Introduction
Renal cell carcinoma (RCC), which originates from
renal parenchyma urinary tubular epithelial cells, is one of the
most common malignant tumors of the urinary system (1–3).
Clear cell RCC (ccRCC) accounts for ~80% of all RCC cases (1,3–6). As
ccRCC is not sensitive to chemoradiotherapy (7), surgical resection is the main
treatment strategy for the disease (1–3,8).
However, the rate of recurrence and distant metastasis remains as
high as ~30% (3,5,8).
Therefore, the identification of novel therapeutic targets is
urgently required to prevent the progression of ccRCC.
Increasing evidence suggests that long non-coding
RNAs (lncRNAs) are involved in the occurrence and progression of
ccRCC (2–4,6).
lncRNAs, defined as transcribed RNA molecules >200 nt in length
(9–12), are an important class of non-coding
RNAs involved in several biological functions (12–17).
Antisense lncRNAs (AS lncRNAs), a subclass of
lncRNAs, are transcribed from complex genetic loci on the opposite
strands of sense protein-coding genes (13–15,18–20).
AS lncRNAs may overlap exons and/or introns of their associated
sense protein-coding transcripts to regulate epigenetic silencing,
transcription and mRNA stability by forming sense-antisense pairs
(18–24). The genomic arrangement of AS lncRNA
genes also suggests that they may be involved in pathways that
allow genes to regulate their own expression (23).
In the present study, RNA-sequencing (seq) was
applied to detect changes in the transcriptome of five paired
surgically resected ccRCC and para-cancerous (PC) tissues. The five
AS lncRNAs with the most significant differences in expression were
selected and AS lncRNA ZNF710-AS1-202 was subsequently chosen for
further experimentation. ZNF710-AS1-202 is transcribed from the
ZNF710-AS1 gene, which is antisense to zinc finger protein 710
(ZNF710). The ZNF710-AS1 gene has two transcripts, ZNF710-AS1-201
and ZNF710-AS1-202. The ZNF710-AS1-202 transcript is wholly mapped
to the fourth intron of the ZNF710-201 transcript. The present
study revealed that ZNF710-AS1-202 negatively regulated ZNF710 mRNA
expression and positively regulated ZNF710 protein expression.
Furthermore, Uniprot analysis revealed that ZNF710 is involved in
transcription regulation.
Reverse transcription-quantitative PCR (RT-q)PCR
verified that ZNF710-AS1-202 expression was significantly decreased
in ccRCC tissues and was associated with the pathological grade,
tumor size, local invasion and TNM stage, suggesting that
ZNF710-AS1-202 exhibited antitumor effects. However, ZNF710-AS1-202
overexpression significantly increased the proliferation of RCC
cells. Opposite results were observed when ZNF710-AS1-202 was
knocked down by small interfering (si) RNA. Furthermore, the levels
of ZNF710-AS1-202 expression in ccRCC cells was significantly
increased compared with in normal cells.
Materials and methods
Patient samples
A total of 34 pairs of ccRCC tissues and
corresponding PC tissues, as well an additional 12 ccRCC tissues,
were obtained from surgeries performed between May 2016 and
December 2018 at the First Affiliated Hospital of Zhengzhou
University (Zhengzhou, China). The age of the included patients
ranged from 28–72 years old, and the ratio of male to female was
32:14. All samples were immediately frozen in liquid nitrogen and
stored at −80°C until RNA extraction. The postoperative pathology
of all tumor specimens in the present study was ccRCC. Tumor
specimens whose postoperative pathological results were not ccRCC
were excluded along with their corresponding adjacent non-tumorous
tissues.
Databases
Ensembl database (http://asia.ensembl.org/index.html) was used to query
the gene sequences of znf710-as1 and ZNF710. The GEPIA website
(http://gepia2.cancer-pku.cn/#analysis) was used to
analyze TCGA data using its preset program. Moreover, the Uniprot
database (https://www.uniprot.org/) was used to
search for information on the ZNF710 protein.
Gene expression profile analysis
A total of five paired ccRCC and PC tissues were
analyzed by high throughout RNA-seq. A paired Student's t-test was
used to compare the AS lncRNA levels between ccRCC and PC
tissues.
Cell culture and reagents
The human RCC cell lines (786-O, ACHN and 769-P) and
the human renal tubular epithelial cell line (HK-2) were obtained
from The Cell Bank of the Type Culture Collection of the Chinese
Academy of Sciences. 786-O and 769-P cells were cultured in
RPMI-1640 medium (GE Healthcare Life Sciences) supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.). ACHN cells were cultured in Dulbecco's modified Eagle's
medium (DMEM; GE Healthcare Life Sciences) containing 10% FBS. HK-2
cells were cultured in DMEM/F12 medium (GE Healthcare Life
Sciences) with 10% FBS. Penicillin (100 U/ml) and streptomycin (100
µg/ml) were used in the culture of all of these cell lines. All
cells were maintained at 37°C in a humidified atmosphere containing
5% CO2.
Cell transfection
pcDNA3.1-ZNF710-AS1-202 and pcDNA3.1-NC vectors were
designed and synthesized by Hanbio Biotechnology Co., Ltd. and
transfected into 786-O and ACHN cells using Lipofectamine™ 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) adding 2.5 µg plasmids
into each well of the 6-well plates or 0.1 µg per well of the
96-well plates according to the manufacturer's protocol. Small
interfering (si)RNAs were designed and synthesized by Guangzhou
RiboBio Co., Ltd. Then, 75 pmol siRNAs were added into each well of
the 6-well plates and 3 pmol each well of the 96-well plates,
according to the manufacturer's protocol of Lipofectamine™ 3000.
The following sequences were used: SiZNF710-AS1-202#1,
5′-CCCAACATGCAGCTGATTT-3′; SiZNF710-AS1-202#2,
5′-TCCCATACACTTATTTGAA-3′; and Si negative control,
5′-TTCTCCGAACGTGTCACGT-3′. Cells were harvested for RNA and protein
extraction 48 and 72 h after transfection, respectively.
RNA extraction and strand-specific
RT-qPCR
Total RNA from ccRCC tissues and cells was extracted
using TRIzol® reagent (Beijing Leagene Biotech Co.,
Ltd.) according to the manufacturer's protocol. Total RNA was
treated with gDNA Remover (Toyobo Life Science) at 37°C for 15 min
and then reverse transcribed into cDNA separately with
strand-specific primers (25)
using the ReverTra Ace qPCR RT Master Mix (Toyobo Life Science)
with the following conditions: 37°C for 15 min, and 98°C for 5 min.
qPCR was performed using a SYBR Green Mix (Roche Diagnostics) and a
QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, Inc.)
in a 20 µl reaction mixture using the following conditions: Initial
denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for
10 sec, 60°C for 10 sec and 72°C for 45 sec, then followed by one
cycle of 95°C for 15 sec, 60°C for 1 min. The heating rate of all
the above steps was 1.6°C/sec. Then, the temperature was increased
to 95°C at 0.15°C/sec from 60°C, remaining at 95°C for 1 sec. The
following primer pairs were used for qPCR: ZNF710-AS1-202 forward,
5′-CTTCCACTACCGCAGCCAGTTG-3′ and reverse,
5′-CCGCTTCATGTTCGCCTGTAGG-3′; ZNF710-AS1-201 forward,
5′-AAAGACGTGGGAAGAGGTAGTTG-3′ and reverse,
5′-GGCAGCCGGAAAGATAAGG-3′; ZNF710 forward,
5′-TTCCACTACCGCAGCCAGTT-3′ and reverse,
5′-GCTCAGGTTGCCCTTGAGATT-3′; β-actin forward,
5′-CCTGGCACCCAGCACAAT-3′ and reverse, 5′-GGGCCGGACTCGTCATAC-3′; and
GAPDH forward, 5′-CAGGAGGCATTGCTGATGAT-3′ and reverse,
5′-GAAGGCTGGGGCTCATTT-3′. mRNA levels were quantified using the
2−ΔΔCq method (26) and
normalized to the internal reference genes GAPDH or β-actin.
Western blot analysis
RCC cells were lysed with RIPA buffer (Beyotime
Institute of Biotechnology) and protein concentrations were
determined with a bicinchoninic acid protein assay kit (Beijing
Leagene Biotech Co., Ltd.). Protein samples (25 µg/lane) were
separated by SDS-PAGE on 12% gels and transferred onto PVDF
membranes. The membranes were blocked by 10% skim milk for 1 h at
25°C and then were incubated with primary antibodies against ZNF710
(cat. no. bs-4373R; 1:1,000; BIOSS), cyclin B1 (cat. no. ab32053;
1:10,000; Abcam) and β-actin (cat. no. ab8227; 1:5,000; Abcam)
overnight at 4°C. Following primary antibody incubation, the
membranes were incubated with a horseradish peroxidase-conjugated
anti-rabbit secondary antibody (cat. no. ab6721; 1:10,000; Abcam)
at 25°C for 1 h. Protein bands were subsequently visualized using
an enhanced chemiluminescence reagent kits (Beyotime Institute of
Biotechnology) and scanned by an imaging system (Bio-Rad
Laboratories, Inc.). The densitometry was quantified using ImageJ
software v1.8.0 (National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
RCC cells were seeded in 96-well plates at a density
of 3,000 cells/well in sextuplicate and cultured in 100 µl complete
media with 10% FBS. Cells were subsequently transfected with the
plasmid vectors or siRNAs using Lipofectamine™ 3000 according to
the manufacturer's protocol. At the indicated time points (0, 24,
48, 72, 96, 120 and 144 h post-transfection), 10 µl CCK-8 reagent
(Dojindo Molecular Technologies, Inc.) was added to each well and
incubated for 90 min according to the manufacturer's instructions.
Then, the absorbance at a wavelength of 450 nm was measured using a
microplate reader.
Cell cycle analysis
Cells were collected 48 h after transfection and
washed twice with PBS. The cells were fixed with ice-cold 75%
ethanol and incubated at 4°C overnight. Fixed cells then were
washed twice with PBS and incubated with 500 µl propidium iodide
(PI) solution with RNase A (BD Biosciences; Becton, Dickinson and
Company) for 30 min at room temperature in the dark. The cells were
subsequently analyzed using a flow cytometer (FACScan®;
BD Biosciences; Becton, Dickinson and Company) and data were
processed with ModFit LT 5.0 (Verity Software House).
Apoptosis assay
The cells were trypsinized without EDTA and washed
twice with cold PBS. Subsequently, the cells were stained using the
Annexin V-FITC/PI kit (BD Biosciences; Becton, Dickinson and
Company) at room temperature for 15 min in the dark according to
the manufacturer's protocol. The cells were subsequently analyzed
using a flow cytometer and data were processed using FlowJo
software (version 10; FlowJo LLC).
RNA fluorescent in situ hybridization
(FISH)
The Cy3-labeled ZNF710-AS1-202 probes were designed
and synthesized by Guangzhou RiboBio Co., Ltd. The probes were ~27
nucleotides in length. ZNF710-AS1-202 was hybridized in situ
with the Cy3-labeled ZNF710-AS1-202 probes in RCC cells according
to the manufacturer's protocol of Ribo™ Fluorescent In Situ
Hybridization kit (RiboBio Co., Ltd.). RCC cells were fixed in 4%
paraformaldehyde at room temperature for 10 min. For
permeabilization, RCC cells were incubated in 1X PBS with 0.5%
Triton X-100 at 4°C for 5 min. For pre-hybridization, RCC cells
were incubated in pre-hybridization buffer at 37°C for 30 min.
Enhanced antifade mounting medium (Beijing Leagene Biotech Co.,
Ltd.) was used at room temperature in the dark after hybridization,
according to the manufacturer's protocol. Subcellular localization
of ZNF710-AS1-202 was detected by laser scanning confocal
microscopy (Carl Zeiss AG; ZEN System).
Statistical analysis
Data are expressed as the mean ± standard deviation
of three independent experiments. Statistical analyses were
performed using GraphPad Prism software (version 8.0; GraphPad
Software, Inc.). The groups were analyzed using the Student's
t-test, Mann Whitney test, one-way analysis of variance followed by
Student-Neuman-Keuls test, the χ2 test, or the log-rank
test as applicable. P<0.05 was considered to indicate a
statistically significant difference.
Results
ZNF710-AS1-202 is downregulated in
ccRCC tissues and is associated with clinicopathological
features
RNA-seq was used to detect changes in the
transcriptome of ccRCC and PC tissues. The five AS lncRNAs with the
most significant differences in expression were identified
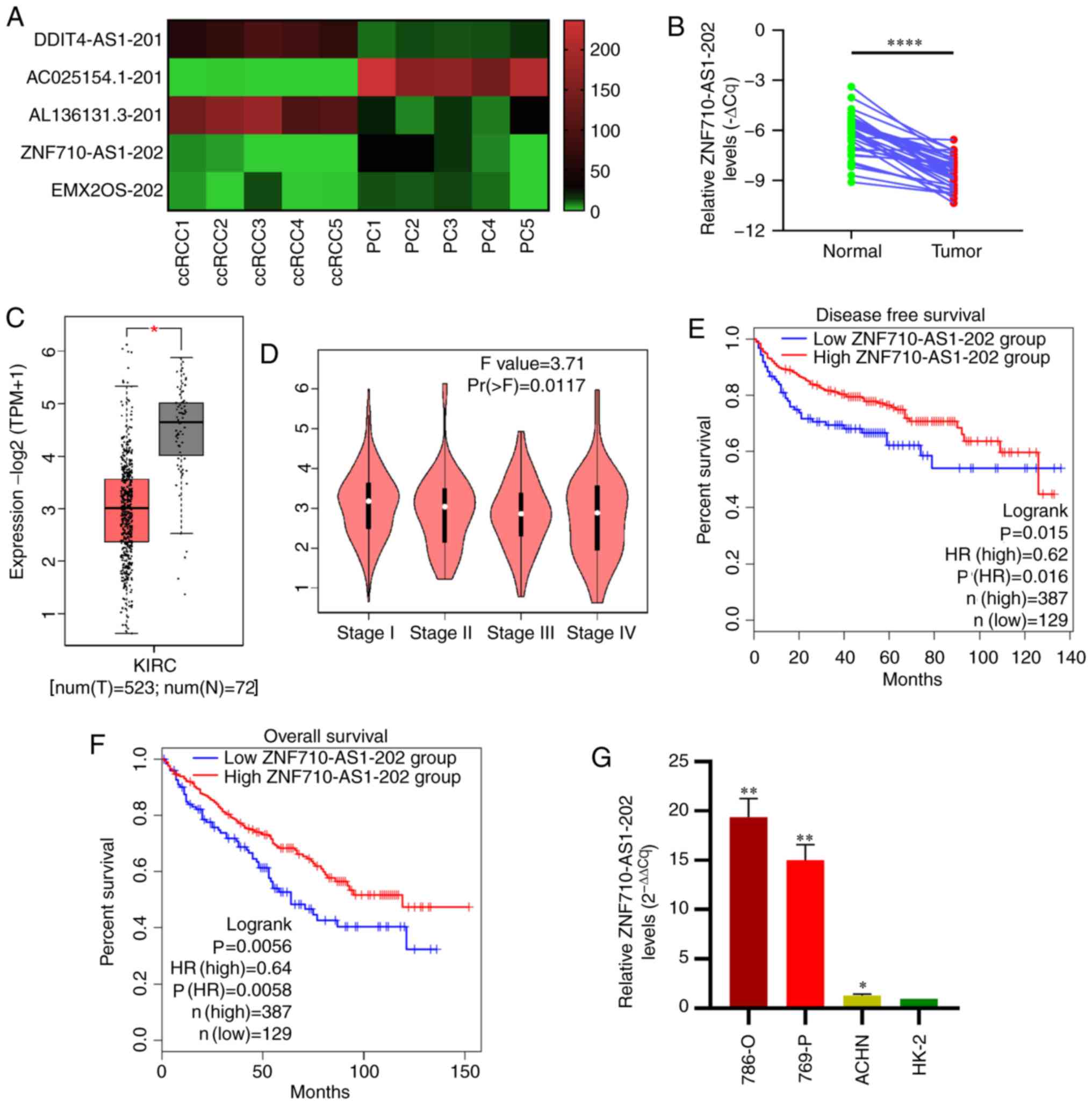
(Fig. 1A) and lncRNA
ZNF710-AS1-202 was subsequently selected for further
experimentation.
ZNF710-AS1-202 expression was analyzed in a cohort
of 34 pairs of ccRCC tissues and corresponding PC tissues as well
an additional 12 ccRCC tissues using RT-qPCR. ZNF710-AS1-202 was
significantly downregulated in human ccRCC tissues (n=34) compared
with their corresponding PC tissues (n=34), which was consistent
with the results of RNA-seq and The Cancer Genome Atlas (TCGA)
analysis (Fig. 1A-C). Furthermore,
a total of 46 human ccRCC tissue samples was used for the analysis
between clinicopathological data and expression level of
ZNF710-AS1-202. As the results show, the levels of ZNF710-AS1-202
were associated with the pathological grade, tumor size, local
invasion and TNM stage, but not with lymph node metastasis or
distant metastasis (Table I).
These results were validated with TCGA data (27) (Fig.
1D). What's more, TCGA analysis revealed that the overall
survival time as well as the disease-free survival time of patients
with higher ZNF710-AS1-202 levels tended to be longer than that of
patients with lower ZNF710-AS1-202 levels (Fig. 1E and F). However, the expression
levels of ZNF710-AS1-202 were higher in the ccRCC cell lines
compared with the HK-2 cells in vitro (Fig. 1G).
 | Table I.Relevance analysis of
LncZNF710-AS1-202 expression in patients with ccRCC. |
Table I.
Relevance analysis of
LncZNF710-AS1-202 expression in patients with ccRCC.
|
|
|
LncZNF710-AS1-202 |
|
|---|
|
|
|
|
|
|---|
| Variable | Patients | Low | High | P-value |
|---|
| Cases | 46 | 23 | 23 |
|
| Age, years |
|
|
| 0.536 |
|
<60 | 30 | 16 | 14 |
|
|
≥60 | 16 | 7 | 9 |
|
| Sex |
|
|
| 1 |
|
Male | 32 | 16 | 16 |
|
|
Female | 14 | 7 | 7 |
|
| Tumor diameter,
cm |
|
|
| 0.044a |
|
<5 | 12 | 3 | 9 |
|
| ≥5 | 34 | 20 | 14 |
|
| T Stage |
|
|
| 0.018a |
| T1 | 22 | 7 | 15 |
|
|
T2-4 | 24 | 16 | 8 |
|
| N Stage |
|
|
| 0.369 |
| N0 | 27 | 12 | 15 |
|
| N1,
Nx | 19 | 11 | 8 |
|
| M Stage |
|
|
| 0.475 |
| M0 | 36 | 17 | 19 |
|
| M1 | 10 | 6 | 4 |
|
| TNM Stage |
|
|
| 0.006a |
| I | 17 | 4 | 13 |
|
|
II–IV | 29 | 19 | 10 |
|
| Grade |
|
|
| 0.044a |
| I | 12 | 3 | 9 |
|
|
II–IV | 34 | 20 | 14 |
|
ZNF710-AS1-202 overexpression promotes
ccRCC cell proliferation and survival, and inhibits apoptosis
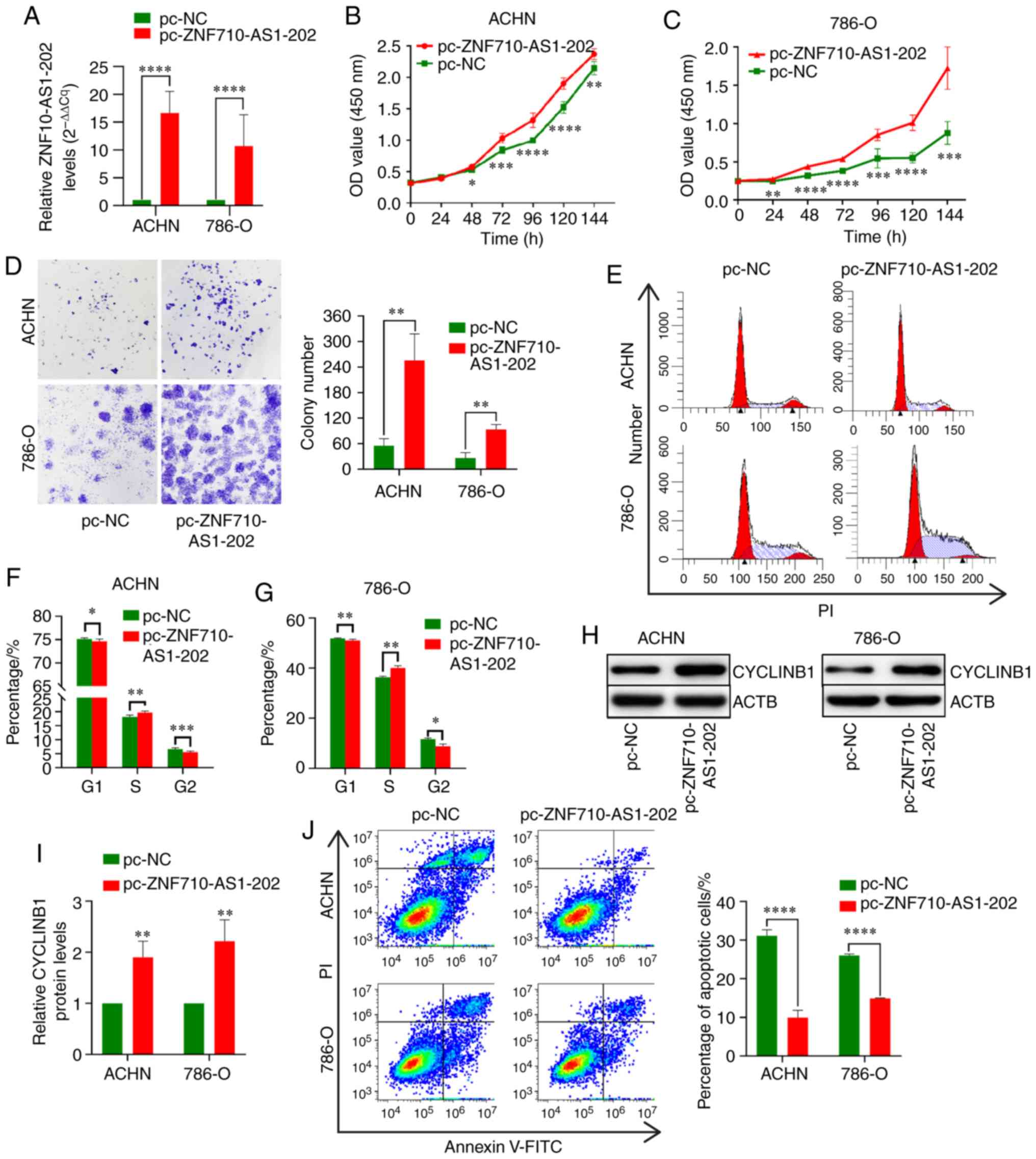
RT-qPCR analysis revealed that ZNF710-AS1-202
expression was significantly increased after transient transfection
with pcDNA3.1-ZNF710-AS1-202 compared with the control cells
(Fig. 2A). The CCK-8 assay
revealed that the proliferation of ACHN and 786-O cells transfected
with pc-ZNF710-AS1-202 was significantly increased compared with
the control cells (Fig. 2B and C).
Similarly, ZNF710-AS1-202 overexpression significantly enhanced the
colony formation ability of RCC cells (Fig. 2D). A significant and reproducible
increase in the S phase and a decrease in the G2/M phase were
observed following ZNF710-AS1-202 overexpression in RCC cells
(Fig. 2E-G) (20). Additionally, western blot analysis
revealed a significant increase in cyclin B1 expression in ACHN and
786-O cells overexpressing ZNF710-AS1-202 (Fig. 2H and I). This suggested that
ZNF710-AS1-202 overexpression promoted cell proliferation.
Moreover, ACHN and 786-O cells transfected with the ZNF710-AS1-202
overexpression plasmid exhibited lower levels of apoptosis compared
with the control cells (Fig.
2J).
ZNF710-AS1-202 is mainly expressed in
the cytoplasm of RCC cells
ZNF710-AS1-202 transcripts were mainly expressed in
cytoplasm of ACHN, 786-O and 769-P cells (Fig. 3).
Depletion of ZNF710-AS1-202 inhibits
ccRCC cell proliferation and survival, and promotes apoptosis
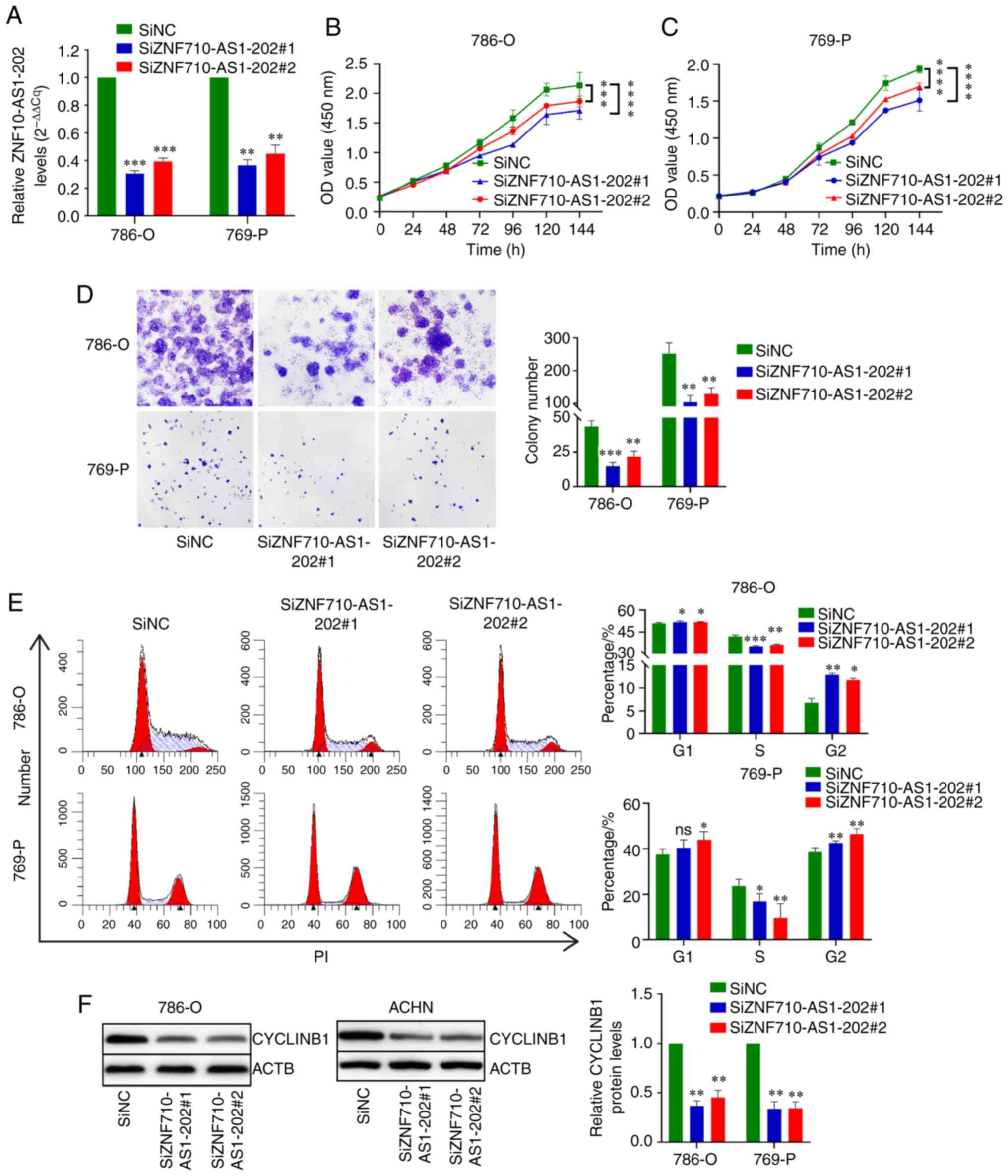
After siZNF710-AS1-202 transfection, ZNF710-AS1-202
expression was significantly decreased compared with control cells
(Fig. 4A). Furthermore,
ZNF710-AS1-202 knockdown significantly decreased the proliferation
rate in 786-O and 769-P cells (Fig. 4B
and C). The colony formation assay revealed that the colony
formation ability of ccRCC cells was impaired after ZNF710-AS1-202
knockdown (Fig. 4D). Furthermore,
ZNF710-AS1-202 knockdown increased the number of cells in the G2/M
phase and decreased the number of cells in the S phase (Fig. 4E). Western blotting revealed that
ZNF710-AS1-202 knockdown downregulated cyclin B1 protein expression
in 786-O and 769-P cells (Fig.
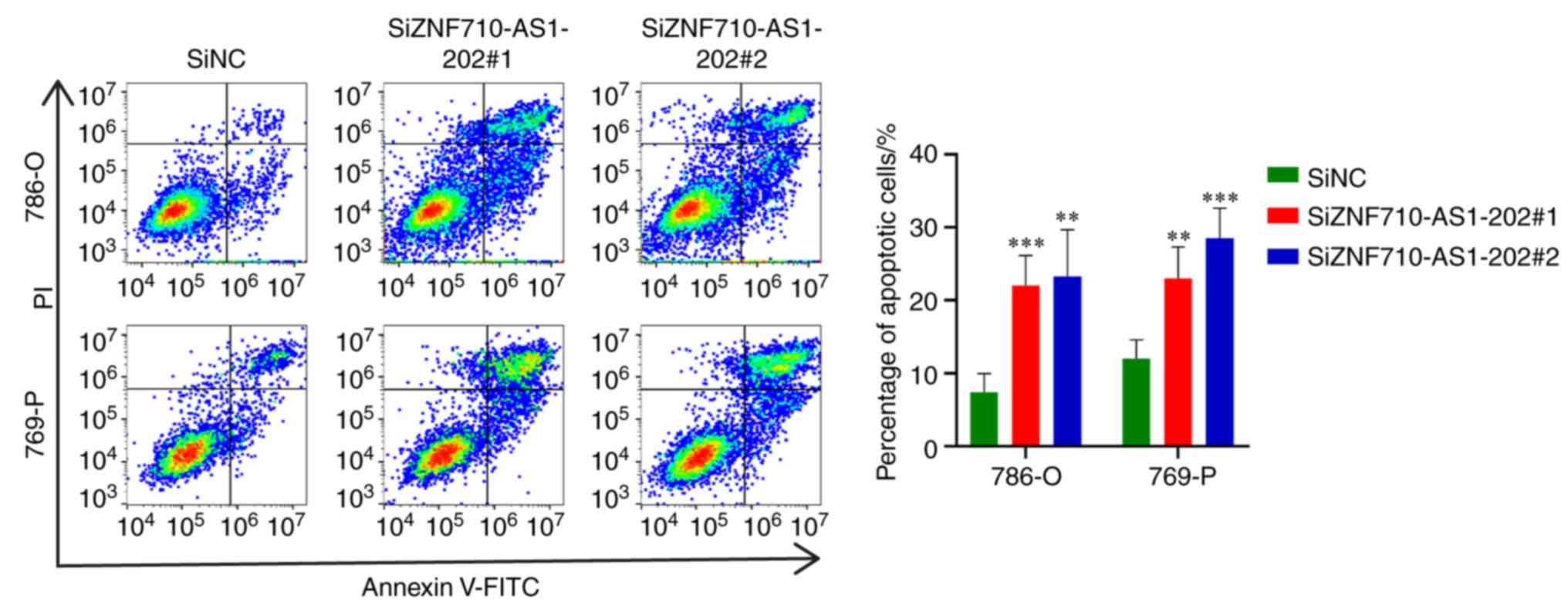
4F). Moreover, 786-O and 769-P cells transfected with
siZNF710-AS1-202 exhibited higher levels of apoptosis compared with
the control cells (Fig. 5).
Dysregulation of ZNF710-AS1-202 causes
significant changes in ZNF710 mRNA and protein expression
ZNF710 is the sense protein-coding gene relative to
ZNF710-AS1 (Fig. 6A).
ZNF710-AS1-202 downregulated the expression of ZNF710 mRNA and
upregulated the expression of ZNF710 protein (Fig. 6B-E and K).
 | Figure 6.Dysregulation of ZNF710-AS1-202
causes significant changes in ZNF710 expression at both the mRNA
and protein levels. (A) Schematic of the lncRNA-ZNF710-AS1 (green)
and ZNF710-201 transcript (yellow) from ENSEMBL. The levels of
ZNF710 expression in RCC cells was detected by (B) reverse
transcription-qPCR or (C) western blotting following ZNF710-AS1-202
overexpression. The levels of ZNF710 expression in RCC cells was
detected by (D) reverse transcription-qPCR or (E) western blotting
ZNF710-AS1-202 suppression. (F) The levels of the ZNF710-201
transcript in different pathological grade ccRCC tissue samples
obtained from TCGA (from GPEIA). (G) Overall survival and (H)
disease free survival of patients with ccRCC in TCGA according to
ZNF710-201 expression (from GPEIA). (I) The expression levels of
ZNF710-AS1-201 in RCC cells following ZNF710-AS1-202
overexpression. (J) Overall survival of patients with ccRCC in TCGA
according to ZNF710-AS1-201 transcript expression (from GPEIA). (K)
Model of ZNF710-AS1-202-dependent regulation of ZNF710 mRNA and
protein. In the nucleus of RCC cells, ZNF710-AS1-202 tethered to
the fourth intron of ZNF710 pre-mRNA and formed complimentary
strands that inhibited ZNF710-201 pre-mRNA splicing. In the
cytoplasm, ZNF710-AS1-202 induced the expression of ZNF710 protein.
**P<0.01 and ***P<0.001 vs. corresponding pc-NC or SiNC.
ZNF710, zinc finger protein 710; TCGA, The Cancer Genome Atlas;
ccRCC, clear cell renal cell carcinoma; GEPIA, Gene Expression
Profiling Interactive Analysis; lnc, long noncoding; OD, optical
density; ns, not significant; siNC, small interfering negative
control. |
TCGA analysis revealed that there was a significant
difference in ZNF710 mRNA expression levels in ccRCC tissues with
different pathological grades (Fig.
6F). Furthermore, the low level of ZNF710 mRNA expression was
associated with poor prognosis (Fig.
6G and H).
The expression of ZNF710-AS1-201, which is the other
isoform of ZNF710-AS1, was not significantly changed after
ZNF710-AS1-202 overexpression in RCC cells (Fig. 6I). Unlike ZNF710-AS1-202, the
expression of ZNF710-AS1-201 tended to be upregulated in ccRCC
tissues. Furthermore, increased expression of ZNF710-AS1-201 was
significantly associated with poor overall survival in patients
with ccRCC (Fig. 6J).
Discussion
The results from the present study revealed that
ZNF710-AS1-202 promoted the proliferation and inhibited apoptosis
of RCC cells in vitro. However, ZNF710-AS1-202 expression
was downregulated in ccRCC tissues compared with PC tissues. These
contradictory results may be explained if the downregulation of
ZNF710-AS1-202 expression is considered as negative feedback
regulation in response to the body's endogenous antitumor activity.
This phenomenon also suggests that not all downregulated genes in
tumor tissues are tumor suppressor genes. Furthermore, in the ccRCC
cell lines investigated in vitro, the cell lines with high
ZNF710-AS1-202 expression exhibited faster proliferation rates.
The ZNF710-AS1-202 transcript plays an important
role in regulating ZNF710 expression. The effect of ZNF710-AS1-202
on RCC cells may be achieved through the ZNF710 protein (19,23,28).
Therefore, further research is required to elucidate the mechanism
by which ZNF710-AS1-202 downregulated ZNF710 mRNA expression and
upregulated ZNF710 protein expression.
The present study used FISH to reveal that lncRNA
ZNF710-AS1-202 is distributed in both the cytoplasm and the nucleus
of RCC cells. ZNF710-AS1-202 in the nucleus may bind to the fourth
intron of ZNF710-201 pre-mRNA to form complimentary strands,
thereby inhibiting ZNF710-201 pre-mRNA processing such as RNA
splicing. In the cytoplasm of RCC cells, lncRNA ZNF710-AS1-202 may
recruit polyribosomes to induce the expression of the ZNF710
protein. As ZNF710-AS1-202 is mainly distributed in the cytoplasm,
the expression level of ZNF710 protein was upregulated.
A hypothesis for the downregulation of
ZNF710-AS1-202 expression in ccRCC tissues observed in the present
study is that only the expression of the ZNF710-AS1-202 in the
nucleus was downregulated (19).
Therefore, further research is required to verify whether the
expression of ZNF710 protein was upregulated in ccRCC tissues
compared with adjacent normal tissues using techniques such as
immunohistochemistry.
While there was no significant difference in the
levels of ZNF710 mRNA expression between ccRCC and PC tissues from
TCGA database, low levels of ZNF710 mRNA expression were associated
with poor prognosis in patients with ccRCC.
To the best of our knowledge, this is the first
study to investigate the AS lncRNA ZNF710-AS1-202, the ZNF710
protein and their associations. The Uniprot database revealed that
ZNF710 is a transcription factor/cofactor that belongs to the
Kruppel C2H2-type zinc-finger protein family and is closely related
to gene transcription by binding to DNA (29). RIP technology may be used to
identify the gene sequence ZNF710 binds to and to explore the
downstream molecules in this pathway (28).
Exogenous ZNF710-AS1-202 was thought to promote the
ZNF710-AS1 gene to express ZNF710-AS1-201 rather than
ZNF710-AS1-202. However, the RT-qPCR results in the present study
revealed that there was no significant change in the expression of
ZNF710-AS1-201 after ZNF710-AS1-202 overexpression. Furthermore,
TCGA analysis demonstrated that there was no significant difference
in the expression of ZNF710-AS1-201 between ccRCC and adjacent
healthy tissues. However, ZNF710-AS1-201 expression was closely
related to the prognosis of patients with ccRCC. In contrast to
ZNF710-AS1-202, the high expression level of ZNF710-AS1-201 in
ccRCC tissues is associated with poor prognosis in patients with
ccRCC. The first exon of ZNF710-AS1-201 overlaps the last 769
nucleotides of the fifth exon of ZNF710-201 mRNA and the second
exon of ZNF710-AS1-201 overlaps 399 nucleotides of the fourth
intron of ZNF710-201 pre-mRNA. Therefore, ZNF710-AS1-201 may also
be relevant to the expression of ZNF710 (28).
In conclusion, the results of the present study
suggested that ZNF710-AS1-202 and ZNF710 may be used as promising
therapeutic targets for ccRCC. However, one drawback of the present
study was that the in vitro results were not validated by
in vivo experiments. Additionally, rescue experiments may be
helpful to clarify the regulatory effects of ZNF710-AS1-201 and
ZNF710-AS1-202 on ZNF710. Furthermore, the function of the ZNF710
protein in ccRCC cells was not investigated and the mechanisms by
which ZNF710-AS1-202 regulated the ZNF710 mRNA and protein levels
were not explored. Future studies are required to investigate the
mechanism of the ZNF710-AS1-ZNF710 axis to identify specific
markers and targets for the diagnosis and treatment of ccRCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Zhengzhou
Science and Technology Bureau (grant no. 131PCXTD627).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GL and MX performed the experiments and analyzed the
data, and were major contributors in writing the manuscript. ZH
performed high throughout RNA-sequencing. PL performed the western
blot assay. HL, ZZ, YD, ZJ and JY conceived and supervised the
project. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of the First Affiliated Hospital of Zhengzhou
University and written informed consent was obtained from all the
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhou B, Zheng P, Li Z, Li H, Wang X, Shi Z
and Han Q: CircPCNXL2 sponges miR-153 to promote the proliferation
and invasion of renal cancer cells through upregulating ZEB2. Cell
Cycle. 17:2644–2654. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Su Y, Lu J, Chen X, Liang C, Luo P, Qin C
and Zhang J: Long non-coding RNA HOTTIP affects renal cell
carcinoma progression by regulating autophagy via the
PI3K/Akt/Atg13 signaling pathway. J Cancer Res Clin Oncol.
145:573–588. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang G, Zhang ZJ, Jian WG, Liu PH, Xue W,
Wang TD, Meng YY, Yuan C, Li HM, Yu YP, et al: Novel long noncoding
RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell
renal cell carcinoma proliferation via the Wnt/β-catenin signaling
pathway. Mol Cancer. 18:152019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dong D, Mu Z, Wei N, Sun M, Wang W, Xin N,
Shao Y and Zhao C: Long non-coding RNA ZFAS1 promotes proliferation
and metastasis of clear cell renal cell carcinoma via targeting
miR-10a/SKA1 pathway. Biomed Pharmacother. 111:917–925. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang FQ, Zhang JQ, Jin JJ, Yang CY, Zhang
WJ, Zhang HM, Zheng JH and Weng ZM: HOXA11-AS promotes the growth
and invasion of renal cancer by sponging miR-146b-5p to upregulate
MMP16 expression. J Cell Physiol. 233:9611–9619. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu Y, Tong Y, Zhu J, Lei Z, Wan L, Zhu X,
Ye F and Xie L: An increase in long non-coding RNA PANDAR is
associated with poor prognosis in clear cell renal cell carcinoma.
BMC Cancer. 17:3732017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu G, Zhao X, Zhou J, Cheng X, Ye Z and
Ji Z: LncRNA TP73-AS1 promotes cell proliferation and inhibits cell
apoptosis in clear cell renal cell carcinoma through repressing
KISS1 expression and inactivation of PI3K_Akt_mTOR signaling
pathway. Cell Physiol Biochem. 48:371–384. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fang L, Zhang Y, Zang Y, Chai R, Zhong G,
Li Z, Duan Z, Ren J and Xu Z: HP-1 inhibits the progression of
ccRCC and enhances sunitinib therapeutic effects by suppressing
EMT. Carbohydr Polym. 223:1151092019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Misawa A, Takayama K, Urano T and Inoue S:
Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes
cell growth and inhibits apoptosis in prostate cancer cells. J Biol
Chem. 291:17861–17880. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deng SJ, Chen HY, Zeng Z, Deng S, Zhu S,
Ye Z, He C, Liu ML, Huang K, Zhong JX, et al: Nutrient
stress-dysregulated antisense lncRNA GLS-AS impairs GLS-mediated
metabolism and represses pancreatic cancer progression. Cancer Res.
79:1398–1412. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu
Q, Tong X, Yang W, Xu Q, Huang D and Tu K: A novel lncRNA
MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by
targeting miR-194-5p/FOXA1 axis. Mol Cancer. 18:282019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Q, Feng Y, Peng W, Ji D, Zhang Z,
Qian W, Li J, Gu Q, Zhang D, Tang J, et al: Long noncoding RNA
Linc02023 regulates PTEN stability and suppresses tumorigenesis of
colorectal cancer in a PTEN-dependent pathway. Cancer Lett.
451:68–78. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Goyal A, Fiškin E, Gutschner T,
Polycarpou-Schwarz M, Groß M, Neugebauer J, Gandhi M,
Caudron-Herger M, Benes V and Diederichs S: A cautionary tale of
sense-antisense gene pairs: Independent regulation despite inverse
correlation of expression. Nucleic Acids Res. 45:12496–12508. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
d'Ydewalle C, Ramos DM, Pyles NJ, Ng SY,
Gorz M, Pilato CM, Ling K, Kong L, Ward AJ, Rubin LL, et al: The
antisense transcript SMN-AS1 regulates SMN expression and is a
novel therapeutic target for spinal muscular atrophy. Neuron.
93:66–79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang MH, Zhao L, Wang L, Ou-Yang W, Hu SS,
Li WL, Ai ML, Wang YQ, Han Y, Li TT, et al: Nuclear lncRNA HOXD-AS1
suppresses colorectal carcinoma growth and metastasis via
inhibiting HOXD3-induced integrin beta3 transcriptional activating
and MAPK/AKT signalling. Mol Cancer. 18:312019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kathuria H, Millien G, McNally L, Gower
AC, Tagne JB, Cao Y and Ramirez MI: NKX2-1-AS1 negatively regulates
CD274/PD-L1, cell-cell interaction genes, and limits human lung
carcinoma cell migration. Sci Rep. 8:144182018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z,
Deng X, Chen H, Shen B, Peng C, et al: Amplification of long
noncoding RNA ZFAS1 promotes metastasis in hepatocellular
carcinoma. Cancer Res. 75:3181–3191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kraus P, Sivakamasundari V, Lim SL, Xing
X, Lipovich L and Lufkin T: Making sense of Dlx1 antisense RNA. Dev
Biol. 376:224–235. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Carrieri C, Cimatti L, Biagioli M, Beugnet
A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C,
et al: Long non-coding antisense RNA controls Uchl1 translation
through an embedded SINEB2 repeat. Nature. 491:454–457. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li C, Li W, Zhang Y, Zhang X, Liu T, Zhang
Y, Yang Y, Wang L, Pan H, Ji J and Wang C: Increased expression of
antisense lncRNA SPINT1-AS1 predicts a poor prognosis in colorectal
cancer and is negatively correlated with its sense transcript. Onco
Targets Ther. 11:3969–3978. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Postepska-Igielska A, Giwojna A,
Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D and Grummt I:
LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via
triplex-mediated changes in chromatin structure. Mol Cell.
60:626–636. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qian W, Cai X, Qian Q, Peng W, Yu J, Zhang
X, Tian L and Wang C: lncRNA ZEB1-AS1 promotes pulmonary fibrosis
through ZEB1-mediated epithelial-mesenchymal transition by
competitively binding miR-141-3p. Cell Death Disease. 10:1292019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Blank-Giwojna A, Postepska-Igielska A and
Grummt I: lncRNA KHPS1 activates a poised enhancer by
triplex-dependent recruitment of epigenomic regulators. Cell Rep.
26:2904–2915.e4. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jadaliha M, Gholamalamdari O, Tang W,
Zhang Y, Petracovici A, Hao Q, Tariq A, Kim TG, Holton SE, Singh
DK, et al: A natural antisense lncRNA controls breast cancer
progression by promoting tumor suppressor gene mRNA stability. PLoS
Genet. 14:e10078022018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ho EC, Donaldson ME and Saville BJ:
Detection of antisense RNA transcripts by strand-specific RT-PCR.
Methods Mol Biol. 630:125–138. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yuan JH, Liu XN, Wang TT, Pan W, Tao QF,
Zhou WP, Wang F and Sun SH: The MBNL3 splicing factor promotes
hepatocellular carcinoma by increasing PXN expression through the
alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 19:820–832.
2017. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
UniProt Consortium, . UniProt: A worldwide
hub of protein knowledge. Nucleic Acids Res. 47:D506–D515. 2019.
View Article : Google Scholar : PubMed/NCBI
|