Introduction
Articular cartilage tissue has a poor healing
potential. Partial-thickness defects of cartilage tissue do not
heal, and full-thickness defects are difficult to heal (1). Moreover, articular cartilage tissue
subjected to traumatic damage has a low healing potential, and
undergoes cartilage degeneration and osteoarthritis (2). Previous studies have focused on
developing methods to promote cartilage tissue healing or
regeneration (1,3–6). It
has been identified that the MRL/MpJ strain of mice can heal ear
hole punches seamlessly within a few weeks, and these mice can
undergo rapid repair of cartilage tissue damage compared with a
control strain, with increasing levels of angiogenesis, cell
proliferation and matrix formation (3).
The cyclin-dependent kinase inhibitor p21 was
initially identified as a potent inhibitor of cell cycle
progression (7). Moreover, p21
knockout (KO) induced a regenerative response following an ear
punch procedure in an appendage of the non-regenerating B6129SF2/J
mouse strain, which was comparable to MRL and B6 ear hole closure
(5). Previous studies have
reported that p21 regulates cytostasis and cell death (8,9).
Furthermore, p21 is activated by p53, and is involved in a negative
feedback mechanism that regulates p53 activity during apoptosis
(9,10).
Currently, the combination of mesenchymal stem cells
(11), growth factors (12) and 3D scaffolds by cartilage tissue
engineering (13) is the most
prevalent strategy for cartilage repair. However, gene therapy also
provides methods to stimulate molecules to enhance bone
regeneration or cartilage repair (14).
Bedelbaeva et al (5) identified that MRL cells do not
express p21, thus suggesting that deletion of p21 converts a
non-healer mouse into a regeneration-competent mouse. Moreover,
Arthur and Heber-Katz (6)
demonstrated that the deletion of p21 may enhance the regenerative
response by altering DNA damage and checkpoint responses via
various pathways, thus leading to enhanced proliferation. Deletion
of p21 further inhibited transforming growth factor-β signaling,
which leads to reduced scar formation and altered differentiation
patterns (6). In addition,
Premnath et al (14)
demonstrated that p21-/- mice exhibit enhanced bone regeneration
after injury (14). However, to
the best of our knowledge, there are no previous studies focusing
on cartilage regeneration in p21-/- mice.
Therefore, it was hypothesized that deletion of p21
may induce rapid healing in damaged articular cartilage. The
present study investigated the healing potential of damaged
articular cartilage to elucidate the mechanisms underlying p21
function and articular cartilage regeneration in vivo.
Materials and methods
Mouse strains and breeding
All procedures were approved by the Animal Studies
Committee at Kobe University. All mice were housed in cages in
pathogen-free conditions at 23±2°C with 55±10% humidity, 12-h
light/dark cycles and ad libitum access to water and standard
laboratory chow. Mice were bred in The Animal Facility at Kobe
University Graduate School of Medicine. A total of four p21 KO male
mice [age, 8 weeks; weight 22.8±0.8 g; B6.129S6
(Cg)-Cdkn1atm1Led/J] were obtained from The Jackson Laboratory.
Mice were backbred against a C56BL/6J (CLEA Japan, Inc.)
background, and a total of 126 male mice (age, 8 weeks; weight
21.4±0.7 g) were used in the experiments. All mice used in the
present study were backcrossed >10 generations. p21-/-
littermates were used as p21 KO mice and p21+/+ littermates were
used as wild-type (WT) controls. In total, six mice were used for
each experiment.
Mouse genotyping
Genotypes were assessed via PCR analysis of
tail-derived DNA. Genomic DNA was extracted using the DNeasy Blood
& Tissue kit (Qiagen, Inc.), according to the manufacturer's
protocol. p21 deletion was confirmed on the basis of a 447-bp
fragment unique to the mutant genotype, which was amplified with a
p21-specific forward primer (5′-GTTGTCCTCGCCCTCATCTA-3′) and
reverse primer (5′-CTGTCCATCTGCACGAGACTA-3′; sequences provided by
The Jackson Laboratory). WT alleles were confirmed on the basis of
a 240-bp fragment amplified with the WT reverse primer
(5′-GCCTATGTTGGGAAACCAGA-3′) and the aforementioned p21-specific
forward primer. The thermocycling conditions were as follows:
Initial denaturation at 94°C for 5 min, followed by 40 cycles at
94°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec; and a final
extension at 72°C for 2 min. KOD FX Neo (Toyobo Life Science) was
used as the DNA polymerase. The DNA products were run on a 2%
agarose gel. DNA bands were visualized using the Luminescent Image
Analyzer LAS-3000UV mini (Fujifilm Holdings Corporation; Fig. 1).
Articular cartilage repair model
The present study was carried out in strict
accordance with the recommendations of The Guide for the Care and
Use of Laboratory Animals of the National Institutes of Health. All
procedures were approved by The Animal Studies Committee of Kobe
University (approval no. P160309). p21 KO and WT mice (age, 8
weeks) were used in these experiments. Mice were anesthetized via
intraperitoneal administration of ketamine (100 mg/kg) and
sacrificed by cervical dislocation (9,15).
Full-thickness injuries were generated as previously described
(16). This surgical procedure was
a modified version of a previously reported full-thickness injury
model in mice (4). A medial
parapatellar skin incision >1 cm in length was made using a
microsurgical scalpel. The joint capsule was opened, and the
patella was dislocated laterally to expose the articular surface of
the trochlear groove. With the femoral epicondyles fixed with
surgical tweezers, a longitudinal full-thickness injury was made in
the patellar groove using the tip of the needle. Penetration to the
subchondral bone was confirmed by bleeding from the cartilage
injury site. After irrigation with normal saline to remove the
debris, the knee dislocation was reduced. The joint capsule and
skin were sutured in separate layers. Postoperatively, mice were
warmed until they recovered from anesthesia (16). All mice were subjected to
weight-bearing following post-anesthesia recovery. Mice were
sacrificed at two different time points (4 and 8 weeks) after
surgery, and the tissues were subjected to histological evaluation.
In total, 24 mice were used, with six mice in each group: p21 KO (4
and 8 weeks) and WT (4 and 8 weeks).
Histological evaluation for cartilage
repair
To investigate the in vivo function of p21 in
cartilage repair, histological analysis was performed 4 and 8 weeks
post-surgery in mouse models of full-thickness injuries at the
patellar groove.
Mouse knee joint tissue was fixed with 4%
paraformaldehyde at 4°C for 24 h, decalcified with 14% EDTA at 25°C
for 7 days and embedded in paraffin. Axial histological sections
(thickness, 6 µm) were obtained from the joint at 60-µm intervals,
and stained with Safranin O and Fast Green at 25°C for 5 min.
Cartilage repair was evaluated in accordance with the Wakitani
score (17). The maximum Wakitani
score is 14 points. The score measures cartilage defects based on
cell morphology, matrix staining, surface regularity, thickness of
cartilage and integration of donor with host (17). A higher Wakitani score corresponds
with increased cartilage damaged. Scoring was performed
independently by two blinded observers.
Immunohistochemical analysis
Deparaffinized sections (thickness, 6 µm) were
digested with proteinase (Dako Retrieval Solution Ready-to-Use;
Agilent Technologies, Inc.) for 20 min and treated with 3% hydrogen
peroxide (Wako Pure Chemical Industries, Ltd.) at 25°C for 10 min
to block endogenous peroxidase. Subsequently, the sections were
blocked with 10% FBS (Sigma-Aldrich; Merck KGaA) for 1 h at room
temperature. Sections were treated with anti- proliferating cell
nuclear antigen (PCNA; cat. no. ab18197; 1:100; Abcam), anti-Sox9
(cat. no. ab26414; 1:100; Abcam) and anti-type II collagen (cat.
no. ab34712; 1:50; Abcam) antibodies at 4°C overnight. Sections
were subsequently treated with peroxidase-labeled anti-rabbit IgG
(cat. no. 424141; Histofine Simple Stain MAX PO; Nichirei
Bioscience Corporation) at 25°C for 30 min. The signal was
developed as a brown reaction product using the peroxidase
substrate. Subsequently, sections were incubated with
3,3′-diaminobenzidine at 25°C for 2 min (Histofine Simple Stain DAB
Solution; Nichirei Bioscience Corporation) and tissue sections were
examined using a BZ-X700 fluorescent microscope (Keyence
Corporation). The average percentage of PCNA, Sox9 and type II
collagen positive cells/total cells was manually calculated. The
number of cells and the positive cell rate per visual field
centered on the defect were examined (magnification, ×400). For
each mouse, one section was selected and analyzed. Six mice were
used for each group, and one section per mouse was selected and
analyzed.
Cell culture
Mouse ATDC5 prechondrocytes are widely used as an
in vitro model of cartilage differentiation (18–20).
Therefore, ATDC5 cells were cultured in the absence of insulin, and
were induced to chondrogenic cells using differentiation medium
containing insulin as previously described (18–20).
For chondrogenic induction, ATDC5 cells (Riken Bioresource Center)
were cultured in a 1:1 mixture of DMEM and Ham's F12 medium (Flow
Laboratories, Ltd.) containing 5% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 10 µg/ml bovine insulin (Wako Pure Chemical
Industries, Ltd.), 10 µg/ml human transferrin (Roche Diagnostics
GmbH) and 3×108 M sodium selenite (Sigma-Aldrich; Merck KGaA) at
37°C in a humidified atmosphere of 5% CO2 for 3 weeks. Cells were
cultured at 2×104 cells/well in a 24-multiwell plate, 4×104
cells/well in a 12-multiwell plate or 6×104 cells/well in a
6-multiwell plate (Corning, Inc.). The medium was replaced every
alternate day (18).
Small interfering RNA (siRNA)
transfection
Lipofectamine® 2000 reagent (Thermo
Fisher Scientific, Inc.) was used to transfect siRNA targeting p21
(Silencer Select Pre-designed siRNA; ID s63813; Ambion; Thermo
Fisher Scientific, Inc.) and non-specific control siRNA (Silencer™
Select Negative Control No.1 siRNA; Invitrogen; Thermo Fisher
Scientific, Inc.) into ATDC5 cell monolayers, in accordance with
the manufacturer's instructions. In brief, 7 days before
transfection, cells were plated (6×104 cells/well) in a 6-well
plate in growth medium without antibiotics to attain 80–100%
confluence at the time of transfection. Subsequently, 100 pmol
siRNA and Lipofectamine® 2000 complexes were prepared
and added to each well. After 48 h of transfection, the complexes
were removed and cells were harvested.
Reverse transcription-quantitative PCR
(RT-qPCR)
Chondrocytes were cultured (6×104 cells/well) in
6-well plates, and RNA was extracted using a QIAshredder and RNeasy
Mini kit (Qiagen GmbH) in accordance with the manufacturer's
protocol. Then, 1 µg total RNA was reverse transcribed to
first-strand cDNA with 1.25 µM oligo(dT) primers (Invitrogen;
Thermo Fisher Scientific, Inc.) in 40 µl PCR buffer II [containing
2.5 mM MgC12, 0.5 mM deoxyribonucleotide triphosphate mix, 0.5 U
RNase inhibitor and 1.25 U of murine leukemia virus reverse
transcriptase (Applied Biosystems; Thermo Fisher Scientific, Inc.)]
at 42°C for 60 min.
Relative mRNA expression levels of p21, collagen
type II α 1 chain (Col2a1) and Sox9 were analyzed using the SYBR
Green RT PCR kit (Thermo Fisher Scientific, Inc.) using an ABI
Prism 7700 sequence detection system (Applied Biosystems; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
The following primer pairs were used for qPCR: GAPDH forward,
5′-AACTTTGGCATTGTGGAAGG-3′ and reverse, 5′-ACACATTGGGGGTAGGAACA-3′;
p21-specific forward, 5′-GACCTGGGAGGGGACAAGAG-3′ and reverse,
5′-TTCTCTTGCAGAAGACCAATC-3′; Col2a1-specific forward,
5′-ACTGGTGGAGCAGCAAGAGC-3′ and reverse, 5′-TCTGGACGTTAGCGGTGTTG-3′;
and sox9-specific forward, 5′-AATGCTATCTTCAAGGCGCTG-3′ and reverse,
5′-GGACCCTGCGATTGCCCAG-3′. The thermocycling conditions used for
qPCR were as follows: Initial denaturation at 50°C for 2 min and at
95°C for 10 min; followed by 40 cycles at 95°C for 15 sec and 60°C
for 1 min; and a melt curve at 95°C for 15 sec, 60°C for 1 min,
95°C for 30 sec and 60°C for 15 sec. Expression levels were
normalized to GAPDH using the comparative Cq method. The difference
between the mean Cq values of the gene of interest and the
housekeeping gene is denoted as ΔCq, and the difference between ΔCq
and the Cq value of the calibrator sample is denoted as ΔΔCq. The
log2 (ΔΔCq) value yields the relative level of gene expression
(21). Data are presented as the
average values of six individual experiments (9).
Statistical analysis
Statistical analysis was performed using one-way or
one-way repeated ANOVA, followed by Tukey's post hoc test for
multiple comparisons of paired samples. Mann-Whitney U test was
used for comparisons between two groups. P<0.05 was considered
to indicate a statistically significant difference. Data are
presented as the mean with 95% CI. Statistical analyses were
performed using the Bell Curve for Excel software (Social Survey
Research Information Co., Ltd.).
Results
Outcome of articular cartilage
repair
It was observed that p21 KO mice had enhanced
healing compared with WT mice at both 4 and 8 weeks post-surgery
(Fig. 2A-H). Furthermore, p21 KO
mice displayed increased cell accumulation around the defect site
at both 4 and 8 weeks post-surgery (Fig. 2C-H). Safranin O staining results
identified reduced staining in the repair area at the site of the
defect in WT mice at 4 weeks (Fig. 2A
and B). Moreover, p21 KO mice had a significantly lower
Wakitani score compared with KO mice (4 weeks: p21 KO, 9.2±2.0
points; WT, 13.7±0.5 points; P=0.005; 8 weeks: p21 KO, 4.3±1.1
points; WT, 8.2±2.0 points; P=0.007; Fig. 2I).
 | Figure 2.Outcome of articular cartilage repair.
WT 4 weeks (A) magnification, ×40 and (B) magnification, ×100. p21
KO 4 weeks (C) magnification, ×40 and (D) magnification, ×100. WT 8
weeks (E) magnification, ×40 and (F) magnification, ×100. p21 KO 8
weeks (G) magnification, ×40 and (H) magnification, ×100. Scale
bar, 200 µm. (I) Average Wakitani scores with 95% CIs. Low scores
indicate improved repair. Six mice were analyzed in each group.
*P<0.05. WT, wild-type; KO, knockout. |
p21 KO mice have more PCNA-positive
cells compared with WT mice
To investigate the in vivo function of p21 in
cell proliferation, immunohistochemical analysis of cartilage
tissue was performed. Analysis of PCNA expression identified
greater cell accumulation in p21 KO mice compared with WT mice (p21
KO, 112.3±9.0 cells/magnification, ×200; WT, 61.2±4.7
cells/magnification, ×200; P=0.005 at 4 weeks; p21 KO, 102.3±8.8
cells/magnification, ×200; WT, 54.8±3.0 cells/magnification, ×200;
P=0.005 at 8 weeks). In addition, it was demonstrated that the
percentage of immunostaining-positive cells was significantly
higher in p21 KO mice compared with WT mice at each time point (p21
KO, 56.9±2.2%/magnification, ×200; WT, 33.8±3.2%/magnification,
×200; P=0.005 at 4 weeks; p21 KO, 53.1±2.4%/magnification, ×200;
WT, 27.8±6.4%/magnification, ×200; P=0.005 at 8 weeks; Fig. 3A and B).
Col2a1 and Sox9 expression levels are
similar in p21 KO and WT mice, but cell accumulation is greater in
p21 KO mice
To investigate the in vivo function of p21 in
extracellular matrix (ECM) production, immunohistochemical analysis
of cartilage tissue was performed. While the rate of positive
immunostaining was comparable between p21 KO and WT mice (p21 KO,
51.4±2.9%/x200 magnification; WT, 49.8±1.6%/x200 magnification;
P=0.230 at 4 weeks; p21 KO, 29.9±3.3%/x200 magnification; WT,
27.8±1.5%/x200 magnification; P=0.298 at 8 weeks), Col2a1 was
expressed around the defect site. Moreover, it was found that cell
accumulation was greater in p21 KO mice compared with WT mice (p21
KO, 109.5±7.5 cells/x200 magnification; WT, 59.5±6.4 cells/x200
magnification; P=0.005 at 4 weeks; p21 KO, 109.0±6.3 cells/x200
magnification; WT, 55.0±4.5 cells/x200 magnification; P=0.005 at 8
weeks; Fig. 3C and D).
In addition, it was demonstrated that the
Sox9-positive cell rate was similar between p21 KO and WT mice (p21
KO, 45.6±4.3%/magnification, ×200; WT, 45.6±3.1%/magnification,
×200; P=0.689 at 4 weeks; p21 KO, 46.1±2.9%/magnification, ×200;
WT, 43.5±3.5%/magnification, ×200; P=0.379 at 8 weeks). However,
the present results suggested that cell accumulation was
significantly greater in p21 KO mice compared with WT mice (p21 KO,
110.2±8.6 cells/magnification, ×200; WT, 61.5±6.1
cells/magnification, ×200; P=0.005 at 4 weeks; p21 KO, 119.7±8.0
cells/magnification, ×200; WT, 56.5±6.4 cells/magnification, ×200;
P=0.005 at 8 weeks; Fig. 3E and
F).
Gene expression during cartilage
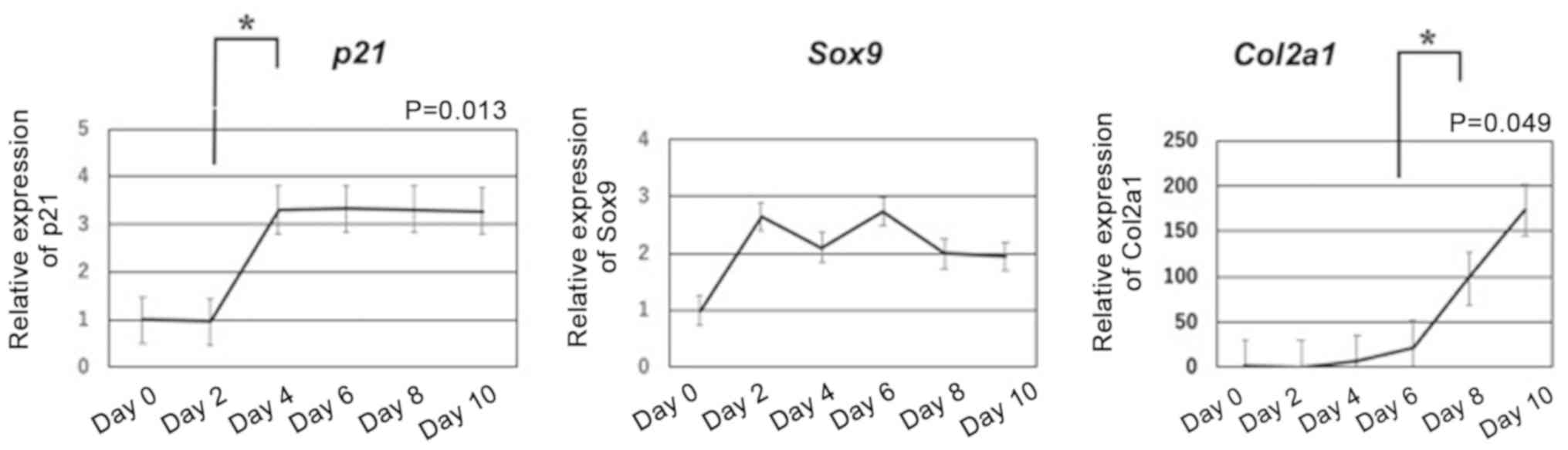
differentiation in ATDC5 cells
To evaluate gene expression during cartilage
differentiation in ATDC5 cells, the present study assessed p21,
Sox9 and Col2a1 mRNA expression levels. It was identified that p21
was significantly upregulated from day 2 to day 4, and then levels
stabilized (P=0.013). Furthermore, Sox9 expression in ATDC5 cells
was similar to baseline levels. In addition, the present results
indicated that Col2a1 was significantly upregulated in ATDC5 cells
from day 6 (P=0.049; Fig. 4).
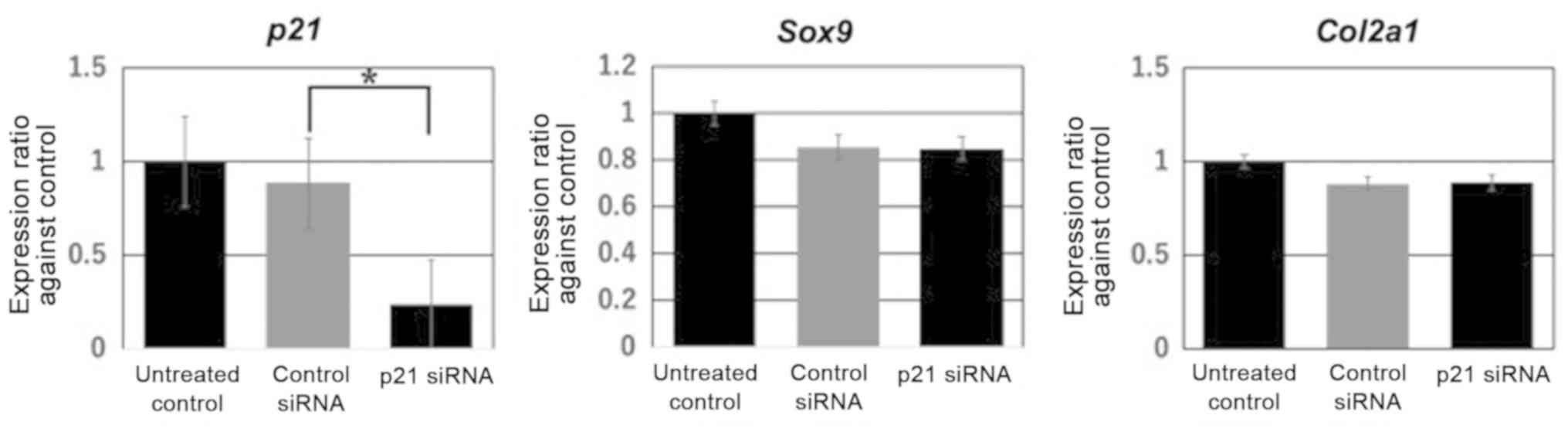
p21 downregulation does not affect
Sox9 and Col2a1 expression levels in vitro
To assess the function of p21 during cartilage
differentiation in ATDC5 cells, the present study quantified Sox9
and Col2a1 expression levels in p21-KO ATDC5 cells. RT-qPCR results
identified that p21 was downregulated to 23.3% after transfection
with p21-siRNA in comparison with non-specific control siRNA
(Fig. 5). However, it was
demonstrated that p21 downregulation did not affect Sox9 and Col2a1
expression levels (Sox9, P=0.89; Col2a1, P=0.51; Fig. 5).
Discussion
The present results suggested that p21 KO enhanced
the healing of damaged articular cartilage, with regards to cell
proliferation, in an in vivo experimental mouse model.
Furthermore, it was identified that p21 deficiency increased
chondrocyte proliferation, but not differentiation in
vitro.
Matsuoka et al (16) established a novel mouse model of
cartilage repair using C57Bl/6 mice, and induced full-thickness
injuries at the patellar groove in 3-week-old (young), 4-week-old
(juvenile) and 8-week-old (adult) C57Bl/6 mice. However, the
present results indicated that healing of damaged cartilage was
more rapid in p21 KO mice compared with C57Bl/6 mice.
p21 is known to regulate the cell cycle (7). Bedelbaeva et al (5) demonstrated that p21 plays a role in
cell cycle regulation and tissue regeneration. Moreover, Bedelbaeva
et al (5) demonstrated that
p21 deficiency enhances cell cycle progression from G1 to G2
checkpoint, increases cell cycle profiles, induces aberrant entry
into S-phase and enhances proliferation. Therefore, DNA damage is
increased in cells lacking p21, possibly as a result of
proliferative and replicative stress. The inability to enter
quiescence at the G0 stage in response to stress leads to G2
arrest, which was also observed in MRL mice (5). The present study identified cartilage
repair in p21 KO mice, with elevated PCNA levels and increased cell
accumulation, compared with WT mice at each time point. PCNA
expression is often distributed in the S-phase (22), thus potentially leading to aberrant
entry into the S-phase. Therefore, the present results suggested
that p21 deficiency upregulates PCNA.
The present results demonstrated that Col2a1 and
Sox9 expression levels remained unchanged in p21 KO and WT mice.
However, the number of immunostaining-positive cells was increased
in vivo in p21 KO mice. Furthermore, it was identified that
p21 was expressed in the early phase (days 2–4) during cartilage
differentiation in ATDC5 cells. Sox9 is a master gene in cartilage
differentiation (23–25), and the present results suggested
that it was upregulated immediately after induction of
differentiation, and its expression remained unchanged 8 days after
induction. Moreover, it was demonstrated that Col2a1 was gradually
upregulated after the induction of differentiation until day 10. In
addition, the present results suggested that p21 downregulation did
not affect Sox9 or Col2a1 expression levels in vitro, which
is in line with previous studies (9,26).
Our previous study also showed that downregulation of p21 does not
change aggrecan expression (9).
Moreover, p21 deficiency does not affect cell morphology, ECM
formation, chondrogenic marker protein expression, chondrocyte
proliferation or cell cycle regulators during embryonic
endochondral ossification in mice (26). It has been demonstrated that p21-/-
mice have enhanced bone-healing capabilities (14). However, the differences observed
may not result from the function of osteoblasts or osteoclasts.
Furthermore, no differences were observed in the differentiation of
progenitor cells, mesenchymal or monocytic, into osteoblasts or
osteoclasts (14). Therefore,
these previous studies support the present results, whereby p21 did
not affect the expression levels of Sox9 or Col2a1 during cartilage
differentiation in the repair of cartilage defects. However, the
present results suggested that p21 deficiency increased chondrocyte
proliferation. Thus, p21 may have functions under stress such as
mechanical loading (15), but does
not have a role in the absence of mechanical stress such as
embryonic environments (26).
A limitation of the present study is the lack of
direct evidence on the mechanisms of injury healing in p21 KO mice.
Therefore, chondrocyte proliferation assays or lineage reporter
mice are required to identify specific cell types after injury to
address this issue.
In conclusion, the present results suggested that
p21 deficiency enhanced healing in damaged articular cartilage
in vivo, in relation to cell proliferation, and increased
chondrocyte proliferation, but not differentiation in
vitro.
Acknowledgements
The authors would like to thank Ms. Minako Nagata
and Ms. Maya Yasuda (Department of Orthopedic Surgery, Kobe
University Graduate School of Medicine) for their technical
assistance.
Funding
The present study was supported by the Japan Society
for the Promotion of Science (grant no. 23791640).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SHay designed the study. KI, SHay, SK, KT, MH, NK
and SHas performed the experiments. KI, SHay, TN and RK analyzed
and interpreted the data. KI drafted the manuscript. RK and SHay
critically reviewed the manuscript for important intellectual
information. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was carried out in strict
accordance with the recommendations of The Guide for the Care and
Use of Laboratory Animals of the National Institutes of Health. All
procedures were approved by The Animal Studies Committee of Kobe
University (approval no. P160309).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rai MF and Sandell LJ: Regeneration of
articular cartilage in healer and non-healer mice. Matrix Biol.
39:50–55. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Buckwalter JA, Rosenberg LC and Huniker
EB: Articular cartilage: composition, structure, response to
injury, and methods of facilitating repair. Articular Cartilage and
Knee Joint Function: Basic Science and Arthroscopy. Ewing JW: Raven
Press Ltd.; New York, NY: 1990
|
|
3
|
Fitzgerald J, Rich C, Burkhardt D, Allen
J, Herzka AS and Little CB: Evidence for articular cartilage
regeneration in MRL/MpJ mice. Osteoarthritis Cartilage.
16:1319–1326. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eltawil NM, De Bari C, Achan P, Pitzalis C
and Dell'accio F: A novel in vivo murine model of cartilage
regeneration. Age and strain-dependent outcome after joint surface
injury. Osteoarthritis Cartilage. 17:695–704. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bedelbaeva K, Snyder A, Gourevitch D,
Clark L, Zhang XM, Leferovich J, Cheverud JM, Lieberman P and
Heber-Katz E: Lack of p21 expression links cell cycle control and
appendage regeneration in mice. Proc Natl Acad Sci USA.
107:5845–5850. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Arthur LM and Heber-Katz E: The role of
p21 in regulating mammalian regeneration. Stem Cell Res Ther.
2:302011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harper JW, Adami GR, Wei N, Keyomarsi K
and Elledge SJ: The p21 Cdk-interacting protein Cip1 is a potent
inhibitor of G1 cyclin-dependent kinases. Cell. 75:805–816. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki A, Tsutomi Y, Akahane K, Araki T
and Miura M: Resistance to Fas-mediated apoptosis: Activation of
caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene
family ILP. Oncogene. 17:931–939. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hayashi S, Fujishiro T, Hashimoto S,
Kanzaki N, Chinzei N, Kihara S, Takayama K, Matsumoto T, Nishida K,
Kurosaka M, et al: p21 deficiency is susceptible to osteoarthritis
through STAT3 phosphorylation. Arthritis Res Ther. 17:3142015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Seoane J, Le HV and Massagué J: Myc
suppression of the p21(Cip1) Cdk inhibitor influences the outcome
of the p53 response to DNA damage. Nature. 419:729–734. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kondo S, Nakagawa Y, Mizuno M, Katagiri K,
Tsuji K, Kiuchi S, Ono H, Muneta T, Koga H and Sekiya I:
Transplantation of aggregates of autologous synovial mesenchymal
stem cells for treatment of cartilage defects in the femoral
condyle and the femoral groove in microminipigs. Am J Sports Med.
47:2338–2347. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qasim M, Chae DS and Lee NY:
Bioengineering strategies for bone and cartilage tissue
regeneration using growth factors and stem cells. J Biomed Mater
Res A. 108:394–411. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nakagawa Y, Fortier LA, Mao JJ, Lee CH,
Goodale MB, Koff MF, Uppstrom TJ, Croen B, Wada S, Carballo CB, et
al: Long-term evaluation of meniscal tissue formation in
3-dimensional-printed scaffolds with sequential release of
connective tissue growth factor and TGF-β3 in an ovine model. Am J
Sports Med. 47:2596–2607. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Premnath P, Jorgenson B, Hess R, Tailor P,
Louie D, Taiani J, Boyd S and Krawetz R: p21-/- mice exhibit
enhanced bone regeneration after injury. BMC Musculoskelet Disord.
18:4352017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kihara S, et al: Cyclin-dependent kinase
inhibitor-1-deficient mice are susceptible to osteoarthritis
associated with enhanced inflammation. JMBR. 32:991–1001. 2017.
|
|
16
|
Matsuoka M, Onodera T, Sasazawa F, Momma
D, Baba R, Hontani K and Iwasaki N: An articular cartilage repair
model in common C57Bl/6 Mice. Tissue Eng Part C Methods.
21:767–772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wakitani S, Goto T, Pineda SJ, Young RG,
Mansour JM, Caplan AI and Goldberg VM: Mesenchymal cell-based
repair of large, full-thickness defects of articular cartilage. J
Bone Joint Surg Am. 76:579–592. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shukunami C, Shigeno C, Atsumi T, Ishizeki
K, Suzuki F and Hiraki Y: Chondrogenic differentiation of clonal
mouse embryonic cell line ATDC5 in vitro: Differentiation-dependent
gene expression of parathyroid hormone (PTH)/PTH-related peptide
receptor. J Cell Biol. 133:457–468. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shukunami C, Ishizeki K, Atsumi T, Ohta Y,
Suzuki F and Hiraki Y: Cellular hypertrophy and calcification of
embryonal carcinoma-derived chondrogenic cell line ATDC5 in vitro.
J Bone Miner Res. 12:1174–1188. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Atsumi T, Miwa Y, Kimata K and Ikawa Y: A
chondrogenic cell line derived from a differentiating culture of
AT805 teratocarcinoma cells. Cell Differ Dev. 30:109–116. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schönenberger F, Deutzmann A, Ferrando-May
E and Merhof D: Discrimination of cell cycle phases in
PCNA-immunolabeled cells. BMC Bioinformatics. 16:1802015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bi W, Deng JM, Zhang Z, Behringer RR and
de Crombrugghe B: Sox9 is required for cartilage formation. Nat
Genet. 22:85–89. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Akiyama H, Chaboissier MC, Martin JF,
Schedl A and de Crombrugghe B: The transcription factor Sox9 has
essential roles in successive steps of the chondrocyte
differentiation pathway and is required for expression of Sox5 and
Sox6. Genes Dev. 16:2813–2828. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ng LJ, Wheatley S, Muscat GE,
Conway-Campbell J, Bowles J, Wright E, Bell DM, Tam PP, Cheah KS
and Koopman P: SOX9 binds DNA, activates transcription, and
coexpresses with type II collagen during chondrogenesis in the
mouse. Dev Biol. 183:108–121. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chinzei N, Hayashi S, Hashimoto S, Kanzaki
N, Iwasa K, Sakata S, Kihara S, Fujishiro T, Kuroda R and Kurosaka
M: Cyclin dependent kinase inhibitor p21 does not impact embryonic
endochondral ossification in mice. Mol Med Rep. 11:1601–1608. 2015.
View Article : Google Scholar : PubMed/NCBI
|