Broeckel U,
Hunt SC and Rao DC: Variants on chromosome 6p22.3 associated with
blood pressure in the HyperGEN study: Follow-up of FBPP
quantitative trait loci. Am J Hypertens. 24:1227–1233. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Pei G, Lan Y, Chen D, Ji L and Hua ZC: FAK
regulates E-cadherin expression via p-SrcY416/p-ERK1/2/p-Stat3Y705
and PPARγ/miR-125b/Stat3 signaling pathway in B16F10 melanoma
cells. Oncotarget. 8:13898–13908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yao R, Sun X, Xie Y, Sun X, Yao Y, Li H,
Li Z, Gao J and Xu K: Identification of a novel c-Myc inhibitor
with anti-tumor effects on multiple myeloma cells. Biosci Rep.
38(pii): BSR201810272018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wang H, Ding Q, Wang M, Guo M and Zhao Q:
miR-29b inhibits the progression of multiple myeloma through
downregulating FOXP1. Hematology. 24:32–38. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ronchetti D, Manzoni M, Todoerti K, Neri A
and Agnelli L: In Silico characterization of miRNA and long
non-coding RNA interplay in multiple myeloma. Genes (Basel).
7(pii): E1072016. View Article : Google Scholar : PubMed/NCBI
|