Introduction
Hepatitis viruses can cause infections that lead to
liver cirrhosis, which then increases the risk of liver cancer;
thus, hepatitis-cirrhosis-liver cancer is considered to be a
‘trilogy’ of events (1). Clinical
data have shown that most patients with liver cancer had a
background of liver fibrosis or cirrhosis (2). In a previous study, the liver
stiffness measurement was evaluated in a group of patients with
chronic hepatitis C, it was found that the stiffness of the liver
could be an independent risk factor for the occurrence of liver
cancer (1). It has also been shown
that the higher the degree of hepatic fibrosis, the higher the
intrahepatic metastasis rate of liver cancer, indicating a
significant association between hepatic fibrosis and proliferation,
invasion and metastasis of liver cancer cells (3). An in vivo experiment, in which
liver cancer cells were injected into mice with induced liver
fibrosis, demonstrated that liver fibrosis was associated with the
development of tumors in these mice (4).
Cirrhosis of the liver can lead to sinusoidal portal
hypertension, in which the pressure in the hepatic portal vein
increases, which can cause these veins to become significantly
dilated (5). Hepatocytes are
epithelial cells that line the sinusoids, which are directly and
indirectly affected by biomechanical factors, such as the pressure
of the hepatic portal vein and the tension strain after the
dilation of hepatic sinusoids (6).
A number of studies have shown the important roles of biomechanical
factors in the regulation of hepatocyte function. Our previous
experiments found that mechanical pressure promotes the
proliferation, migration and invasion of liver cancer cells (HepG2
and Huh-7) (7). A total of five
pressure-responsive microRNAs (miRNAs/miRs) were screened from the
results of the mRNA and miRNA microarray, which were uploaded to
the Gene Expression Omnibus database (chip nos. GSE119881 and
GSE120194). Our previous and present results have both indicated
that mechanical stimulation may affect the growth and metastasis of
liver cancer (7). Moreover,
mechanical stretching has been found to significantly increase the
expression of transforming growth factor β (TGF-β) mRNA and related
proteins in liver cells (8). In
the early stages of portal hypertension, mechanical stretching
increased the expression level of matrix metalloproteinase-1 in
hepatic stellate cells, reduced the expression levels of tissue
inhibitor of metalloproteinases (TIMP)1 and TIMP2, and inhibited
the degradation of the extracellular matrix (9). However, the effect of tensile strain
on liver cancer cells and its related miRNAs have not yet been
reported.
miRNAs are 21-to-23-nucleotide-long noncoding RNA
molecules that have important roles in regulating cell
proliferation, differentiation and apoptosis (10). Some miRNAs are related to both
tensile strain and liver cancer, including miR-29 and miR-21. For
example, previous studies demonstrated that the expression of the
miR-29 family was downregulated in periodontal ligament cells
loaded with tensile strain for 24 h (11) and in liver cancer cells (12). Additionally, the expression of
miR-21 in vascular smooth muscle cells (VSMCs) increased
significantly after tensile strain loading and in liver cancer
tissues (13). These results
indicated that certain tension-responsive miRNAs may be important
in the development of liver cancer. Therefore, in the present
study, the effects of strain force on the proliferation of a human
liver cancer cell line (HepG2) were measured by flow cytometry,
Cell Counting Kit-8 (CCK-8) and 5-bromodeoxyuridine (BrdU) assays.
An Agilent monochromatic marker chip was used to screen
differentially expressed miRNAs, and reverse
transcription-quantitative (RT-q)PCR was used to verify the
results. Gene Ontology (GO) and pathway analyses were used to
analyze genes and provide information on how the mechanical
microenvironment of liver cancer affects the behavior of liver
cancer cells.
The present results suggested that the expression
levels of miRNAs in HepG2 cells was significantly changed after
loading under the optimal conditions (amplitude of 15% at 1 Hz for
24 h). In total, seven differentially expressed miRNAs and 224
target genes were screened. Among them, SMAD7 and SP1 were further
examined, with regards to their related functions and mechanisms.
GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
suggested that the biological functions of the target genes were
mainly related to various cancer types, including
TGF-β/mitogen-PI3K/Akt and other signaling pathways, closely
related to different cancer types. Moreover, the possible mechanism
was identified to be via the regulation of SMAD7 and TGF-β
signaling pathway by hsa-miR-503-5p.
Materials and methods
Cell culture
HepG2 cells were purchased from American Type
Culture Collection (cat. no. HB 8065). HL-7702 cells were purchased
from cell bank of typical culture preservation Committee of Chinese
Academy of Sciences. The cells were cultured at 37°C in an
atmosphere containing 5% CO2 in Dulbecco's modified
Eagle's medium (DMEM; Jiangsu Kaiji Biotechnology Co., Ltd.)
containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) for different durations (2, 6, 12, 24 and 48 h).
The cell line was authenticated by short tandem repeat
analysis.
Cyclic stretching device
In this experiment, the FX-4000T™ Tension system
(Flexcell International Corporation) purchased by the Biomechanics
Laboratory of Shanghai Jiao Tong University was used. The tensile
strain principle of the device was to use a vacuum pump to draw
air, make space under an elastic film in the vacuum environment and
produce downward tension strain. When the vacuum pump was relaxed,
the elastic membrane retracted under its own elastic force and
circulated, thus producing periodic tensile strain (14).
BioFlex amino-culture plate
coating
Numerous routine treatment methods were tested in
the laboratory, such as soaking in hydrochloric acid (plate) for 24
h, followed by soaking in water for 24 h, then drying, gelatin
coating or cleaning. Moreover, drying after soaking in 75% alcohol
for 24 h, then gelatin coating was performed. However, when using
these methods cells did not adherent to the wells and grew poorly.
Later, the plate was washed and dried. Then, the gelatin was
changed into rat tail collagen for coating, it was found that the
cells could adhere to the wall and grow efficiently. The BioFlex
amino-culture plates (Flexcell International Corporation) were
coated with 2 mg/ml rat tail collagen solution (Roche Diagnostics).
Then, 20 µl of collagen solution was added to each well and smeared
evenly with an aseptic cell scraper. Subsequently, the plates were
blow-dried on a clean bench and irradiated with ultraviolet light
for 1 h.
Cyclic stretch application
The cells were seeded on BioFlex amino-culture
plates at a density of 1×105 cells/well. After 24 h, the
culture medium was replaced with serum-free medium for another 24 h
to synchronize the cells in 37°C incubator. The cells in the
serum-free medium were then exposed to cyclic stretch provided by
the FX-4000T Tension system.
Flow cytometry analysis
In the periodic strain-loaded cells, three main
parameters were used; these were tensile strain amplitude, tension
strain frequency and time. By fixing two of the parameters, the
other parameter was changed to observe the effect of tension strain
on HepG2 cells (except these three parameters, all the other
experimental conditions were the same). After synchronizing HepG2
cells implanted on BioFlex amino-culture plates for 24 h, three
groups of experiments were set up: i) Tension strain time (tensile
strain frequency fixed to 1 Hz; amplitude to 15%; and tension
strain time set to 2, 6, 12, 24 or 48 h, a total of five
experimental groups); ii) tensional strain frequency (tensile
strain time fixed to 24 h; amplitude to 15%; and tensile strain
frequency set to 0.5 and 1 Hz, a total of two experimental groups);
and iii) tensile strain amplitude (tensile strain time fixed to 24
h; frequency to 1 Hz; and tensile strain amplitude set to 5 and
15%, a total of two experimental groups). All the control groups
were nontension loaded groups. After tension loading, the cells
were digested with trypsin (containing EDTA; Jiangsu Kaiji
Biotechnology Co., Ltd.), resuspended with 0.5 ml 1X PBS containing
50 µg/ml propidium iodide (PI) and 200 µg/ml ribonuclease (Roche
Diagnostics) and incubated at 37°C for 30 min. PI is an insertion
nucleic acid fluorescent dye; its binding amount is proportional to
the content of DNA in the cell. Using flow cytometry and FlowJo V10
software (FlowJo LLC) DNA distribution status at various stages of
the cell cycle can be obtained, and the percentage of each cell
cycle can be calculated. After propidium iodide staining, if the
fluorescence intensity of G0/G1 phase cells is 1, the theoretical
value of the fluorescence intensity of G2/M phase cells that
contain double genomic DNA is 2, and the fluorescence intensity of
S phase cells undergoing DNA replication is 1–2 (15). Then, flow cytometry was used to
detect the cell cycle with a standard program. In the analysis, the
proliferation index (PI) = (S + G2/M) / (G0/G1 + S + G2/M) was used
to evaluate the effect of different tensile strain loading
conditions on cell proliferation, and the ideal tension strain
conditions for HepG2 cells were determined. The experiment was
repeated three times.
BrdU assay
The HepG2 cells and HL-7702 cells seeded on the
BioFlex amino-culture plates were synchronized for 24 h and then
connected to the Flexcell FX-4000T Tension system. The experimental
group was treated for 24 h with a tensile strain amplitude of 15%
and a frequency of 1 Hz, while the control group was the
non-tension loaded group. BrdU (2 µl/well; Sigma-Aldrich; Merck
KGaA) was added to the tension strain and control groups
simultaneously at 8 h before the end of the tension strain
treatment in the 37°C incubator. At the end of the strain, the
cells were collected by trypsin digestion, and the cell
concentration was adjusted to 1×105/ml in DMEM
(containing 10% FBS). The cells were cultured on the 96-cell plate
(100 µl of cell suspensions/well) at 37°C in an atmosphere
containing 5% CO2 for 4 h until they adhered. Finally,
the medium was dried and put in a 4°C refrigerator overnight.
Furthermore, FixDenat solution (200 µl/well; Sigma-Aldrich; Merck
KGaA) was added and incubated for 30 min at room temperature. The
anti-BrdU-Peroxidase storage solution was diluted 100 times with
antibody diluent, added to the cells (100 µl/well) and incubated at
room temperature for 90 min. The supernatant was removed, and PBS
was used to wash the cells three times. Next, tetramethylbenzidine
substrate (100 µl/well; Sigma-Aldrich; Merck KGaA) was added at
room temperature for 5–30 min to complete color development.
Subsequently, sulfuric acid (1 M) was added to terminate the
reaction immediately (25 µl/well). The 96-cell plate was vibrated
for 60 sec and the absorbance was measured at 450 nm using a
microplate reader (Bio-Rad Laboratories, Inc.), with a reference
wavelength of 630 nm.
CCK-8 assay
The HepG2 cells seeded on the BioFlex amino-culture
plates were synchronized for 24 h and then connected to the
Flexcell FX-4000T Tension system. The experimental group was
treated for 24 h with a tensile strain amplitude of 15% and a
frequency of 1 Hz, while the control group was the nontension
loaded group. Then, 0.5 h before the end of the tension strain
treatment, CCK-8 reagent (Sangon Biotech, Co., Ltd.) was added to
the tension strain and control groups at the same time (200
µl/well), and the assay was operated according to the
manufacturer's protocols. After the tension strain treatment, the
cell culture medium in each well was transferred into a 96-well
plate. Subsequently, the 96-cell plate was put in the microplate
reader (Bio-Rad Laboratories, Inc.). After vibration for 60 sec,
the absorbance at 450 nm was measured, with a reference wavelength
of 630 nm.
miRNA expression analysis using a
miRNA array
Total RNA was extracted by TRIzol®
reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol, purified using miRNeasy Mini kit (Qiagen
GmbH) and quantified using the NanoDrop™ 2000 (Thermo Fisher
Scientific, Inc.), and the RNA integrity was assessed using an
Agilent 2100 Bioanalyzer instrument (Agilent Technologies, Inc.).
The sample labeling, microarray hybridization and washing were
performed based on the manufacturer's standard protocols.
Differentially expressed miRNAs were then identified based on
fold-change and P-value, as calculated using the t-test. The
threshold set for up- and downregulated genes was a fold-change
≥2.0 and P<0.05.
RT-qPCR
RT-qPCR was performed to validate the microarray
data. The extracted total RNAs were reverse transcribed using
Moloney Murine Leukemia Virus reverse transcriptase (Thermo Fisher
Scientific, Inc.) with a special stem-loop primer (RT primer). The
thermocycling conditions were as follows: 42°C for 1 h, followed by
70°C for 5 min. Then, RT-qPCR was performed with a SYBR Green PCR
kit (Takara Bio, Inc.) on an ABI PRISM 7500 Real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocols. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 15 min, followed by 40
cycles at 62°C for 40 sec, 95°C for 1 min and 55°C for 1 min. The
sequences of RT and PCR primers are listed in Table I. The expression levels of miRNAs
were normalized to U6 expression. miRNA expression levels were
calculated using the 2−ΔΔCq method (16).
 | Table I.Primer design. |
Table I.
Primer design.
| Gene name | Primer sequence
(5′→3′) |
|---|
|
hsa-miR-7845-5p | F:
GCGAAGGGACAGGGAGGG |
|
| R:
AGTGCAGGGTCCGAGGTATT |
|
| RT primer:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCACGA |
|
hsa-miR-6889-5p | F:
GTCGGGGAGTCTGGGGTC |
|
| R:
AGTGCAGGGTCCGAGGTATT |
|
| RT primer:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATTCCG |
|
hsa-miR-6794-5p | F:
CGCAGGGGGACTGGGG |
|
| R:
AGTGCAGGGTCCGAGGTATT |
|
| RT primer:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTCAC |
|
hsa-miR-6752-5p | F:
GGGGGGTGTGGAGCCA |
|
| R:
AGTGCAGGGTCCGAGGTATT |
|
| RT primer:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCCCC |
| hsa-miR-503-5p | F:
CGTAGCAGCGGGAACAGTT |
|
| R:
AGTGCAGGGTCCGAGGTATT |
|
| RT primer:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGCAG |
| hsa-miR-4428 | F:
CGCAAGGAGACGGGAACA |
|
| R:
AGTGCAGGGTCCGAGGTATT |
|
| RT primer:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTCCA |
| hsa-miR-296-5p | F:
GAGGGCCCCCCCTCAA |
|
| R:
AGTGCAGGGTCCGAGGTATT |
|
| RT primer:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAGGA |
| U6 | F:
GCTTCGGCAGCACATATACTAAAAT |
|
| R:
CGCTTCACGAATTTGCGTGTCAT |
|
| RT primer:
CGCTTCACGAATTTGCGTGTCAT |
Target genes of differentially
expressed miRNAs
Target genes of differentially expressed miRNAs were
determined as the intersection of the predictions of the three
databases: TargetScan V7.2 (http://www.targetscan.org/), microRA.org
(http://www.microrna.org; November 2010 version)
and PITA (https://genie.weizmann.ac.il/) (17). All data were obtained from the
databases in January 2019.
GO and pathway analyses of predicted
mRNA targets
GO analysis (http://geneontology.org/docs/go-citation-policy/)
and pathway analysis were performed on predicted mRNA targets of
miRNAs using Database for Annotation, Visualization, and Integrated
Discovery (18) and KEGG database
(19,20), respectively. Specific biological
process categories and pathways were enriched. The threshold of
significance was defined by P<0.05 and Benjamini-Hochberg
(21) false discovery rate
(FDR-bh) <0.25.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 6 software (GraphPad Software, Inc.). Each experiment was
performed at least three times, and all values were expressed as
the mean ± standard deviation. Data from ≥3 were analyzed using
one-way ANOVA followed by Fisher's Least Significant Difference or
Dunnett's post hoc comparison test as applicable. P<0.05 was
considered to indicate a statistically significant difference.
Results
BioFlex amino-culture plate
coating
Due to the lack of related reports on HepG2 cells
cultured on BioFlex plates, several methods were tried in this
experiment. Finally, type I rat tail collagen was selected. The
results showed that type I rat tail collagen coating led to
increased cell adherence and growth of HepG2 cells in the BioFlex
plates (Fig. 1).
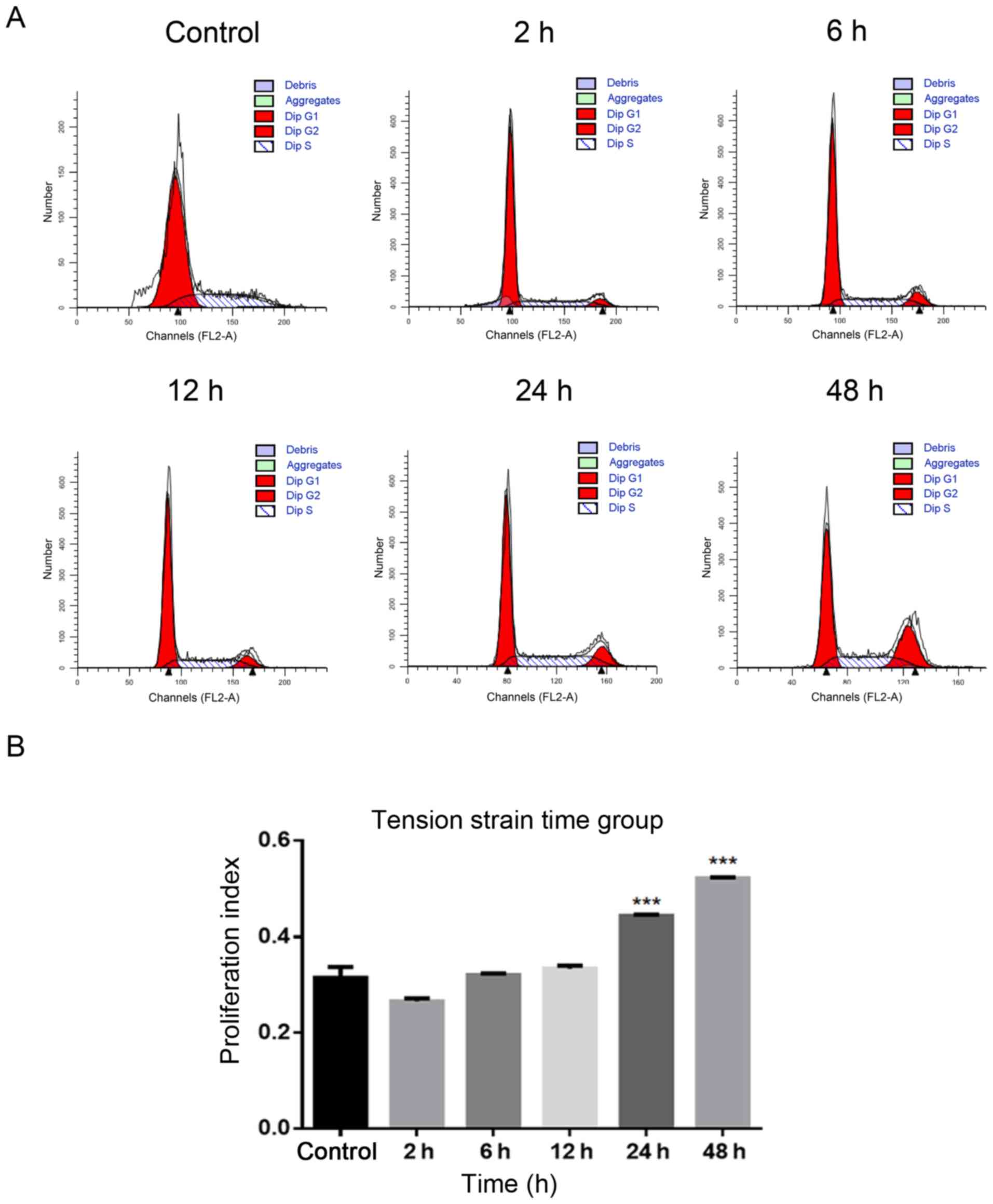
Using flow cytometry to determine the
most suitable strain condition
i) Tension strain time: The PI increased
significantly in 24 and 48 h groups compared with the control group
without tension (P<0.0001; Fig. 2A
and B). ii) Tensional strain frequency: The PI increased
significantly in the 1 Hz group compared with the control group
without tension (P<0.001; Fig. 2C
and D). iii) Tensile strain amplitude: The PI increased
significantly in the 15% group compared with the control group
without tension (P<0.0001; Fig. 2E
and F).
Based on the experimental results of the
aforementioned three groups of tensile strain loading conditions, a
tensile strain amplitude of 15%, frequency of 1 Hz and time of 24 h
were selected as the ideal conditions for the following
experiments.
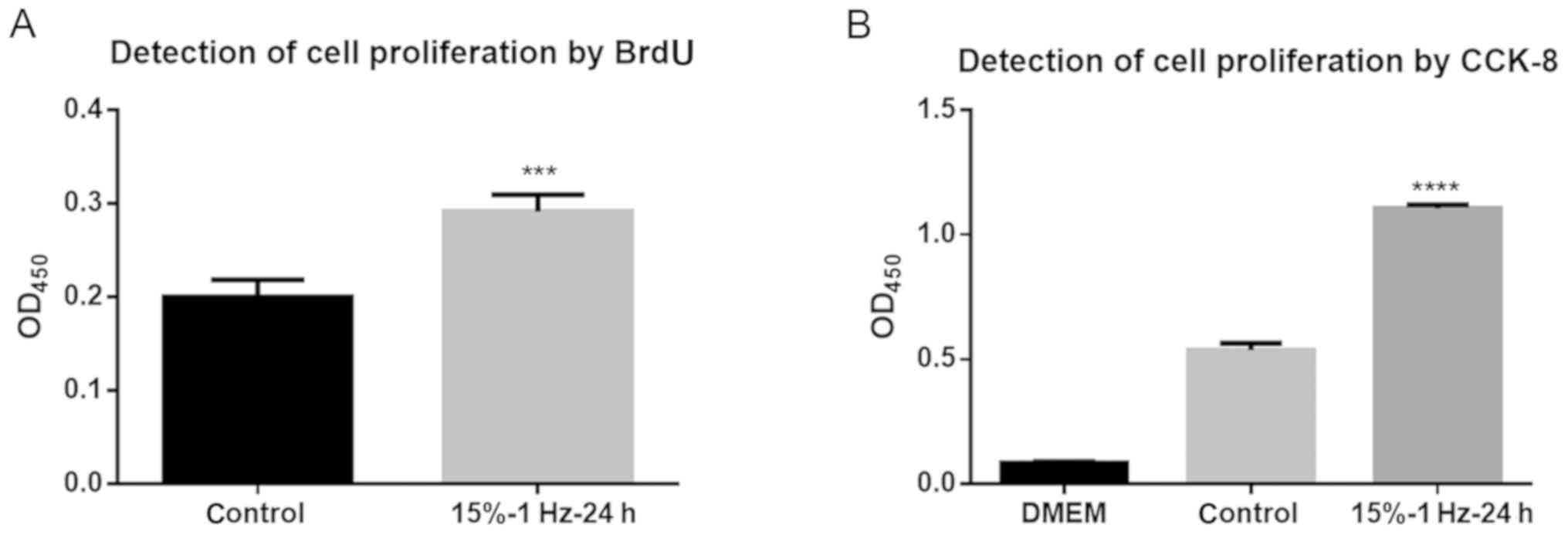
Proliferation of liver cancer cells is
promoted by tensor strain
The results of the BrdU assay showed that the
tensile strain amplitude of 15% at 1 Hz for 24 h significantly
promoted DNA synthesis in cells compared with the control group
(P<0.001; Fig. 3A).
Additionally, the CCK-8 assay also demonstrated that these
conditions significantly promoted DNA synthesis in the cells
(P<0.0001; Fig. 3B). Thus, this
indicated that tensile strain could lead to increased liver cancer
cell proliferation.
Differentially expressed miRNAs in
response to tensile strain
To investigate the effects of tensile strain on the
miRNA profile of HepG2 cells, tensile strain was applied to the
activated HepG2 cells, and RNA was extracted and subjected to
microarray analysis. As shown in Table II, seven significantly
differentially expressed miRNAs showed a fold-change threshold of
≥2.0 and P<0.05. Of these seven miRNAs in the tensile
strain-treated group, five were upregulated (hsa-miR-296-5p,
hsa-miR-6752-5p, hsa-miR-6794-5p, hsa-miR-6889-5p and
hsa-miR-7845-5p) and two were downregulated (hsa-miR-4428 and
hsa-miR-503-5p) compared with the control group.
 | Table II.Differentially expressed miRNAs. |
Table II.
Differentially expressed miRNAs.
| miRNA | P-value | Regulation | FC |
|---|
| hsa-miR-296-5p | 0.00094 | up | 6.387268 |
| hsa-miR-4428 | 0.015763873 | down | −2.8974507 |
| hsa-miR-503-5p | 0.043706737 | down | −4.2659197 |
|
hsa-miR-6752-5p | 0.015388427 | up | 9.723028 |
|
hsa-miR-6794-5p | 0.000131 | up | 20.911135 |
|
hsa-miR-6889-5p | 0.027508989 | up | 11.433076 |
|
hsa-miR-7845-5p | 0.000946 | up | 9.37658 |
Validation of miRNA microarray with
RT-qPCR
RT-qPCR was performed to validate the microarray
results of the five upregulated and two downregulated miRNAs. As
shown in Fig. 4, the changes in
the seven miRNAs detected by RT-qPCR were consistent with those of
the miRNA microarray.
GO and KEGG pathway analyses
Potential target genes for the aforementioned seven
differentially expressed miRNAs were searched using three
bioinformatics databases, miRNAorg, PITA and TargetScan, which
predicted 2,306, 1,277 and 16,205 target genes, respectively. The
false results were removed by taking the intersection of the
results from the three databases for 224 target genes (Fig. 5). The 16,205 predictive target
genes refer to the sum of all seven predictive target genes that
differentially express miRNAs. The components are shown in the
Table III.
 | Table III.Predictive target genes for
differentially expressed miRNAs. |
Table III.
Predictive target genes for
differentially expressed miRNAs.
| miRNA | Predictive target
genes |
|---|
| hsa-miR-296-5p |
2,613 |
| hsa-miR-4428 |
3,043 |
|
hsa-miR-6752-5p |
2,572 |
|
hsa-miR-6794-5p |
2,521 |
|
hsa-miR-6889-5p |
1,279 |
|
hsa-miR-7845-5p |
2,327 |
| hsa-miR-503-5p |
1,850 |
| Total | 16,205 |
GO and pathway analyses were used to understand the
biological characteristics of the aforementioned target genes.
Using a threshold of FDR-bh <0.25, 132 GO terms were considered
to be significant. Additionally, 6, 25 and 101 GO terms belonged to
the ‘cellular component’ category (Fig. 6A), ‘molecular function’ category
(Fig. 6B) and ‘biological process’
category (Fig. 6C), respectively.
The x-axis of Fig. 6 shows the
P-value corrected by FDR-bh algorithm, which is converted by
-log10. The longer the column is, the higher the enrichment degree
is. The target genes were concentrated in ‘cyclin-dependent protein
kinase holoenzyme complex’, ‘neuromuscular junction’, ‘microtubule
cytoskeleton’, ‘protein kinase binding’, ‘protein binding’,
‘positive regulation of transcription from RNA polymerase II
promoter’, ‘positive regulation of transcription, DNA-templated’,
‘negative regulation of cell migration’, ‘positive regulation of
skeletal muscle tissue development’ and ‘patterning of blood
vessel’ categories.
Using KEGG pathway analysis, 52 significant KEGG
pathways were identified at a cutoff FDR-bh <0.25. These
pathways included diseases, such as ‘breast cancer’, ‘melanoma’,
‘prostate cancer’, ‘endocrine resistance’ and ‘bladder cancer’, and
signaling pathways, such as ‘mitogen-activated protein kinase
(MAPK) signaling pathway’, ‘PI3K-Akt signaling pathway’, ‘forkhead
box O signaling pathway’ and ‘Ras signaling pathway’ (Fig. 6D).
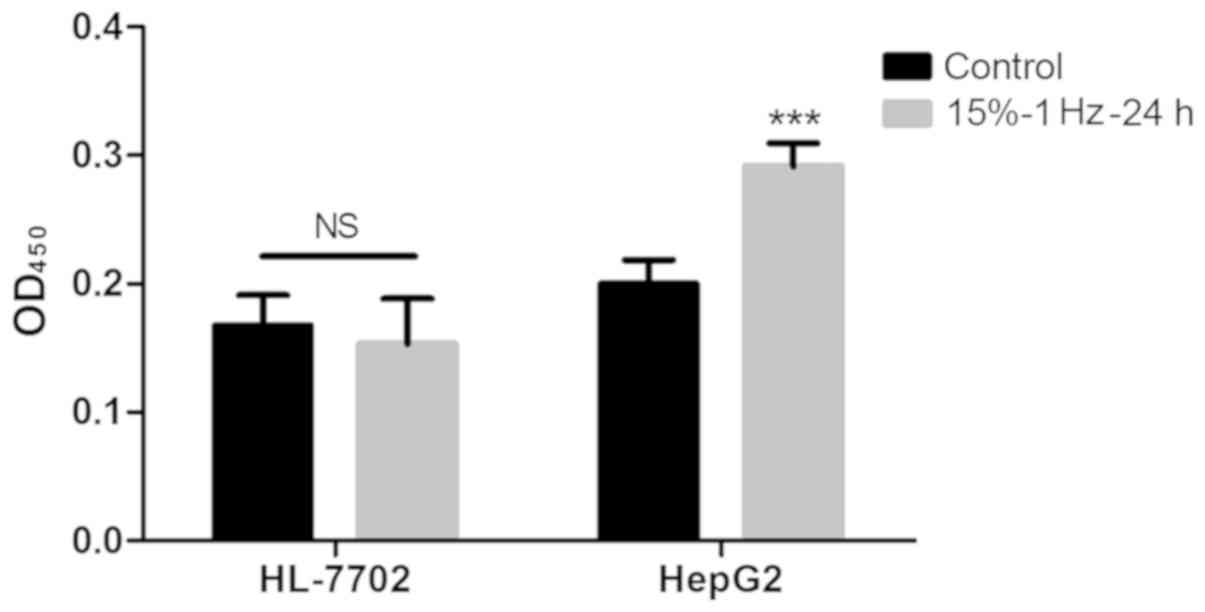
Strain has no effect on the
proliferation of normal liver cells
Using BrdU experiments, it was demonstrated that the
proliferative ability of HepG2 cells after tension intervention was
significantly improved (P<0.05), while the proliferative ability
of normal hepatocyte HL-7702 did not change significantly (Fig. 7).
Flexcell-5000T was also used to performed traction
experiments on Huh-7 cells and MHCC97H cells, but the two cells
could not adhere to the membrane on the traction plate, and thus
HepG2 cells were selected for subsequent experimentation.
Discussion
Human liver cancer cells grow in a complex internal
environment where the mechanical factors should not be ignored. The
present study demonstrated that tensile strain promoted the growth
of liver cancer cells and accelerated their proliferation. To some
extent, this could explain why cirrhosis can significantly increase
the occurrence and development of liver cancer, but the exact
underlying mechanism is still unclear. It has been suggested that
other biomechanical forces, such as fluid shear stress, may affect
the cytoskeleton of liver cancer cells (22). Subsequently, this fluid shear
stress could activate signaling pathways, affect the gene
expression and influence cellular features (23). Numerous studies have shown that
strain can promote the proliferation of various types of cells,
such as VSMCs (24), osteoblasts
(25) and dermal keratinocytes
(26). In the present study, type
I rat tail collagen was chosen to coat BioFlex plates for effective
adherence and cell growth. Then, suitable tensile strain loading
conditions were identified for human liver cancer cell line HepG2,
and seven differentially expressed miRNAs were screened after
tensile strain loading.
Previous studies have shown that miR-296-5p is
upregulated and plays a role in the progression of various tumors,
such as esophageal and laryngeal cancer (27–29).
Additionally, it is also overexpressed in a number of tumor cells
and induces carcinogenesis in human cells by downregulating the
p53-p21 (wild-type p53 activated fragment 1) pathway (30). It has been demonstrated that
miR-6889-5p has a catalytic role in type 1 autoimmune liver disease
(31). It can also inhibit the
proliferation of human cells (32). miR-4428 is associated with
malignant B-cell lymphoma (33).
These previous results suggest that a number of differentially
expressed miRNAs are closely related to the development of
cancer.
In the present study, it was shown that miR-503-5p
was downregulated in tensile strain-treated HepG2 cells. Previous
reports have indicated that miR-503-5p is related to the
proliferation and invasion of various cancer cells. For example, as
a cell migration and invasion suppressor, miR-503-5p can inhibit
the metastasis of ovarian cancer cells by inhibiting the
CD97-mediated Janus kinase 2/STAT3 pathway (34). It was recently found to inhibit
cell epithelial-to-mesenchymal transition and metastasis of liver
cancer by inhibiting WEE1 G2 checkpoint kinase, thus predicting the
prognosis of liver cancer (35).
As a cell proliferation suppressor, miR-503-5p inhibited the
proliferation of T24 and EJ bladder cancer cells by interfering
with the retinoblastoma gene/E2F signaling pathway (36). However, whether miR-503-5p affects
the growth of liver cancer cells by regulating their proliferation
has not yet been reported, and it is worth further study. It is
worth noting that SMAD7, as one of the target genes of miR-503-5p,
is closely related to the expression of TGF-β. TGF-β is a key
factor in the process of hepatic fibrosis (8). Previous studies have demonstrated
that SMAD7 has roles in various types of cancer; it is
overexpressed in endometrial carcinoma (37), it can enhance the tumorigenicity of
pancreatic cancer (38) and it is
closely associated with colon cancer (39). miR-92b (40) and long noncoding RNA snail family
zinc finger 3-antisense 1 (41)
could promote liver cancer proliferation and metastasis by
targeting SMAD7. These results suggested that mechanical factors
may have a biological role by regulating the expression of SMAD7
gene in the development of liver cancer. The specific mechanism
underlying this process needs to be identified and studied
further.
To understand the specific function of the
differentially expressed miRNAs, the target mRNAs of the
differentially expressed miRNAs were also analyzed by GO analysis.
The results showed that the target mRNAs were involved in numerous
physiological processes, including ‘cyclin-dependent protein kinase
holoenzyme complex’, which could indicate that they may exert an
anticancer effect by inhibiting the effect of this complex
(42). Furthermore, the genes were
enriched in ‘transcription factor complex’, ‘positive regulation of
transcription from RNA polymerase II promoter’ and ‘positive
regulation of transcription, DNA templated’, which are also related
to the DNA transcription and RNA translation of the cells,
indicating that the function of miRNAs is closely related to the
proliferation and viability of cancer cells. Also, enrichment in
‘patterning of blood vessels’ may be associated with tumor cells
stimulating angiogenesis to provide nutrition. To reveal the
regulation of pathways by these miRNAs, KEGG pathway analysis was
performed based on the predicted target mRNAs of these miRNAs.
Notably, some pathways related to cancers, such as ‘breast cancer’,
‘melanoma’, ‘prostate cancer’ and ‘bladder cancer’, were identified
as significant. The significant ‘MAPK signaling pathway’ has been
reported to be associated with multiple cancers (43–45).
Also, ‘PI3K-Akt signaling pathway’ components were found to be
frequently altered in human cancers (46). The results suggested that these
pathways might be involved in the proliferation and development of
liver cancer, but the specific mechanisms need to be further
explored.
In conclusion, a series of changes in tensile
strain-treated HepG2 cells were explored in this study. Tensile
strain was found to promote the proliferation of HepG2 cell line.
Some differentially expressed miRNAs were screened, and their
possible target genes and related pathways were preliminarily
discussed. There were two major limitations of this research. The
first one is that only a single cell line was used in this study.
The second is that no experiments were performed to confirm these
findings with liver cancer tissues, such as small RNA-seq datasets
of low degree hepatic fibrosis and a high degree of hepatic
fibrosis liver cancer samples. The follow-up studies may need to be
completed by other research groups. The findings could provide new
information for the study of carcinogenesis and development of
liver cancer and contribute to improving the outcomes for patients
with liver cancer.
Acknowledgements
Not applicable.
Funding
This study was funded by the National Natural
Science Foundation of China (grant no. 11472300).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL, SS, SY and LZ conceived the study, performed the
research and wrote the first draft. JH, YS, KG and LZ collected and
analyzed the data. All authors contributed to the design and
interpretation of the study, and to further drafts. LZ was the
guarantor and made substantial contributions to conception and
design of the study, and the interpretation of data. LZ supervised
and reviewed the results of the experiment and all aspects of the
research work and revised the content of the manuscript prior to
submission. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
LZ is a professor of the Second Affiliated Hospital
of the Second Military Medical University, a member of the Shanghai
Biomechanics Professional Committee and a peer-reviewed expert of
the National Natural Science Foundation of China (NSFC). His main
research direction is biomechanical study of digestive system
diseases and tumors, with emphasis on liver hemodynamics and
gastrointestinal dynamics. For numerous years, our team performed
research into the hemodynamics of portal hypertension before and
after TIPS, and completed the NSFC project: The role and mechanism
of microRNAs in the biomechanical response of hepatic stellate
cells. The research reported by this manuscript is one part of
another NSFC project: The role of microRNAs regulating the
differentiation and invasion of liver cancer in response to
mechanical force.
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
HepG2, tensile strain, liver cancer,
mechanotransduction
|
References
|
1
|
Masuzaki R, Tateishi R, Yoshida H, Goto E,
Sato T, Ohki T, Imamura J, Goto T, Kanai F, Kato N, et al:
Prospective risk assessment for liver cancer development in
patients with chronic hepatitis C by transient elastography.
Hepatology. 49:1954–1961. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fattovich G, Stroffolini T, Zagni I and
Donato F: Hepatocellular carcinoma in cirrhosis: Incidence and risk
factors. Gastroenterology. 127 (Suppl 1):S35–S50. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Osada S, Kanematsu M, Imai H, Goshima S
and Sugiyama Y: Hepatic fibrosis influences the growth of liver
cancer. Hepatogastroenterology. 55:184–187. 2008.PubMed/NCBI
|
|
4
|
Kornek M, Raskopf E, Tolba R, Becker U,
Klöckner M, Sauerbruch T and Schmitz V: Accelerated orthotopic
liver cancers growth is linked to increased expression of
pro-angiogenic and prometastatic factors in murine liver fibrosis.
Liver Int. 28:509–518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qi F, Hu JF, Liu BH, Wu CQ, Yu HY, Yao DK
and Zhu L: miR-9a-5p regulates proliferation and migration of
hepatic stellate cells under pressure through inhibition of Sirt1.
World J Gastroenterol. 21:9900–9915. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yi S, Qin X, Luo X, Zhang Y, Liu Z and Zhu
L: Identification of miRNAs associated with the mechanical response
of hepatic stellate cells by miRNA microarray analysis. Exp Ther
Med. 16:1707–1714. 2018.PubMed/NCBI
|
|
7
|
Shen S, Luo X, Gao K, Sun Y, Yao D and Zhu
L: Identification and integrative analysis of microRNAs and mRNAs
involved in proliferation and invasion of pressure treated human
liver cancer cell lines. Mol Med Rep. 20:375–387. 2019.PubMed/NCBI
|
|
8
|
Sakata R, Ueno T, Nakamura T, Ueno H and
Sata M: Mechanical stretch induces TGF-beta synthesis in hepatic
stellate cells. Eur J Clin Invest. 34:129–136. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goto T, Mikami KI, Miura K, Ohshima S,
Yoneyama K, Nakane K, Watanabe D, Otaka M and Watanabe S:
Mechanical stretch induces matrix metalloproteinase 1 production in
human hepatic stellate cells. Pathophysiology. 11:153–158. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kozomara A, Birgaoanu M and
Griffiths-Jones S: miRBase: From microRNA sequences to function.
Nucleic Acids Res. 47(D1): D155–D162. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Y, Mohammed A, Oubaidin M, Evans CA,
Zhou X, Luan X, Diekwisch TG and Atsawasuwan P: Cyclic stretch and
compression forces alter microRNA-29 expression of human
periodontal ligament cells. Gene. 566:13–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Parpart S, Roessler S, Dong F, Rao V,
Takai A, Ji J, Qin LX, Ye QH, Jia HL, Tang ZY, et al: Modulation of
miR-29 expression by α-fetoprotein is linked to the liver cancer
epigenome. Hepatology. 60:872–883. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vanderploeg EJ, Imler SM, Brodkin KR,
García AJ and Levenston ME: Oscillatory tension differentially
modulates matrix metabolism and cytoskeletal organization in
chondrocytes and fibrochondrocytes. J Biomech. 37:1941–1952. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Davies D and Allen P: DNA Analysis by Flow
Cytometry. Flow Cytometry. Macey MG: Humana Press; pp. 165–179.
2007, View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yi M, Horton JD, Cohen JC, Hobbs HH and
Stephens RM: WholePathwayScope: A comprehensive pathway-based
analysis tool for high-throughput data. BMC Bioinformatics.
7:302006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Goto S, Kawashima S, Okuno Y
and Hattori M: The KEGG resource for deciphering the genome.
Nucleic Acids Res. 32:D277–D280. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
22
|
Yan Z, Su G, Gao W, He J, Shen Y, Zeng Y
and Liu X: Fluid shear stress induces cell migration and invasion
via activating autophagy in HepG2 cells. Cell Adhes Migr.
13:152–163. 2019. View Article : Google Scholar
|
|
23
|
Wang X, Zhang Y, Feng T, Su G, He J, Gao
W, Shen Y and Liu X: Fluid Shear Stress Promotes Autophagy in
Hepatocellular Carcinoma Cells. Int J Biol Sci. 14:1277–1290. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
de Waard V, Arkenbout EK, Vos M, Mocking
AI, Niessen HW, Stooker W, de Mol BA, Quax PH, Bakker EN, VanBavel
E, et al: TR3 nuclear orphan receptor prevents cyclic
stretch-induced proliferation of venous smooth muscle cells. Am J
Pathol. 168:2027–2035. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weyts FA, Bosmans B, Niesing R, van
Leeuwen JP and Weinans H: Mechanical control of human osteoblast
apoptosis and proliferation in relation to differentiation. Calcif
Tissue Int. 72:505–512. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nishimura K, Blume P, Ohgi S and Sumpio
BE: The effect of different frequencies of stretch on human dermal
keratinocyte proliferation and survival. J Surg Res. 155:125–131.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hong L, Han Y, Zhang H, Li M, Gong T, Sun
L, Wu K, Zhao Q and Fan D: The prognostic and chemotherapeutic
value of miR-296 in esophageal squamous cell carcinoma. Ann Surg.
251:1056–1063. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Maia D, de Carvalho AC, Horst MA, Carvalho
AL, Scapulatempo-Neto C and Vettore AL: Expression of miR-296-5p as
predictive marker for radiotherapy resistance in early-stage
laryngeal carcinoma. J Transl Med. 13:2622015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vaira V, Faversani A, Dohi T, Montorsi M,
Augello C, Gatti S, Coggi G, Altieri DC and Bosari S: miR-296
regulation of a cell polarity-cell plasticity module controls tumor
progression. Oncogene. 31:27–38. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoon AR, Gao R, Kaul Z, Choi IK, Ryu J,
Noble JR, Kato Y, Saito S, Hirano T, Ishii T, et al: MicroRNA-296
is enriched in cancer cells and downregulates p21WAF1 mRNA
expression via interaction with its 3′ untranslated region. Nucleic
Acids Res. 39:8078–8091. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Migita K, Komori A, Kozuru H, Jiuchi Y,
Nakamura M, Yasunami M, Furukawa H, Abiru S, Yamasaki K, Nagaoka S,
et al: Circulating microRNA Profiles in Patients with Type-1
Autoimmune Hepatitis. PLoS One. 10:e01369082015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Polioudakis D, Abell NS and Iyer VR:
miR-503 represses human cell proliferation and directly targets the
oncogene DDHD2 by non-canonical target pairing. BMC Genomics.
16:402015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jima DD, Zhang J, Jacobs C, Richards KL,
Dunphy CH, Choi WW, Au WY, Srivastava G, Czader MB, Rizzieri DA, et
al Hematologic Malignancies Research Consortium, : Deep sequencing
of the small RNA transcriptome of normal and malignant human B
cells identifies hundreds of novel microRNAs. Blood. 116:e118–e127.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park GB and Kim D: MicroRNA-503-5p
Inhibits the CD97-Mediated JAK2/STAT3 Pathway in Metastatic or
Paclitaxel-Resistant Ovarian Cancer Cells. Neoplasia. 21:206–215.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiang SP and Li ZR: miR-503-5p regulates
cell epithelial-to-mesenchymal transition, metastasis and prognosis
of liver cancer through inhibiting WEE1. Eur Rev Med Pharmacol Sci.
23:2028–2037. 2019.PubMed/NCBI
|
|
36
|
Li X, Han X, Yang J, Sun J and Wei P:
miR-503-5p inhibits the proliferation of T24 and EJ bladder cancer
cells by interfering with the Rb/E2F signaling pathway. Xi Bao Yu
Fen Zi Mian Yi Xue Za Zhi. 33:1360–1364. 2017.(In Chinese).
PubMed/NCBI
|
|
37
|
Dowdy SC, Mariani A, Reinholz MM, Keeney
GL, Spelsberg TC, Podratz KC and Janknecht R: Overexpression of the
TGF-beta antagonist Smad7 in endometrial cancer. Gynecol Oncol.
96:368–373. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kleeff J, Ishiwata T, Maruyama H, Friess
H, Truong P, Büchler MW, Falb D and Korc M: The TGF-beta signaling
inhibitor Smad7 enhances tumorigenicity in pancreatic cancer.
Oncogene. 18:5363–5372. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Broderick P, Carvajal-Carmona L, Pittman
AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K,
Fielding S, et al CORGI Consortium, : A genome-wide association
study shows that common alleles of SMAD7 influence colorectal
cancer risk. Nat Genet. 39:1315–1317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhuang LK, Yang YT, Ma X, Han B, Wang ZS,
Zhao QY, Wu LQ and Qu ZQ: MicroRNA-92b promotes liver cancer
progression by targeting Smad7 and is mediated by long non-coding
RNA XIST. Cell Death Dis. 7:e22032016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Y, Guo D, Ren M, Zhao Y, Wang X, Chen
Y, Liu Y, Lu G and He S: Long non-coding RNA SNAI3-AS1 promotes the
proliferation and metastasis of liver cancer by regulating the
UPF1/Smad7 signalling pathway. J Cell Mol Med. 2019.
|
|
42
|
Mani S, Wang C, Wu K, Francis R and
Pestell R: Cyclin-dependent kinase inhibitors: Novel anticancer
agents. Expert Opin Investig Drugs. 9:1849–1870. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fang JY and Richardson BC: The MAPK
signalling pathways and colorectal cancer. Lancet Oncol. 6:322–327.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim EK and Choi EJ: Pathological roles of
MAPK signaling pathways in human diseases. Biochim Biophys Acta.
1802:396–405. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ling MT, Wang X, Ouyang XS, Lee TK, Fan
TY, Xu K, Tsao SW and Wong YC: Activation of MAPK signaling pathway
is essential for Id-1 induced serum independent prostate cancer
cell growth. Oncogene. 21:8498–8505. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fresno Vara JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and González-Barón M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|