Introduction
The cell cytoskeleton provides shape and structural
support (1,2) and consists of microfilaments,
intermediate filaments and microtubules. Microtubules and actin
filaments cooperate to facilitate the assembly, organization and
remodeling of the cytoskeleton, which is critical for different
stages of cellular development (3,4).
Microtubules perform a variety of functions, including tracking
intracellular trafficking, localizing organelles, mediating
intracellular signals and maintaining cellular structure (5). Microtubule defects cause various
abnormalities and disorders of microtubule-mediated processes;
moreover, studies have illustrated that several developmental
defects are associated with mutations in microtubule-related genes,
such as those of the microtubule-associated proteins,
motor-associated regulators, microtubule-severing proteins and
microtubule-based motor proteins (6–8).
However, a number of the underlying molecular mechanisms regarding
the severing of microtubule-based structures have yet to be
elucidated.
Fidgetin (Fign), a member of the ATPase associated
with diverse cellular activities (AAA) protein family, belongs to
the same subfamily as microtubule-severing proteins, such as
spastin and katanin (9–14). AAA proteins are involved in a
diverse range of cellular activities, including membrane fusion,
peroxisome biogenesis, proteasome function, vesicle-mediated
transport and microtubule dynamics (15–17).
Fign mutations frequently result in developmental abnormalities of
the brain, eyes, inner ear and specific bones (12). It is predominantly localized in the
nucleus due to the presence of a bipartite nuclear localization
signal (NLS) in its central region (11). As reported, the function of Fign is
closely associated with its subcellular localization (18); however, the mechanism of nuclear
import during these cellular events is poorly understood. Although
the majority of microtubule-severing proteins preferentially act on
the stable domain of the microtubule, Fign acts at the labile
domains (19). However, whether an
alteration in its cellular distribution affects its
microtubule-severing function remains to be elucidated.
Based on the phenotype of Fign-mutant mice, Fign was
revealed to be involved in the regulation of signaling pathways,
particularly those involved in developmental processes (11). Since the specific function of an
AAA protein depends on its interacting partners and precise
subcellular location (9), it is
important to assess the distribution of Fign and the mechanism of
its regulation between the cytoplasm and the nucleus.
In the present study, a Fign mutant was used to
confirm its nuclear translocation and microtubule-severing
function. An NLS motif was also identified to mediate the nuclear
import of the protein. Furthermore, the results indicated that Fign
did not sever microtubules into visibly shorter fragments; rather,
that Fign and its mutants decreased the number of tyrosinated
microtubules (tyr-MTs) at the cell periphery. Collectively, the
findings of the present study indicate that Fign is a nuclear
protein, and that the basic amino acids K317 and R318 within its
NLS are key to its nuclear localization and microtubule-severing
function.
Materials and methods
Reagents
The pEGFP-N1 (cat. no. 6085-1; Addgene, Inc.)
plasmid was maintained within our laboratory. Normal Donkey serum
(cat. no. D9663) was purchased from Sigma-Aldrich; Merck KGaA.
Opti-MEM, DMEM high glucose medium, trypsin and FBS were purchased
from Gibco; Thermo Fisher Scientific, Inc., and
Lipofectamine® 2000 and TRIzol® were
purchased from Invitrogen; Thermo Fisher Scientific, Inc.
Anti-α-tubulin and anti-tyrosinated tubulin antibodies were
supplied by Sigma-Aldrich; Merck KGaA. The homologous recombination
kit (cat. no. C122-02; Vazyme Biotech Co., Ltd.), DNA Polymerase
(cat. no. P508-d1; Vazyme Biotech Co., Ltd.) and PrimeScript™ IV
1st strand cDNA synthesis mix (cat. no. 6215A; Takara Biotechnology
Co., Ltd.) were used for construction of the recombinant plasmid,
and the water used in the present study was ultra-pure (R≥18 M
Ω/cm).
Cell culture and transfection
293T and HeLa cells were purchased from Shanghai
Institutes for Biological Sciences. The cells were routinely
resuscitated, seeded into T25 cell culture flasks and cultured in
DMEM high-glucose medium (containing 10% FBS and 100 U/ml
penicillin and streptomycin) at 37°C in a 5% CO2
humidified atmosphere. At 70–80% confluency, 293T and HeLa cells
were transfected with 5 µg GFP, Fign-GFP wild-type and mutant
plasmids using a Lipofectamine® 2000 transfection kit.
Following a 30-h incubation period at 37°C in a 5% CO2
humidified atmosphere, the cells were harvested for western
blotting.
Plasmid construction and
mutagenesis
A construct containing the full-length coding
sequence of Fign was synthesized from rat hippocampi using general
PCR with gene-specific primers (listed in Table I) designed from the rat Fign gene
(NM_001106484.1). PCR was performed as follows: 95°C for 3 min,
followed by 34 cycles of 95°C for 30 sec, 59°C for 30 sec and 72°C
for 1 min, then 72°C for 7 min. The hippocampus was dissociated
from four 1-day-old Sprague-Dawley rats. The rats were purchased
from the animal experimental center of Sun Yat-sen University and
immediately sacrificed upon arrival. Briefly, the newborn rats were
exposed to carbon dioxide at 20% chamber replacement rate, and
declared dead when the heart stopped beating, following which brain
tissue was dissected. All animal procedures were performed in
strict accordance with the recommendations in the Guide for the
Care and Use of Laboratory Animals produced by the National
Institutes of Health. The protocol was approved by the
Institutional Animal Care and Use Committee at Jinan University.
The gene was cloned into the BglII and EcoRI sites of
pEGFP-N1 to generate the Fign-GFP plasmid. To construct the
Fign-mNLS-GFP and Fign-DNLS-GFP plasmids, two-step PCR method was
used to substitute or delete the basic amino acid residues in Fign.
During the first PCR reaction step, complementary oligonucleotide
primers were used to generate two DNA fragments with overlapping
ends that contained the mutated or deleted bases. The overlapping
ends were annealed, and the resulting fusion product served as a
template for the second PCR reaction. Ligation of the mutant and
deleted Fign fragments into the PEGFP-N1 vector was conducted using
the homologous recombination kit, according to the manufacturer's
protocols; colony PCR and agarose gel electrophoresis (1% w/v
agarose gel) experiments verified the fragments were successfully
ligated to PEGFP-N1. Automated sequencing was used to confirm
successful mutagenesis or deletion, and plasmid DNA was purified
using the Plasmid Maxi kit (cat. no. 12163; Qiagen, Inc.),
according to the manufacturer's instructions.
 | Table I.Primer sequences for PCR. |
Table I.
Primer sequences for PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| Fign-GFP | F:
GATTCTAGAGCTAGCGAATTCGCCACCATGCCGTACATCTTTGCCTTTT |
| Fign-GFP | R:
GATCGCAGATCCTTCGCGGCCGCTTACAATCCTGTGGCTCCCAA |
| Fign-mNLS-GFP | F:
CGGCAAGTTCTCTCAATAATAAAGCTTTCTATATGG |
| Fign-mNLS-GFP | R:
CCATATAGAAAGCTTTATTATTGAGAGAACTTGCCG |
| Fign-ΔNLS-GFP | F:
ACCAACAGTTCGGCAGCAGGGCAAGGAGAC |
| Fign-ΔNLS-GFP | R:
GTCTCCTTGCCCTGCTGCCGAACTGTTGGT |
Western blotting
After transfection, the 293T and HeLa cells were
lysed in lysis buffer [150 mM NaCl, 20 mM Tris, 1 mM EDTA, 1%
Triton X-100 and protease inhibitors cocktail (Sigma-Aldrich; Merck
KGaA)] on ice for 30 min. The lysates were centrifuged at 13,000 ×
g for 15 min at 4°C. A bicinchoninic acid protein assay was used to
quantify the total protein concentration of each sample. The
protein samples (~30 µg /lane) were separated by SDS-PAGE on a 10%
gel, and transferred to PVDF membranes (EMD Millipore). The
membranes were blocked with 5% non-fat milk in TBS, 0.1% Tween-20
(TBST) at room temperature for 1 h, and incubated overnight at 4°C
with the following primary antibodies: Anti-α-tubulin (1:2,000;
cat. no. ab18251; rabbit polyclonal; Abcam); anti-tyrosinated
tubulin (1:2,000; cat. no. ab6160; rat monoclonal; Abcam); anti-GFP
(1:5,000; cat. no. ab290; rabbit polyclonal; Abcam); anti-GAPDH
(1:5,000; cat. no. ab181602; rabbit monoclonal; Abcam) and
anti-Fign (1:400; cat. no. sc-68343; rabbit polyclonal; Santa Cruz
Biotechnology, Inc.). After three washes in TBST, the membranes
were incubated with horseradish peroxidase-conjugated donkey
anti-rabbit (1:5,000; cat. no. AS038; ABclonal Biotech Co., Ltd.)
or goat anti-mouse secondary antibodies (1:5,000; cat. no. AS003;
ABclonal Biotech Co., Ltd.) at room temperature for 1 h, and
visualized using enhanced chemiluminescence reagents. Western blot
bands were quantified by densitometric analysis using Image-Pro
Plus 7.0 (Media Cybernetics, Inc.)
Immunocytochemistry assays
The HeLa cells were seeded onto glass cover slips
(Thermo Fisher Scientific, Inc.) in 24-well cell culture plates
(Corning Inc.) at a density of 1×105/ml, 24 h prior to
transfection. At a confluence of 40–50%, HeLa cells were
transfected with 1 µg/well GFP, Fign-GFP wild-type and mutant
plasmids using a Lipofectamine 2000 transfection kit. The
transfection rate of HeLa cells in each group was ~70%. At 24 h
post-transfection, the cells were fixed in 4% paraformaldehyde for
40 min at 4°C, blocked with blocking buffer (TBST in 3% normal
donkey serum) at room temperature for 1 h, and incubated overnight
with an anti-α-tubulin (1:200) or anti-tyrosinated tubulin (1:500)
primary antibody at 4°C. Next, the cells were incubated with a goat
anti-mouse FITC secondary antibody (1:1,000; cat. no. ab150115;
Abcam) for 2 h at room temperature. After staining, the cells were
mounted on glass slides using Fluoro Gel II with DAPI (cat. no.
17985-50; Electron Microscopy Sciences) for confocal microscopy
(LSM 700; Zeiss GmbH). Images were acquired with the same optical
slice thickness in every channel using magnification ×63 oil
objective and a resolution of 1,024×1,024 pixels.
NLS prediction
Online prediction software (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi)
was used to predict the Fign NLS. cNLS Mapper software accurately
predicts NLSs specific to the importin αβ pathway by calculating
NLS scores (level of NLS activity); higher scores of Fign indicates
stronger NLS activity.
Statistical analysis
Comparisons between different experimental groups
were analyzed using one-way ANOVA. All data were tested for normal
distribution; if the data were normally distributed and had similar
variances, then one-way ANOVA was performed to compare the means
among all measured variables followed by Bonferroni's post hoc
test, P<0.05 was considered to indicate a statistically
significant difference. All statistical analyses were performed
using SPSS version 12.0 software (SPSS, Inc.). Data are expressed
as the mean ± SEM of 3–5 separate experiments for each assay.
Results
Fign is predominantly located in the
nucleus
In order to determine the cellular distribution of
Fign, a eukaryotic expression vector containing the Fign coding
sequence was constructed, which was amplified from rat cDNA. Colony
PCR was used to determine successful insertion of the Fign sequence
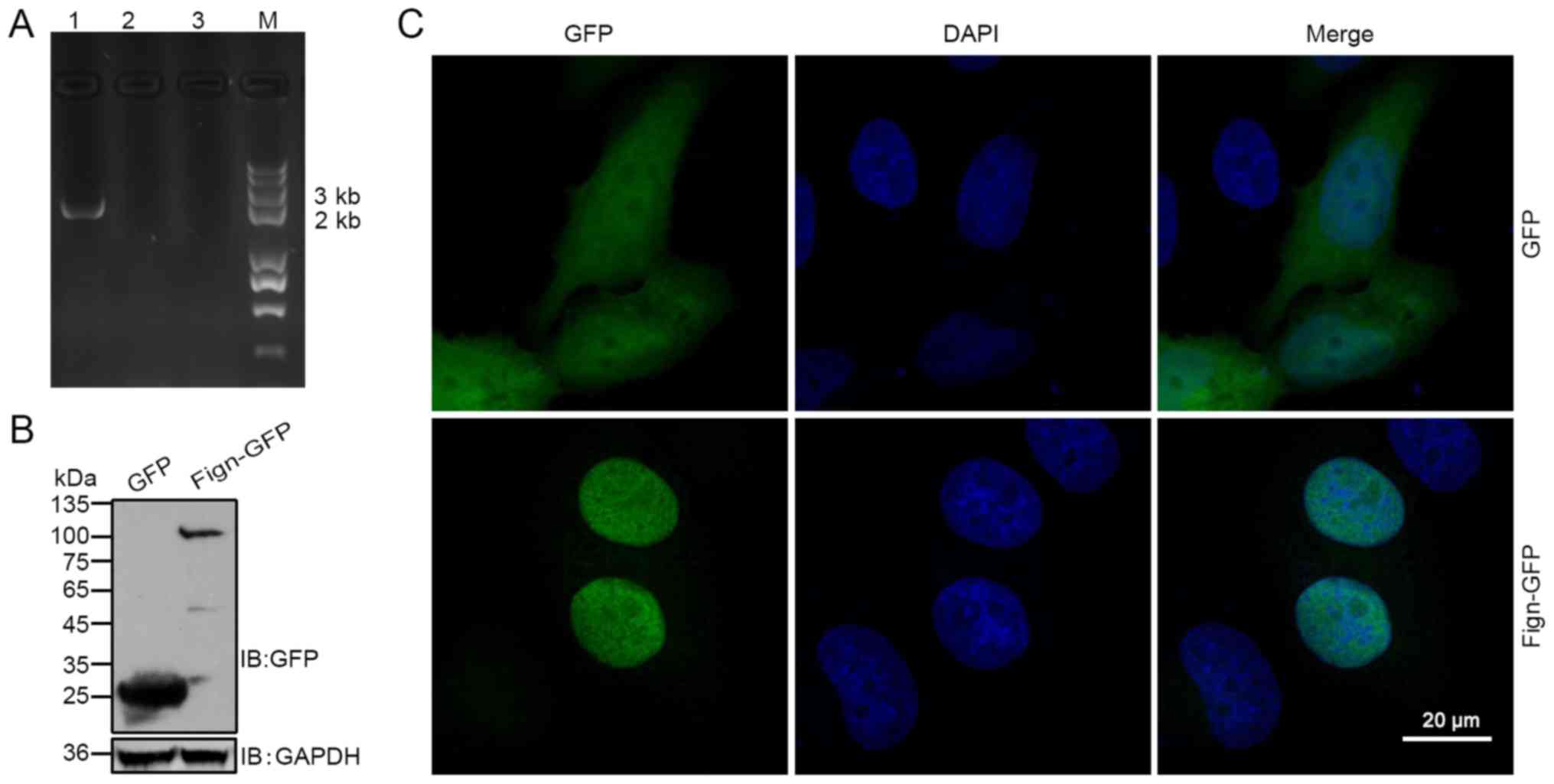
into the pEGFP-N1 plasmid. Fig. 1A
shows the result of agarose gel electrophoresis of the
representative colony PCR products. To further clarify whether
Fign-GFP proteins were expressed from the corresponding plasmids,
293T cells were transfected with the Fign-GFP plasmids; western
blotting with a GFP antibody revealed a major dominant band of the
expected size (~105 kDa; Fig. 1B).
Next, the GFP epitope-tag was detected to delineate the subcellular
localization of Fign in HeLa cells. With the exception of a small
degree of cytoplasmic localization, the fusion protein was
predominantly detected in the nucleus (Fig. 1C). However, no signal was
detectable in the nucleoli of the GFP-positive cells, indicating
that the overexpression of Fign-GFP fusion protein was localized to
the nucleoplasm. Collectively, the results indicated that Fign is a
nuclear protein.
A signal peptide sequence controls the
translocation of Fign from the cytoplasm into the nucleus
Numerous studies have indicated that Fign is
primarily involved in microtubule severing (19,20),
and the present study indicated that it is localized to the
nucleus. As the NLS is an amino acid sequence that tags a protein
for import into the nucleus, in the present study, the NLS of Fign
was mutated to alter its cellular distribution, and the subsequent
effects on microtubule severing were assessed. Using online NLS
prediction software, the potential NLS of Fign was indicated to
comprise amino acids 314–323, a sequence that is relatively well
conserved in rats, mice and humans (Fig. 2A). To further confirm that this
putative sequence was a nuclear targeting sequence, the integrity
of the NLS was disrupted by substituting amino acids 317 and 318
(KR to NN; Fign-mNLS-GFP), or removing the NLS entirely
(Fign-ΔNLS-GFP; Fig. 2B). The
sequencing results demonstrated that the mutagenesis was
successful, and the colony PCR results showed bands in lanes 4 and
8 that were consistent with the molecular weight of Fign (Fig. 2C). The 293T cells were then
transfected with the mutant plasmids, and Fign expression was
detected by western blotting. Both proteins were successfully
expressed in 293T cells (Fig.
2D).
 | Figure 2.Schematic representation of the
putative NLS and subcellular localization of Fign-mNLS-GFP and
Fign-ΔNLS-GFP proteins. (A) Schematic representation of Fign
protein and the putative NLS (amino acids 314–323). (B) Schematic
representation of the NLS and its mutants. Fign-mNLS-GFP, KR
(317–318) was replaced by NN; Fign-ΔNLS-GFP, the NLS was deleted.
(C) Agarose gel electrophoresis of Fign and its fragments. Lane 4,
Fign-mNLS-GFP colony PCR; lane 8, Fign-ΔNLS-GFP colony PCR; lanes 1
and 2, N-terminal fragments and C-terminal of Fign-mNLS-GFP; lanes
5 and 6, N-terminal and C-terminal fragments of Fign-ΔNLS-GFP;
lanes 3 and 7, mNLS and ΔNLS fragments of Fign. (D) Expression of
mutant proteins was confirmed by western blotting. (E) Ratio of the
average GFP signals in the nucleus and whole-cell. The cell and
nuclear morphology were delineated under white light and DAPI
conditions, respectively, and the gray value and area of the nuclei
and intact cells were calculated by Image-Pro Plus 7.0 software.
Nucleus signal ratio=nuclear grayscale mean/cell grayscale mean.
(F) Fign and its mutant proteins expressed in HeLa cells.
*P<0.05 vs. Control group. #P<0.05 vs. Control
group. Fign, fidgetin; NLS, nuclear localization signal. |
In order to evaluate the potential contribution of
the NLS to nuclear localization, HeLa cells were transfected with
Fign-mNLS-GFP and Fign-ΔNLS-GFP. It was also revealed that the
Fign-mNLS-GFP and Fign-ΔNLS-GFP mutants were expressed in both the
cytoplasm and the nucleus, compared with cells transfected with
wild-type Fign-GFP, whose protein was almost exclusively located in
the nucleus (Fig. 2E and F). Taken
together, the results indicated that the Fign NLS comprises amino
acids 314–323 (N-terminal sequence, SSLKRKAFYM), and that K317 and
R318 are key sites for the nuclear localization process.
Furthermore, the mutation or deletion of this potential NLS
subsequently affected the translocation of Fign into the
nucleus.
Fign preferentially severs
tyr-MTs
To investigate whether Fign regulates microtubule
severing, thus altering their dynamics, HeLa cells were selected to
observe microtubule morphology. As shown in Fig. 3A, both Fign-GFP wild type and
mutant plasmids were successfully expressed in HeLa cells. Because
Fign is primarily expressed in the nucleus, the microtubules of the
transfected cells showed no obvious morphological changes, and were
not cut into short fragments. Also, there were no significant
differences in fluorescence intensity or protein expression
(Fig. 3A and D). Of note, no
significant changes in α-tubulin were observed in cells transfected
with Fign-mNLS-GFP or Fign-ΔNLS-GFP compared with control cells
(Fig. 3B and E). As Fign remained
in the cytoplasm following NLS mutation or deletion, these data
suggested that Fign did not alter microtubule dynamics by cutting
the microtubules into short fragments.
Microtubules undergo various forms of
post-translational modification. Since tyrosine modification
increases microtubule instability, the microtubule-severing
function of Fign in unstable microtubules was assessed.
Immunocytochemistry assays and western blotting were used to detect
tyr-MTs in the transfected cells. Although Fign was not responsible
for cutting tyr-MTs into visibly shorter fragments, the
fluorescence intensity and protein expression levels of these
tyr-MTs were significantly reduced (Fig. 4A and C) compared with those of the
control cells. Moreover, when cells were transfected with
Fign-mNLS-GFP or Fign-ΔNLS-GFP, these parameters were also
significantly reduced (Fig. 4B and
D), collectively suggesting that Fign primarily exerts its
effects on tyr-MTs.
Discussion
In the present study, online software was used to
predict the potential NLS of Fign, and to identify its contribution
to nuclear localization and microtubule severing. Since
epitope-tagged Fign was predominantly localized to the nucleus, it
was revealed to be a nuclear protein. Furthermore, amino acids K317
and R318 of the N-terminal sequence of Fign were identified to be
necessary for its nuclear localization, and mutation of the NLS
affected its nuclear transport. Moreover, Fign was indicated to
preferentially sever tyr-MTs.
AAA proteins contain a highly conserved 230
amino-acid AAA domain, which forms hexameric rings essential for
their ATPase activity (20–22).
Similar to spastin and katanin, Fign efficiently severs
microtubules in an ATP-dependent manner, resulting in their
destruction (9,23). By severing the lattice or removing
tubulins directly from the tubule ends, microtubule-severing
enzymes fulfill a key role in creating and maintaining microtubule
arrays in a variety of cell types; creating new ‘bare ends’ renders
microtubules highly responsive to other regulators in the local
microenvironment (20).
The results of the present study provide important
and complementary insights into the understanding of the nuclear
export of Fign. Firstly, the N-terminal SSLKRKAFYM sequence (amino
acids 314–323) was identified to be an NLS, enabling Fign to return
to the cytoplasm from the nucleus. It was also revealed that Fign
is predominantly localized in the nucleus, and is thus a nuclear
protein.
Nuclear localization is essential for controlling
the transcription of protein-coding genes (24,25).
The majority of NLSs contain basic amino acid sequences of 4–30
residues, which are too short to form independent structures, and
typically exist within nuclear proteins. The NLS is required for
the transport of large proteins (>60 kDa) from or into the
nucleus (26–28). As the NLS loses its capacity to
bind to components within the nucleus, by default, the associated
protein will be exported back into the cytoplasm (29–32).
Since the molecular mass of Fign is ~80 kDa, the Fign-GFP fusion
protein exceeds the size of the nuclear pore complex (40–60 kDa;
utilized for simple diffusion), suggesting that an active transport
mechanism was necessary for the nuclear distribution of the fusion
protein.
In order to identify the basic amino acid residues
responsible for the nuclear localization of Fign, online software
was used to predict its potential NLS. To generate Fign-NLS
mutants, two conserved residues were then replaced, or the putative
NLS was deleted entirely. Disruption of the NLS resulted in Fign
expression in both the cytoplasm and the nucleus, indicating that
the identified NLS was important for its cellular distribution. To
identify the amino acid residues responsible for nuclear transport,
a KR-to-NN mutation was generated in the Fign NLS, and its cellular
distribution was assessed. The results showed that this mutation
ablated nuclear translocation, and suggested that K317 and R318 are
key residues for transportation regulation. Yang et al
(11) have reported that Fign is a
nuclear protein with a binary NLS that reveals two small clusters
of essential amino acids (K366, K369, R378 and K379), similar to
the canonical binary NLS. After comparison, the present study
reported a different NLS than Yang et al (11).
A number of studies have emphasized the importance
of the microtubule-severing function of Fign, which primarily
influences microtubule dynamics (19,20).
It was also identified that Fign preferentially severs tyr-MTs
(33). In addition to maintaining
cell shape and structure, microtubules are involved in
intracellular transport, cellular motility and the positioning of
organelles (34). Microtubules are
heterogeneous and highly dynamic both in vivo and in
vitro, and undergo cycles of polymerization and rapid
depolymerization (35). This
‘dynamic instability’ is an intrinsic property that has a key role
in microtubule functioning (36).
The present study revealed that, unlike spastin, another
microtubule-severing protein, Fign, does not cut microtubules into
short fragments, suggesting that it may not act on stable
microtubules.
Although Fign does not have a significant effect on
the shortening of intact, full-length microtubules, its influence
on post-translationally modified microtubules remained to be
elucidated. In order to determine whether Fign acted on modified
microtubules, intracellular detection of tyr-MTs was conducted in
the present study. The results showed that Fign and its mutants
caused a significant decrease in the expression of tyr-MTs compared
with wild-type Fign. It has been reported that tubulin is
post-translationally modified in various ways, including
phosphorylation, ubiquitination, sumoylation, and palmitoylation
(37,38). However, detyrosination, Δ2-tubulin
generation, polyglutamylation, polyglycylation and acetylation
occur less frequently. Different microtubule post-translational
modifications, alone or in combination, alter the binding state of
tubulin proteins and confer specific cellular functions (39–42).
Detyrosination has been linked to microtubule stability, as stable
microtubules are detyrosinated in multiple different cell types
(43). Because tyrosine
modification increases microtubule instability, the results of the
current study suggested that Fign may preferentially sever
tyrosine-modified and unstable, rather than stable, microtubules.
Hu et al (33) reported
that Fign has an essential role in cultured astrocyte migration by
preferentially targeting tyr-MTs. The present study yielded
consistent results with those of Hu et al (33). Before the experiment, it was
hypothesized that mutations in the Fign NLS might lead to two
outcomes. The first is that the mutation disrupts the function of
Fign targeting the cleavage of tyr-MTs, resulting in an increase in
tyr-MTs. The second is that the mutation does not affect the
function of Fign, and the increased level of Fign in the cytoplasm
enhances the cleavage of tyr-MTs. However, in fact, the Fign
mutation did not result in a significant change in tyr-MTs compared
with wild-type Fign, suggesting that the cellular distribution of
the Fign protein itself does not affect the function of microtubule
severing. Spastin and katanin, two other microtubule-severing
proteins, destabilize local microtubule lattice contacts by pulling
on the disordered and negatively-charged C-terminal tails of
tubulin, which is dependent on the orientation of tubulin (44). As a microtubule-severing protein,
Fign may utilize similar mechanisms.
In conclusion, the present study revealed that Fign
is a nuclear protein that targeted and severed tyr-MTs, and that
K317 and R318 (located at the N-terminus of the Fign gene) are key
residues for its nuclear translocation. Further studies are
required, however, to elucidate the mechanisms of the
K317/R318-associated nuclear entry of Fign and the precise function
of its signal peptide sequence.
Acknowledgements
Not applicable.
Funding
This work was supported by National Natural Science
Foundation of China (grant nos. 81571191 and 81771144), Natural
Science Foundation of Guangdong Province, China (grant nos.
2017B030311002 and 2017A030310342), Medical Research Foundation of
Guangdong Province, China (grant no. A2016343) and Guangzhou
Institute of Pediatrics/Guangzhou Women and Children's Medical
Center (grant no. IP-2018-010).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JFZ, CC and GG conceived and designed the
experiments. JL, FW and JQZ performed the experiments. JL and LFC
analyzed the data. JL wrote the paper, LC and TF performed the
western blotting. JFZ and GG critically revised the manuscript. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
All animal procedures were performed in strict
accordance with the recommendations in the Guide for the Care and
Use of Laboratory Animals produced by the National Institutes of
Health. The protocol was approved by the Institutional Animal Care
and Use Committee at Jinan University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fletcher DA and Mullins RD: Cell mechanics
and the cytoskeleton. Nature. 463:485–492. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guerin CM and Kramer SG: Cytoskeletal
remodeling during myotube assembly and guidance: Coordinating the
actin and microtubule networks. Commun Integr Biol. 2:452–457.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kapitein LC and Hoogenraad CC: Building
the neuronal microtubule cytoskeleton. Neuron. 87:492–506. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lasser M, Tiber J and Lowery LA: The role
of the microtubule cytoskeleton in neurodevelopmental disorders.
Front Cell Neurosci. 12:1652018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Barlan K and Gelfand VI: Microtubule-based
transport and the distribution, tethering, and organization of
organelles. Cold Spring Harb Perspect Biol. 9(pii): a0258172017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lipka J, Kuijpers M, Jaworski J and
Hoogenraad CC: Mutations in cytoplasmic dynein and its regulators
cause malformations of cortical development and neurodegenerative
diseases. Biochem Soc Trans. 41:1605–1612. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reiner O and Sapir T: LIS1 functions in
normal development and disease. Curr Opin Neurobio. 23:951–956.
2013. View Article : Google Scholar
|
|
8
|
Liu Y, Lee JW and Ackerman SL: Mutations
in the microtubule-associated protein 1A (Map1a) gene cause
Purkinje cell degeneration. J Neurosci. 35:4587–4598. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mukherjee S, Diaz Valencia JD, Stewman S,
Metz J, Monnier S, Rath U, Asenjo AB, Charafeddine RA, Sosa HJ,
Ross JL, et al: Human Fidgetin is a microtubule severing the enzyme
and minus-end depolymerase that regulates mitosis. Cell Cycle.
11:2359–2366. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ghosh DK, Dasgupta D and Guha A: Models,
regulations, and functions of microtubule severing by Katanin. ISRN
Mol Biol. 2012:5962892012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Y, Mahaffey CL, Bèrubè N, Nystuen A
and Frankel WN: Functional characterization of fidgetin, an
AAA-family protein mutated in fidget mice. Exp Cell Res. 304:50–58.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cox GA, Mahaffey CL, Nystuen A, Letts VA
and Frankel WN: The mouse fidgetin gene defines a new role for AAA
family proteins in mammalian development. Nat Genet. 26:198–202.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Y, Mahaffey CL, Bérubé N and Frankel
WN: Interaction between fidgetin and protein kinase A-anchoring
protein AKAP95 is critical for palatogenesis in the mouse. J Biol
Chem. 281:22352–22359. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eckert T, Le DT, Link S, Friedmann L and
Woehlke G: Spastin's microtubule-binding properties and comparison
to katanin. PLoS One. 7:e501612012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Grimm I, Saffian D, Platta HW and Erdmann
R: The AAA-type ATPases Pex1p and Pex6p and their role in
peroxisomal matrix protein import in Saccharomyces cerevisiae.
Biochim Biophys Acta. 1823:150–158. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
White SR and Lauring B: AAA+ ATPases:
Achieving diversity of function with conserved machinery. Traffic.
8:1657-16672007.
|
|
17
|
Snider J, Thibault G and Houry WA: The
AAA+ superfamily of functionally diverse proteins. Genome Biol.
9:2162008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Onitake A, Yamanaka K, Esaki M and Ogura
T: Caenorhabditis elegans fidgetin homolog FIGL-1, a
nuclear-localized AAA ATPase, binds to SUMO. J Struct Biol.
179:143–151. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Leo L, Yu W, D'Rozario M, Waddell EA,
Marenda DR, Baird MA, Davidson MW, Zhou B, Wu B, Baker L, et al:
Vertebrate Fidgetin restrains axonal growth by severing labile
domains of microtubules. Cell Rep. 12:1723–1730. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sharp DJ and Ross JL: Microtubule-severing
enzymes at the cutting edge. J Cell Sci. 125:2561–2569. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lupas AN and Martin J: AAA proteins. Curr
Opin Struct Biol. 12:746–753. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vale RD: AAA proteins. J Cell Biol.
150:F13–F19. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang D, Rogers GC, Buster DW and Sharp
DJ: Three microtubule severing enzymes contribute to the
‘Pacman-flux’ machinery that moves chromosomes. J Cell Biol.
177:231–242. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Spit A, Hyland RH, Mellor EJ and Casselton
LA: A role for heterodimerization in nuclear localization of a
homeodomain protein. Proc Natl Acad Sci USA. 95:6228–6233. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hunter CC, Siebert KS, Downes DJ, Wong KH,
Kreutzberger SD, Fraser JA, Clarke DF, Hynes MJ, Davis MA and Todd
RB: Multiple nuclear localization signals mediate nuclear
localization of the GATA transcription factor AreA. Eukaryot Cell.
13:527–538. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
LaCasse EC and Lefebvre YA: Nuclear
localization signals overlap DNA- or RNA-binding domains in nucleic
acid-binding proteins. Nucleic Acids Res. 23:1647–1656. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kosugi S, Hasebe M, Matsumura N, Takashima
H, Miyamoto-Sato E, Tomita M and Yanagawa H: Six classes of nuclear
localization signals specific to different binding grooves of
importin alpha. J Biol Chem. 284:478–485. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Melnikov S, Ben-Shem A, Yusupova G and
Yusupov M: Insights into the origin of the nuclear localization
signals in conserved ribosomal proteins. Nat Commun. 6:73822015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakielny S and Dreyfuss G: Import and
export of the nuclear protein import receptor transportin by a
mechanism independent of GTP hydrolysis. Curr Biol. 8:89–95. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang R and Brattain MG: The maximal size
of protein to diffuse through the nuclear pore is larger than 60
kDa. FEBS Lett. 581:3164–3170. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Freitas N and Cunha C: Mechanisms and
signals for the nuclear import of proteins. Curr Genomics.
10:550–557. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Marfori M, Mynott A, Ellis JJ, Mehdi AM,
Saunders NF, Curmi PM, Forwood JK, Bodén M and Kobe B: Molecular
basis for specificity of nuclear import and prediction of nuclear
localization. Biochim Biophys Acta. 1813:1562–1577. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu Z, Feng J, Bo W, Wu R, Dong Z, Liu Y,
Qiang L and Liu M: Fidgetin regulates cultured astrocyte migration
by severing tyrosinated microtubules at the leading edge. Mol Biol
Cell. 28:545–553. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kaverina I and Straube A: Regulation of
cell migration by dynamic microtubules. Semin Cell Dev Biol.
22:968–974. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Muroyama A and Lechler T: Microtubule
organization, dynamics and functions in differentiated cells.
Development. 144:3012–3021. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mitchison T and Kirschner M: Dynamic
instability of microtubule growth. Nature. 312:237–242. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song Y and Brady ST: Post-translational
modifications of tubulin: Pathways to functional diversity of
microtubules. Trends Cell Biol. 25:125–136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Audagnotto M and Dal Peraro M: Protein
post-translational modifications: In silico prediction tools and
molecular modeling. Comput Struct Biotechnol J. 15:307–319. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Howes SC, Alushin GM, Shida T, Nachury MV
and Nogales E: Effects of tubulin acetylation and tubulin
acetyltransferase binding on microtubule structure. Mol Biol Cell.
25:257–266. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Magiera MM and Janke C: Post-translational
modifications of tubulin. Curr Biol. 24:R351–R354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Janke C and Bulinski JC:
Post-translational regulation of the microtubule cytoskeleton:
Mechanisms and functions. Nat Rev Mol Cell Biol. 12:773–786. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fukushima N, Furuta D, Hidaka Y, Moriyama
R and Tsujiuchi T: Post-translational modifications of tubulin in
the nervous system. J Neurochem. 109:683–693. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Schneider N, Ludwig H and Nick P:
Suppression of tubulin detyrosination by parthenolide recruits the
plant-specific kinesin KCH to cortical microtubules. J Exp Bot.
66:2001–2011. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Roll-Mecak A and Vale RD: Structural basis
of microtubule severing by the hereditary spastic paraplegia
protein spastin. Nature. 451:363–367. 2008. View Article : Google Scholar : PubMed/NCBI
|