Introduction
Glioblastoma (GBM) is one of the commonest solid
tumors and the leading cause of central nervous system-associated
mortality worldwide (1).
Statistical analysis has demonstrated that glioblastoma accounts
for ~75% of all malignant tumors associated with the brain
(2). Conventional therapeutic
approaches, including surgery, chemotherapy and radiotherapy, have
greatly improved patient survival; however, the treatment efficacy
remains poor, with an overall survival of 12–14 months following
surgical resection (3–5). Tumor resistance and recurrence are
particularly problematic and require further investigation
(6–8). It is therefore crucial to determine
the underlying mechanisms of glioma occurrence and progression, in
order to develop novel therapeutic targets.
Long non-coding RNAs (lncRNAs) are defined as
transcripts of 200–1,000 nucleotides in length that are not
translated into proteins (9), and
which modulate various biological processes, including cellular
migration, invasion and apoptosis (10). Increasing evidence has indicated
that lncRNAs are promising biomarkers for tumor diagnosis and
prognosis, including in GBM (11–12).
Matrix metallopeptidase (MMP)-2 and MMP-9 are members of the MMP
family, which promote ECM degradation to allow cancer cells to
migrate out of the primary tumour to form metastases during cancer
progression (13). These studies
revealed the importance of lncRNAs and suggested a novel potential
therapeutic strategy for the treatment of GBM.
The lncRNA Feline Leukemia Virus Subgroup C Cellular
Receptor 1 Antisense RNA 1 (FLVCR1-AS1) is a novel tumor suppressor
located on chromosome 1q32.3 (13). Previous studies have identified
impaired expression of FLVCR1-AS1 in hepatocellular carcinoma, lung
cancer and ovarian cancer (14–17);
however, the underlying mechanism of FLVCR1-AS1 in glioma remains
unknown.
MicroRNAs (miRNAs) are a class of non-coding RNAs
that play pivotal roles in cellular proliferation, migration,
invasion and apoptosis. Numerous miRNAs have been confirmed as
potential biomarkers for glioma development (18,19).
Previous studies reported a decrease in miR-30b-3p expression in
various types of cancer, including glioma (20,21).
However, the molecular mechanism by which miR-30b-3p deregulation
contributes to glioma tumorigenesis remains elusive.
Materials and methods
Cell lines and clinical samples
The human GBM cell lines U251, T98G, LN229 and SHG44
were purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. Normal Human Astrocyte (NHA) cells
were obtained from the American Type Culture Collection. Cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) and maintained at 37°C
(5% CO2) in a humidified atmosphere.
Human GBM samples (n=50) and adjacent normal brain
samples (n=50; 0.5–1 cm from the tumor margin) ere resected from
patients with GBM between June 2013 and June 2017 at The Second
Affiliated Hospital of Harbin Medical University (Harbin, China).
All GBM samples and adjacent normal tissues were confirmed by two
senior pathologists. Then the samples were immediately frozen at
the Department of Pathology, and the clinicopathological
characteristics of all patients were collected. Patients consisted
of 22 men and 28 women with age ranging from 20 to 74 years (median
age, 46 years). None of the patients had received any therapy
before surgery. All GBM samples were confirmed by two senior
pathologists. The present study was approved by The Institutional
Review Board of Harbin Medical University, and all patients
provided written informed consents.
Bioinformatics and Gene Ontology (GO)
term enrichment analysis
The edgeR software package (R Studio3.5.1;
Bioconductor; http://www.rstudio.com) was used to
analyze the aberrantly expressed lncRNAs among normalized gene
expression profile data from The Cancer Genome Atlas (TCGA) GBM
database (http://cancergenome.nih.gov)
(22,23). Samples from moderate to severe
TCGA-GBM cases were compared with those from healthy individuals. A
log fold change >2 and false-discovery rate (P-value) <0.01
were used as the cutoff values for significance, for which
aberrantly expressed candidate lncRNAs were detected. Clinical data
were obtained from the Gene Expression Profiling Interactive
Analysis (GEPIA) dataset (http://gepia.cancer-pku.cn/), and GO term enrichment
analysis was conducted using the Database for Annotation,
Visualization and Integrated Discovery version 6.8 (DAVID;
http://david.ncifcrf.gov/).
Reverse transcription-quantitative
(RT-q) PCR)
RT-qPCR was used to detect the expression levels of
FLVCR1-AS1 and miR-30b-3p. Total RNA was extracted from clinical
samples (250 mg) or cell lines (1×106) using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The concentration
of eluted RNA was determined with the NanoDrop spectrophotometer
(Applied Biosystems; Thermo Fisher Scientific, Inc.). cDNA was
synthesized using the PrimeScript™ RT kit, and qPCR was conducted
using SYBR® Green PCR Master Mix (both Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocols.
The thermocycling conditions were as follows: 95°C for 10 min,
followed by 40 cycles at 95°C for 15 sec, and 60°C for 60 sec.
GAPDH was used as an endogenous control to normalize lncRNA
FLVCR1-AS1 expression level. U6 was used as an endogenous control
to normalize miR-30b-3p expression level and the results were
calculated using the 2−ΔΔCq method (24). The sequences of the primers are
presented in Table I.
 | Table I.Sequences of the primers used for
reverse transcription-quantitative PCR. |
Table I.
Sequences of the primers used for
reverse transcription-quantitative PCR.
| Gene | Sequence |
|---|
|
FLVCR1-AS1-sense |
5′-GTGGCTCTCTCGTTCCC-3′ |
|
FLVCR1-AS1-antisense |
5′-CCGTCCTTCGGTAGTGTC-3′ |
|
miR-30b-3p-sense |
5′-UGUAAACAUCCUACACUCAGCU-3′ |
|
miR-30b-3p-antisense |
5′-ACAUUUGUAGGAUGUAGUCGA-3′ |
| MMP-9-sense |
5′-AGACCTGGGCAGATTCCAAAC-3′ |
|
MMP-9-antisense |
5′-CGGCAAGTCTTCCGAGTAGT-3′ |
| MMP-2-sense |
5′-CAGGACATTGTCTTTGATGG-3′ |
|
MMP-2-antisense |
5′-TGAAGAAGTAGCTATGACCA-3′ |
| GAPDH-sense |
5′-TCCTCTGACTTCAACAGCGACAC-3′ |
|
GAPDH-antisense |
5′-CACCCTGTTGCTGTAGCCAAATTC-3′ |
| U6-sense |
5′-CTCGCTTCGGCAGCACA-3′ |
| U6-antisense |
5′-AACGCTTCACGAATTTGC-3′ |
Cell transfection
Small interfering RNAs (siRNAs) targeting FLVCR1-AS1
and the corresponding negative control (si-NC; both Shanghai
GenePharma, Co., Ltd.) were used to generate the
FLVCR1-AS1-knockdown model. In order to increase or decrease the
level of miR-30b-3p, the miR-30b-3p mimic or inhibitor and
corresponding negative control (miR-NC; all Shanghai GenePharma,
Co., Ltd.) were used to up- and downregulate the expression of
miR-30b-3p, respectively. LN229 and T98G cells (5×105)
were transfected with 20 µM of each construct using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's protocol
and incubated at 37°C for 6 h. The culture medium was then changed
with fresh DMEM containing 10% FBS, and subsequent experimentation
was conducted 24 h post-transfection. The sequences of the siRNA
are listed in Table II.
 | Table II.Sequences of the siRNAs used in the
present study. |
Table II.
Sequences of the siRNAs used in the
present study.
| Gene | Sequence |
|---|
|
si-FLVCR1-AS1-sense |
5′-GGUAAGCAGUGGCUCCUCUAA-3′ |
|
si-FLVCR1-AS1-antisense |
5′-AAUUCUCCGAACGUGUCACGU-3′ |
| miR-30b-3p mimics
sense |
5′-UGUAAACAUCCUACACUCAGCU-3′ |
| miR-30b-3p mimics
antisense |
5′-UCACAACCUCCUAGAAAGAGUA-3′ |
| miR-inhibitor
mimics sense |
5′-AGCUGAGUGUAGGAUGUUUAC-3′ |
| miR-inhibitor
mimics antisense |
5′-GGUAAGCAGUGGCUCCUCUAA-3′ |
| siRNA-NC-sense |
5′-UUCUCCGAACGUGUCACGUUU-3′ |
|
siRNA-NC-antisense |
5′-ACGUGACACGUUCGGAGAAUU-3′ |
| miR-mimics
NC-sense |
5′-ACAGUCGCGUUUGCGACUGUU-3′ |
| miR-mimics
NC-antisense |
5′-UUGUCAGCGCAAACGCUGACC-3′ |
| miR-inhibitor
NC-sense |
5′-CAGUACUUUUGUGUAGUACAA-3′ |
| miR-inhibitor
NC-antisense |
5′-UUAACUAAUAUUUCAUCCAUA-3′ |
Dual-luciferase reporter assay
TargetScan (www.targetscan.org/) and StarBase (http://starbase.sysu.edu.cn/) databases were used to
predict the potential target miRs of FLVCR1-AS1. Among the
statistically relevant miRs, the top three were selected in terms
of their prediction score, and included miR-30b-3p, miR-96-5p and
miR-513. As the highest scoring miR in both databases, miR-30b-3p
was selected for further experimentation. The 3′-untranslated
region of FLVCR1-AS1 was cloned into the human FLVCR1-AS1
Luc-reporter plasmid (GenePharm, Inc.), which was then used to
transfect LN229 cells as aforementioned. Briefly, cells were seeded
into 6-well plates at 3×105 cells/well, and the
recombinant vectors were co-transfected with miR-30b-3p mimics or
miR-NC using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Luciferase activity was measured 48 h post-transfection using the
Dual Luciferase Reporter Assay System (Promega Corporation), and
firefly luciferase activity was normalized to that of
Renilla luciferase.
Cell proliferation and colony
formation assays
LN229 and T98G cells were collected 24 h
post-transfection and were seeded into 96-well plates at the
density of 1×104 cells/well. Cell proliferation was
evaluated using the Cell Counting Kit-8 (CCK-8; Beyotime Institute
of Biotechnology) at days 1, 2 and 3, according to the
manufacturer's protocol. Briefly, 10 µl CCK8 solution was added to
each well. After incubation at 37°C for 4 h, the absorbance at 450
nm was measured using a microplate reader (Bio-Rad laboratories,
Inc.).
LN229 and T98G cells (~300 cells/well) were seeded
into 6-well plates in fresh DMEM with 10% FBS and cultured at 37°C.
The medium was replaced every 3 days. After 14 days, cells were
fixed with 4% polyoxymethylene for 10 min at room temperature and
stained with 10% Giemsa (Sigma-Aldrich; Merck KGaA) at room
temperature for 30 min. Colonies >50 cells were counted under a
light microscope (Nikon Corporation).
Invasion assays
LN229 and T98G cells (5×104) were seeded
into the upper chambers of Transwell inserts pre-coated with
Matrigel® (pore size, 8.0 µm; Corning Life Sciences),
and incubated at 37°C (5% CO2) in a humidified
atmosphere for 24 h. Serum-free DMEM was added to the upper chamber
whereas the lower chamber contained DMEM with 20% FBS as a
chemoattractant. The invasive cells were fixed for 10 min using 4%
paraformaldehyde and stained with hematoxylin and eosin for 5 min,
both at room temperature. The stained cells were counted in five
random fields under a light microscope (Nikon Corporation;
magnification, ×100).
Western blotting
Total protein was extracted from patient tissue
samples (250 mg/sample) and cell lines (LN229 and T98G) using RIPA
lysis buffer (Pierce; Thermo Fisher Scientific, Inc.) containing
Protease Inhibitor Cocktail (Complete™ Mini; Roche Applied Science)
at 4°C for 10 min. Protein concentration was measured using a
bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology). Proteins (40 µg) were separated by 10% SDS-PAGE and
transferred onto PVDF membranes (Beyotime Institute of
Biotechnology). Membranes were blocked with tris-buffered saline
(TBS) containing 5% non-fat milk (w/v) for 1 h at room temperature.
Subsequently, membranes were incubated with primary antibodies
targeted against MMP-2 (rabbit polyclonal; 1:1,000; cat. no.
409494; Cell Signaling Technology, Inc.), MMP-9 (rabbit polyclonal;
1:1,000; cat. no. 13667; Cell Signaling Technology, Inc.) and GAPDH
(mouse monoclonal; 1:1,000; cat. no. SC-47724; Santa Cruz
Biotechnology, Inc.) overnight at 4°C. Horseradish
peroxidase-conjugated anti-mouse or anti-rabbit (cat. no. ab6721
and ab6728; 1:2,000; Abcam) secondary antibodies were added
separately for 1 h at room temperature. The bands were visualized
using enhanced chemiluminescence detection (Beyotime Institute of
Biotechnology) with a ChemiDoc™ MP Imaging System and analyzed by
Image Lab software (version 3.0; Bio-Rad Laboratories, Inc.).
Immunofluorescence staining
LN229 and T98G cells (1×105) were
cultured on glass slides treated overnight with 0.1% poly-L-Lysine,
fixed with 4% paraformaldehyde (5% BSA; cat. no. 9048-46-8; Sigma
Aldrich; Merck KGaA) for 20 min, and washed with 0.1% Triton X-100
for 10 min at room temperature. Slides were incubated with a
primary antibody against MMP-2 (rabbit polyclonal; 1:1,000; cat.
no. 40094s; Cell Signaling Technology, Inc.) at 4°C for 1 h, and a
fluorescence-labeled rabbit secondary antibody [Rhodamine
(TRITC)-conjugated goat anti-rabbit immunoglobulin G (IgG); 1:100;
cat. no. SA00007-2; ProteinTech Group, Inc.] at room temperature
for 1 h. Cell nuclei were stained with DAPI (1 µg/ml; cat. no.
4083s; Cell Signaling Technology, Inc.) for 15 min and images were
captured using a fluorescence microscope (Nikon Corporation;
magnification, ×400).
Anti-AGO2 RNA-binding protein
immunoprecipitation (RIP) assay
RIP assay was conducted by using the Magna RIP
RNA-Binding Protein Immunoprecipitation kit (cat. no. 17-701; EMD
Millipore). As a core element of RISC complex, AGO2 directly
initiates the degradation of target mRNAs via its catalytic
activity in gene silencing processes guided by siRNAs or miRNAs.
AGO2 is associated with tumorgenesis via miRNAs-dependent or
independent pathways (25). LN229
and T98G cells transfected with miR-30b-3p were lysed using RIPA
lysis buffer and 100 µl of the cell lysate was collected for RIP
experiments using an anti-AGO2 antibody (Abcam) according to the
manufacturer's instructions. The RNA fraction isolated by RIP was
subjected to RT-qPCR analysis to detect the direct binding between
FLVCR1-AS1 and miR-30b-3p.
Statistical analysis
Data were expressed as the means ± standard
deviation. Statistical analysis was conducted using SPSS v20.0 (IBM
Corp.). One-way ANOVA with Tukey's post hoc test and Student's
t-test were used to determine the level of significance between
groups. The Kaplan-Meier method and the log-rank test were used for
survival analyses using GraphPad Prism v5.0 (GraphPad Software,
Inc.). The association between FLVCR1-AS1 expression and the
clinicopathological characteristics of patients with GBM was
evaluated using χ2 or Fisher's exact tests. The
correlation between the expression of FLVCR-AS1 and the expression
of miR-30b-3p, MMP-2 and MMP-9 was analyzed using Pearson's
correlation. Experiments were independently performed three times.
P<0.05 was considered to indicate a statistically significant
difference.
Results
FLVCR1-AS1 is upregulated in GBM
tissues and is associated with poor prognosis
The top 20 differentially expressed lncRNAs were
identified from TCGA based on the Benjamini-Hochberg method
(26) [log2-fold change (FC) of
>2 and a false discovery rate, P<0.01; Fig. 1A]. FLVCR1-AS1 was the most highly
expressed differentially expressed lncRNAs among the GBM tissue
samples, and its expression was significantly increased in GBM
tissues compared with adjacent normal brain tissue (Fig. 1A and B). To investigate the
potential function of FLVCR1-AS1, the associated gene expression
profiles collected from the GO database were analyzed. ‘Cell
invasion’, ‘Cell adhesion’ and ‘Zinc-finger’ were the most
prominent biological processes (Fig.
1C). In addition, clinical data collected from the GEPIA
database indicated that increased FLVCR1-AS1 was associated with
worse overall survival in patients with GBM, according to the
median patient survival time (Fig.
1D). Among the clinical samples collected for the present
study, the expression level of FLVCR1-AS1 was significantly
associated with mean tumor diameter (n=50; Table III; P=0.030). Furthermore, the
results from RT-qPCR demonstrated that FLVCR1-AS1 expression level
was significantly increased in GBM tissues compared with adjacent
normal brain tissues (Fig. 1E).
These results suggested that FLVCR1-AS1 may be involved in the
development of GBM.
 | Table III.Clinical characteristics of patients
with glioblastoma patients according to FLVCR1-AS1 tissue level
(n=50). |
Table III.
Clinical characteristics of patients
with glioblastoma patients according to FLVCR1-AS1 tissue level
(n=50).
|
|
| FLVCR1-AS1
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | No. of cases | Low | High | P-value |
|---|
| Age, years |
|
<60 | 23 | 6 | 17 | 0.781 |
|
≥60 | 27 | 8 | 19 |
|
| Sex |
|
Male | 22 | 7 | 15 | 0.594 |
|
Female | 28 | 7 | 21 |
|
| Karnofsky
performance status |
|
<60 | 21 | 5 | 16 | 0.991 |
|
≥60 | 29 | 9 | 20 |
|
| Mean tumor
diameter, cm |
|
<5 | 27 | 11 | 16 | 0.030a |
| ≥5 | 23 | 3 | 20 |
|
| Necrosis on
magnetic resonance imaging |
|
Yes | 31 | 8 | 23 | 0.659 |
| No | 19 | 6 | 13 |
|
| Seizure |
|
Yes | 10 | 4 | 6 | 0.345 |
| No | 40 | 10 | 30 |
|
FLVCR1-AS1-knockdown inhibits GBM cell
proliferation, colony formation and invasive ability
To determine FLVCR1-AS1 function in GBM, its
expression level in GBM (U251, LN229, T98G and SHG44) and NHA cells
was assessed using RT-qPCR (Fig.
2A). The results demonstrated a significant increase in
FLVCR1-AS1 expression level in all GBM cells compared with NHA
cells, in particular in LN229 and T98G cell lines. To determine the
effect of FLVCR1-AS1 on the proliferative and invasive abilities of
GBM cells, FLVCR1-AS1 was knocked down in LN229 and T98G cells. The
transfection efficiency was confirmed by RT-qPCR, where FLVCR1-AS1
was significantly decreased in transfected cells compared with
siRNA-Negative Control (si-NC; Fig.
2B). Furthermore, the results from CCK-8 assay demonstrated
that the proliferation of GBM cells transfected with si-FLVCR1-AS1
was decreased compared with cells transfected with si-NC (Fig. 2C). In addition,
FLVCR1-AS1-knockdown significantly decreased the colony formation
(Fig. 2D) and invasion abilities
of GBM cells (Fig. 2E). These
findings suggested that GBM cell proliferation and invasive ability
may be inhibited by FLVCR1-AS1-knockdown in vitro.
 | Figure 2.Knockdown of FLVCR1-AS1 inhibits the
proliferation and invasive ability of GBM cell lines in
vitro. (A) FLVCR1-AS1 gene expression levels in GBM cell lines
(U251, LN229, T98G and SHG44) vs. normal NHA cells were assessed by
RT-qPCR analysis. (B) FLVCR1-AS1 was efficiently knocked down by
siRNA in LN229 and T98G cells, compared with the si-NC group,
(detected by RT-qPCR). (C) GBM cell proliferation was assessed
using the Cell Counting Kit-8 assay (si-FLVCR1-AS1 vs. si-NC). (D)
GBM cells transfected with si-NC or si-FLVCR1-AS1 were incubated
for 14 days and colonies >50 cells were counted. (E) Transwell
assay assessed invasive ability; magnification, ×100. *P<0.05
and **P<0.01 vs. si-NC. Data are presented as the mean ±
standard deviation of three independent experiments. FLVCR1-AS1,
Feline Leukemia Virus Subgroup C Cellular Receptor 1 Antisense RNA
1; GBM, glioblastoma; NHA, Normal Human Astrocyte; RT-qPCR, reverse
transcription-quantitative PCR; si-, small interfering; NC,
negative control. |
Data from TCGA database were used to confirm whether
reduced FLVCR1-AS1 expression affected the invasive capacity of
GBM. The results suggested a positive correlation between
FLVCR1-AS1 expression and the invasion-associated markers MMP-2
(r=0.3428; P=0.0027) and MMP-9 (r=0.2928; P=0.0004; Fig. 3A). Furthermore, RT-qPCR and western
blotting were performed to assess the mRNA and protein expression
levels of MMP-2 and MMP-9 in samples from patients with GBM. MMP-2
levels were also determined by immunofluorescence detection. The
results demonstrated that FLVCR1-AS1-knockdown decreased MMP-2 and
MMP-9 expression in GBM tissues (Fig.
3B and C). In addition, the results from immunofluorescence
staining confirmed that FLVCR1-AS1-knockdown inhibited the
expression of MMP-2 in GBM cell lines (Fig. 3D). Taken together, these findings
suggested that FLVCR1-AS1 may be associated with GBM cell
proliferation and invasive ability.
 | Figure 3.Expression of proliferation- and
invasion-related markers in GBM cells. (A) Correlation between
FLVCR1-AS1 and MMP-2/MMP-9 expression in sample data from TCGA GBM
database. Following transfection with si-FLVCR1-AS1 or si-NC, the
(B) protein and (C) mRNA expression levels of PCNA, MMP-2 and MMP-9
were determined. (D) Immunofluorescence staining revealed a
decreased level of MMP-2 in si-FLVCR1-AS1-transfected cells
(magnification, ×400). *P<0.05 and **P<0.01 vs. si-NC.
Experiment was repeated three times. GBM, glioblastoma; FLVCR1-AS1,
Feline Leukemia Virus Subgroup C Cellular Receptor 1 Antisense RNA
1; MMP, matrix metalloproteinase; TCGA, The Cancer Genome Atlas;
si-, small interfering; NC, negative control; si, small interfering
RNA; TCGA, The Cancer Genome Atlas. |
Correlation between FLVCR1-AS1 and
miR-30b-3p expression
Using the TargetScan and StarBase databases, a
complementary sequence was identified between FLVCR1-AS1 and
miR-30b-3p (Fig. 4A). Luciferase
reporter assays were subsequently used to confirm this putative
miR-30b-3p target site. miR-30b-3p mimics or inhibitor mimics were
used to assess transfection efficiency in the LN229 and T98G cell
lines (Fig. 4B), and luciferase
reporter vectors carrying either the predicted miR-30b-3p wild-type
binding site (FLVCR1-AS1-WT) or its mutant fragment
(FLVCR1-AS1-MUT) were constructed. The luciferase activity of the
LN229 cells was significantly decreased following co-transfection
with the miR-30b-3p mimics and FLVCR1-AS1-WT, but not with
FLVCR1-AS1-MUT (Fig. 4C).
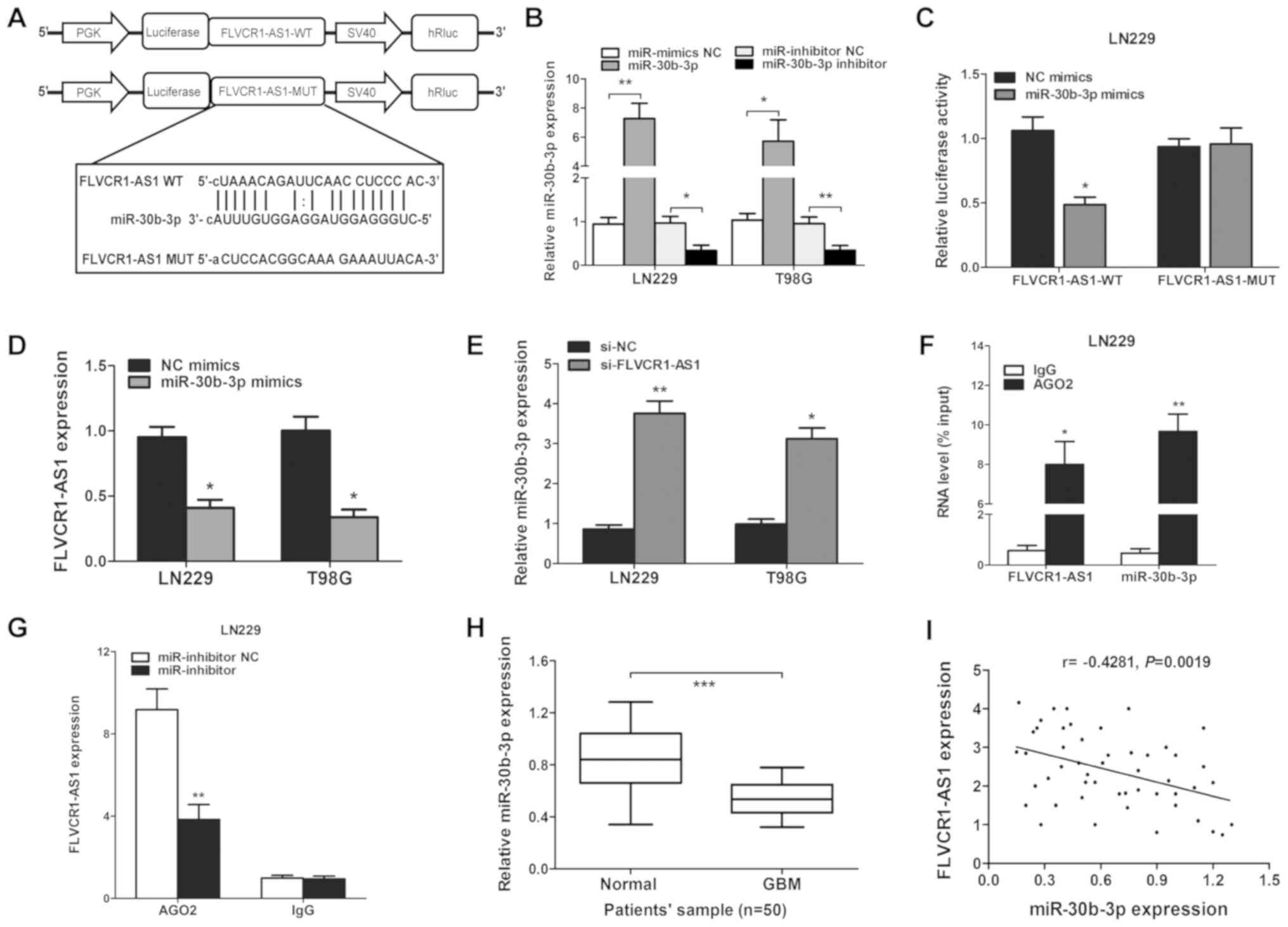 | Figure 4.FLVCR1-AS1 is targeted by miR-30b-3p
at the 3′UTR. (A) Target site of miR-30b-3p in the 3′UTR region of
FLVCR1-AS1. (B) miR-30b-3p expression was upregulated by miR-30b-3p
mimics compared with the miR-mimic NC (**P<0.01 in LN229 and
T98G cells), or inhibited by an miR-30b-3p inhibitor compared with
the miR-inhibitor NC (*P<0.05 and **P<0.01), respectively, in
LN229 and T98G cells. (C) Relative luciferase activity was
determined following co-transfection of LN229 cells with miR-30b-3p
mimics or miR-NC and FLVCR1-AS1-WT or FLVCR1-AS1-MUT. (D) After
transfection with miR-30b-3p mimics, the level of FLVCR1-AS1 was
decreased in LN229 cells, as detected by RT-qPCR. (E) Expression
levels of miR-30b-3p in GBM cell lines following transfection with
si-FLVCR1-AS1 or si-NC. (F) Association between FLVCR1-AS1 and
miR-30b-3p with AGO2. (G) Change in FLVCR1-AS1 level in glioma
cells transfected with an miR-30b-3p inhibitor. AGO2 RNA level
determined by RT-qPCR. (H) Expression level of miR-30b-3p in GBM
compared with adjacent normal tissues. (I) Pearson's correlation
coefficient analysis between FLVCR1-AS1 and miR-30b-3p expression
level. *P<0.05, **P<0.01 and ***P<0.001. Experiments were
repeated three times. FLVCR1-AS1, Feline Leukemia Virus Subgroup C
Cellular Receptor 1 Antisense RNA 1; miR, microRNA; NC, negative
control; UTR, untranslated region; WT, wild-type; MUT, mutant;
siRNA, small interfering RNA; IgG, immunoglobulin G; RT-qPCR,
reverse transcription-quantitative PCR; GBM, glioblastoma. |
The interaction between FLVCR1-AS1 and miR-30b-3p
was subsequently assessed in GBM tissues and cell lines, in
particular whether miR-30b-3p could decrease FLVCR1-AS1 expression.
The results demonstrated that increased miR-30b-3p expression
inhibited FLVCR1AS1 expression in LN229 cells (Fig. 4D). To determine whether miR-30b-3p
was negatively regulated by FLVCR1-AS1, FLVCR1-AS1 was knocked down
in GBM cells. The results demonstrated that miR-30b-3p was
upregulated following FLVCR1-AS1-knockdown in LN229 cells (Fig. 4E).
miRNAs have been reported to function by interacting
with the RNA-induced silencing complex (RISC), which is required
for miRNA-mediated gene silencing. As a core element of RISC
complex, AGO2 directly initiates the degradation of target mRNAs
via its catalytic activity in gene silencing processes guided by
siRNAs or miRNAs. A RIP assay was therefore conducted with LN229
cells and an anti-AGO2 antibody, followed by RT-qPCR analysis. As
demonstrated in Fig. 4F, compared
with the NC (anti-IgG), FLVCR1-AS1 and miR-30b-3p were both
preferentially increased in AGO2 antibody-incubated group. In
addition, the miR-30b-3p inhibitor suppressed the interaction
between AGO2 and FLVCR1-AS1 in LN229 cells (Fig. 4G). The results indicated that
FLVCR1-AS1 and miR-30b-3p reciprocally repressed one another in GBM
cells.
miR-30b-3p inhibits the effect of
FLVCR1-AS1 in GBM cells
The expression level of miR-30b-3p in the 50 GBM and
adjacent normal brain samples was determined by RT-qPCR. A
significant decrease in miR-30b-3p expression was observed in GBM
tissues compared with adjacent normal tissues (P<0.001; Fig. 4H). Furthermore, results from
Pearson's correlation analysis revealed that FLVCR1-AS1 expression
was negatively correlated with miR-30b-3p expression in GBM tissues
(n=50; r=−0.4281; P=0.0019; Fig.
4I). These data suggested that FLVCR1-AS1 may interact with
miR-30b-3p in GBM cells. Furthermore, the results from Kaplan-Meier
analysis demonstrated that miR-30b-3p expression was associated
with the overall survival of patients with glioma from the GEPIA
database (Fig. 5A). Furthermore,
miR-30b-3p expression level in GBM cells was significantly
downregulated compared with that in NHA cells (Fig. 5B). miR-30b-3p expression level was
increased following transfection with si-FLVCR1-AS1, compared with
si-NC. However, following treatment with miR-30b-3p inhibitor,
miR-30b-3p level was decreased in the si-FLVCR1-AS1-transfected
group compared with the miR-inhibitor NC group (Fig. 5C). miR-30b-3p inhibitor also
attenuated the decrease in cell invasive ability induced by
FLVCR1-AS1-knockdown (Fig. 5D).
These findings confirmed that miR-30b-3p may be considered as a
crucial mediator of FLVCR1-AS1-regulated proliferation and invasive
ability in GBM.
Discussion
GBM is one of the most common tumors of the central
nervous system, with a high mortality rate (1). Although great progress has been made,
there is no efficient therapy for GBM. It has been reported that
lncRNAs are key regulators in the progression and development of
various types of cancer type. lncRNAs can act as potential tumor
oncogenes or suppressors, and altered lncRNA expression has been
associated with tumorigenesis (27–30).
The mechanism by which FLVCR1-AS1 functions as an
oncogene in GBM remains unknown. In the present study, FLVCR1-AS1
expression was significantly increased in GBM tissues compared with
adjacent normal tissues. Furthermore, miR-30b-3p expression was
downregulated in GBM tissues, and miR-30b-3p expression was
negatively correlated with FLVCR1-AS1 expression. Results from
bioinformatics analysis was used to validate miR-30b-3p as a
potential target of FLVCR1-AS1. In addition, FLVCR1-AS1-knockdown
inhibited GBM cell proliferation and invasive ability, which was
reversed by miR-30b-3p inhibitor. Taken together, these findings
suggested that FLVCR1-AS1-knockdown may inhibit GBM progression,
indicating a potential therapeutic method for GBM.
Numerous studies reported that the interaction
between lncRNA and miRNA regulates gene expression during
tumorigenesis (31–34). A previous study illustrated that
lncRNA maternally expressed 3 was downregulated in cervical cancer,
and altered cell proliferation and apoptosis by regulating miR-21
(35). Similarly, the lncRNA H19
was demonstrated to act as a competing endogenous RNA (ceRNA) for
miR-29b-3p, which may promote epithelial-mesenchymal transition and
bladder cancer metastasis (36).
In non-small cell lung cancer, the lncRNA small nucleolar RNA host
gene 1 was reported to regulate cell proliferation and invasive
ability by increasing the expression of metadherin via an
interaction with miR-145-5p (37).
It has been reported that aberrant expression of
miRNAs serves a vital role in the biological processes of various
types of cancer (38). For
example, Kumar et al (39)
reported that miR-30b-3p acts as a direct regulator of androgen
receptor signaling in prostate cancer. It was also reported that
FLVCR1-AS1 may interact with miR-513, miR-573 and the Wnt/β-catenin
signaling pathway in hepatocellular carcinoma, lung cancer and
ovarian cancer (14–17). Furthermore, Liu et al
(40) demonstrated that FLVCR1-AS1
can act as a sponge for miR-155, promoting therefore gastric cancer
tumorigenesis by targeting c-Myc. As such, FLVCR1-AS1 may be used
as a ceRNA to indirectly regulate the proliferation, migration and
invasive ability of cholangiocarcinoma cells (41). Yan et al (42) reported that FLVCR1-AS1 aggravates
the biological behaviors of glioma cells via targeting the
miR-4731-5p/E2F2 axis. Bioinformatics analysis identified
FLVCR1-AS1 as a sponge for miR-4731-5p, which resulted in E2F2
upregulation. Rescue assays indicated that FLVCR1-AS1 can modulate
the expression of E2F2 during glioma progression. This study
demonstrated that FLVCR1-AS1 may be considered as a potential
target for tumor therapy. lncRNAs can function as ceRNAs,
sequestering miRNAs and subsequently preventing their expression.
The present study aimed therefore to investigate whether FLVCR1-AS1
could act as a ceRNA for miR-30b-3p in GBM.
In the present study, the putative binding site
between FLVCR1-AS1 and miR-30b-3p was confirmed by using a
luciferase reporter assay system. The results validated FLVCR1-AS1
as a potential tumor suppressor regulating miR-30b-3p and
subsequently inhibiting GBM cell proliferation and invasive
ability. The suppressive effect of si-FLVCR1-AS1 was impeded by
co-transfection with miR-30b-3p inhibitor. Taken together, the
results from the present study described the role and underlying
mechanism of FLVCR1-AS1 in the proliferation and invasive ability
of GBM cells, which may provide some essential information on its
role in GBM tumorigenesis.
In conclusion, the present study demonstrated that
FLVCR1-AS1 may act as an oncogenic lncRNA that could promote the
development and progression of GBM through miR-30b-3p, suggesting
that FLVCR1-AS1 may be considered as a potential biomarker and
therapeutic target for GBM.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WG designed the study, performed experiments,
analyzed the data and wrote the manuscript. HL and YL performed the
in vitro experiments. YZ and FL analyzed the data and
drafted the manuscript. FL and HZ designed and supervised the study
and edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients and the study was approved by the Ethics Committee of
Harbin Medical University (Harbin, China). All procedures were
performed in accordance with national (D.L.n.26, March 4th, 2014)
and international laws and policies (directive 2010/63/EU).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lai NS, Wu DG, Fang XG, Lin YC, Chen SS,
Li ZB and Xu SS: Serum microRNA-210 as a potential noninvasive
biomarker for the diagnosis and prognosis of glioma. Br J Cancer.
112:1241–1246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang K, Kievit FM, Jeon M, Silber JR,
Ellenbogen RG and Zhang M: Nanoparticle-mediated target delivery of
TRAIL as gene therapy for glioblastoma. Adv Healthc Mater.
4:2719–2726. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Delgado-López PD and Corrales-García EM:
Survival in glioblastoma: A review on the treatment modalities.
Clin Transl Oncol. 18:1062–1071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li C, Jing H, Ma G and Liang P: Allicin
induces apoptosis through activation of both intrinsic and
extrinsic pathways in glioma cells. Mol Med Rep. 17:5976–5981.
2018.PubMed/NCBI
|
|
5
|
Nikolov V, Stojanovic M, Kostic A,
Radisavljevic M, Simonovic N, Jelenkovic B and Berilazic L: Factors
affecting the survival of patients with glioblastoma multiforme. J
BUON. 23:173–178. 2018.PubMed/NCBI
|
|
6
|
Han Y: Analysis of the role of the Hippo
pathway in cancer. J Transl Med. 17:1162019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tamura R, Tanaka T, Miyake K, Yoshida K
and Sasaki H: Bevacizumab formalignant gliomas: Current
indications, mechanisms of action and resistance, and markers of
response. Brain Tumor Pathol. 34:62–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li C, Zheng H, Hou W, Bao H, Xiong J, Che
W, Gu Y, Sun H and Liang P: Long non-coding RNA linc00645 promotes
TGF-β-induced epithelial-mesenchymal transition by regulating
miR-205-3p-ZEB1 axis in glioma. Cell Death Dis. 10:7172019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Do H and Kim W: Roles of oncogenic long
non-coding RNAs in cancer development. Genomics Inform. 16:e182018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vital AL, Tabernero MD, Castrillo A,
Rebelo A, Tão H, Gomes F, Nieto AB, Resende Oliveira C, Lopes MC
and Orfao A: Gene expression profiles of human glioblastomas are
associated with both tumor cytogenetics and histopathology. Neuro
Oncol. 12:991–1003. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang K, Zhao Z, Yu J, Chen W, Xu Q and
Chen L: lncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate
cancer cell proliferation, migration, and invasion in
hepatocellular carcinoma. J Cell Biochem. 119:6045–6056. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Westermarck J and Kahari VM: Regulation of
matrix metalloproteinase expression in tumor invasion. FASEB J.
13:781–792. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao X, Zhao S, Yang X, Zang S and Yuan X:
Long non-coding RNA FLVCR1-AS1 contributes to the proliferation and
invasion of lung cancer by sponging miR-573 to upregulate the
expression of E2F transcription factor 3. Biochem Biophys Res
Commun. 505:931–938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin H, Shangguan Z, Zhu M, Bao L, Zhang Q
and Pan S: lncRNA FLVCR1-AS1 silencing inhibits lung cancer cell
proliferation, migration, and invasion by inhibiting the activity
of the Wnt/β-catenin signaling pathway. J Cell Biochem.
120:10625–10632. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan H, Li H, Silva MA, Guan Y, Yang L, Zhu
L, Zhang Z, Li G and Ren C: lncRNA FLVCR1-AS1 mediates miR-513/YAP1
signaling to promote cell progression, migration, invasion and EMT
process in ovarian cancer. J Exp Clin Cancer Res. 38:3562019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li S, Zeng A, Hu Q, Yan W, Liu Y and You
Y: miR-423-5p contributes to a malignant phenotype and temozolomide
chemoresistance in glioblastomas. Neuro Oncol. 19:55–65. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang D, Liu Z, Zheng N, Wu H, Zhang Z and
Xu J: miR-30b-5p modulates glioma cell proliferation by direct
targeting MDTH. Saudi J Biol Sci. 25:947–952. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu ED, Li N, Li BS, Li W, Zhang WJ, Mao
XH, Guo G, Zou QM and Xiao B: miR-30b, down-regulated in gastric
cancer, promotes apoptosis and suppresses tumor growth by targeting
plasminogen activator inhibitor-1. PLoS One. 9:e1060492014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang Da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robinson MD, Mccarthy DJ and Smyth GK:
EdgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate-A practical and powerful approach to
multiple testing. J R Stat Soc. 57:289–300. 1995.
|
|
26
|
Ye Z, Jin H and Qian Q: Argonaute 2: A
novel rising star in cancer research. J Cancer. 6:877–882. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu
S, Zhang A, Jia Z, Wang G, Yu S, et al: lncRNA profile of
glioblastoma reveals the potential role of lncRNAs in contributing
to glioblastoma pathogenesis. Int J Oncol. 40:2004–2012.
2012.PubMed/NCBI
|
|
28
|
Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK,
Ho AS, Lui WM, Fung CF, Wong TS and leung GK: A long non-coding RNA
signature in glioblastoma multiforme predicts survival. Neurobiol
Dis. 58:123–131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhi F, Wang Q, Xue L, Shao N, Wang R, Deng
D, Wang S, Xia X and Yang Y: The use of three long non-coding RNAs
as potential prognostic indicators of astrocytoma. PLoS One.
10:e01352422015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma. Neurobio
Dis. 48:1–8. 2012. View Article : Google Scholar
|
|
31
|
Hu Y, Deng C, Zhang H, Zhang J, Peng B and
Hu C: Long non-coding RNA XIST promotes cell growth and metastasis
through regulating miR-139-5p mediated Wnt/β-catenin signaling
pathway in bladder cancer. Oncotarget. 8:94554–94568. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li C, Wan L, Liu Z, Xu G, Wang S, Su Z,
Zhang Y, Zhang C, Liu X, Lei Z and Zhang HT: Long non-coding RNA
XIST promotes TGF-β-induced epithelial-mesenchymal transition by
regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer.
Cancer Lett. 418:185–195. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo
B: Long noncoding RNA (lncRNA)-mediated competing endogenous RNA
networks provide novel potential biomarkers and therapeutic targets
for colorectal cancer. Int J Mol Sci. 20:E57582019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao J, Li L, Han ZY, Wang ZX and Qin LX:
Long noncoding RNAs, emerging and versatile regulators of
tumor-induced angiogenesis. Am J Cancer Res. 9:1367–1381.
2019.PubMed/NCBI
|
|
35
|
Zhang J, Yao T, Wang Y, Yu J, Liu Y and
Lin Z: Long noncoding RNA MEG3 is downregulated in cervical cancer
and affects cell proliferation and apoptosis by regulating miR-21.
Cancer Biol Ther. 17:104–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lv M, Zhong Z, Huang M, Tian Q, Jiang R
and Chen J: lncRNA H19 regulates epithelial-mesenchymal transition
and metastasis of bladder cancer by miR-29b-3p as competing
endogenous RNA. Biochim Biophys Acta Mol Cell Res. 1864:1887–1899.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu Q, Shan S, Li Y, Zhu D, Jin W and Ren
T: Long noncoding RNA SNHG1 promotes non-small cell lung cancer
progression by up-regulating MTDH via sponging miR-145-5p. FASEB J.
32:3957–3967. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8:45–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kumar B, Khaleghzadegan S, Mears B, Hatano
K, Kudrolli TA, Chowdhury WH, Yeater DB, Ewing CM, Luo J, Isaacs
WB, et al: Identification of miR-30b-3p and miR-30d-5p as direct
regulators of androgen receptor signaling in prostate cancer by
complementary functional microRNA library screening. Oncotarget.
7:72593–72607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Y, Guo G, Zhong Z, Sun L, Liao L, Wang
X, Cao Q and Chen H: Long non-coding RNA FLVCR1-AS1 sponges miR-155
to promote the tumorigenesis of gastric cancer by targeting c-Myc.
Am J Transl Res. 11:793–805. 2019.PubMed/NCBI
|
|
41
|
Bao W, Cao F, Ni S, Yang J, Li H, Su Z and
Zhao B: lncRNA FLVCR1-AS1 regulates cell proliferation, migration
and invasion by sponging miR-485-5p in human cholangiocarcinoma.
Oncol Lett. 18:2240–2247. 2019.PubMed/NCBI
|
|
42
|
Yan Z, Zhang W, Xiong Y, Wang Y and Li Z:
Long noncoding RNA FLVCR1-AS1 aggravates biological behaviors of
gliomacells via targeting miR-4731-5p/E2F2 axis. Biochem Biophys
Res Commun. 521:716–720. 2020. View Article : Google Scholar : PubMed/NCBI
|



















