Introduction
Pancreatic cancer is a highly lethal disease with a
mortality rate that is similar to the incidence rate and is a
worldwide health burden, particularly in China (1,2).
Pancreatic cancer accounts for 5% of all cancer-associated
mortality (3). In addition, the
prognosis of pancreatic cancer is very poor, with an overall 5-year
survival rate of only ~5% (4) and
a median survival time of ≤6 months (5). Despite developments in surgical
treatment and early detection, this situation has not significantly
changed (6,7). The poor prognosis is mainly due to
the fact that nearly 80% of patients present with locally advanced
or metastatic disease (8).
Therefore, improved understanding regarding the molecular
mechanisms involved in pancreatic cancer will be helpful for
providing new insights into effective treatments for patients with
pancreatic cancer.
MicroRNAs (miRNAs/miRs) are a class of small,
non-coding, regulatory RNAs that are ~22 nucleotides in length
(9). Currently, >2,000 miRNAs
have been discovered in humans, which regulate one third of all
genes (10). miRNAs regulate the
expression of their target protein-coding genes by degradation of
mRNA or translational inhibition through binding to the
3′-untranslated regions (3′-UTRs) of target genes (11). Dysregulation of miRNAs is common in
various human cancer types, with upregulated miRNAs often acting as
oncogenes and downregulated miRNAs acting as tumor suppressors
(12). These cancer-associated
miRNAs mediate various biological functions associated with cancer
development and progression, including proliferation,
differentiation, cell signaling, cell survival, apoptosis and
metastasis (13). Due to their
small size and the network of proteins regulated by miRNAs, miRNA
therapeutics is developing; therefore, more miRNAs need to be
identified that could be applied in clinical trials (14). miR-142 serves a critical role in
cancer, virus infection, inflammation and immune tolerance, and
miR-142-5p is the passenger strand generated from the miR-142
hairpin (15). A previous study
revealed that overexpression of miR-142-5p could inhibit tumor
growth of pancreatic cancer in vivo (16). However, the biological function and
molecular mechanism of miR-142-5p in pancreatic cancer remain
largely unknown.
The present study measured the expression of
miR-142-5p in pancreatic cancer tissues and cell lines.
Additionally, the effects of miR-142-5p, as well as its target gene
phosphoinositide-3-kinase catalytic subunit α (PIK3CA), on cell
proliferation, migration and invasion were investigated. The
results demonstrated that miR-142-5p inhibits the migration and
invasion by targeting PIK3CA, which suggests that miR-142-5p may be
a potential target for the treatment of pancreatic cancer.
Materials and methods
Patients and specimens
Pancreatic cancer tissue and paired adjacent normal
tissue samples (n=35, including 24 male and 11 female; age range
from 48–77 years and the average age was 62.17 years) were obtained
from patients who underwent surgery at The Affiliated Huaian No.1
People's Hospital of Nanjing Medical University (Huai'an, China)
from February 2017 to July 2018. All patients received no
preoperative treatment and provided written informed consent prior
to surgery. The fresh tissues were immediately frozen in liquid
nitrogen and stored at −80°C until further use. The present study
was approved by the Ethics Committee of The Affiliated Huaian No.1
People's Hospital of Nanjing Medical University.
Cell lines and culture
Human pancreatic cancer cell lines (PanC1, BxPC3,
SW1990 and CAPAN-1) and the normal human fibroblast WI-38 cell line
were purchased from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. All cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum, 100 U/ml penicillin
and 100 µg/ml streptomycin (all from Gibco; Thermo Fisher
Scientific, Inc.), and incubated in an incubator at 37°C with 5%
CO2.
Prediction of miR-124-5p target
genes
The miR-142-5p targets were predicted using
Targetscan (http://www.targetscan.org/vert_72/).
Dual-luciferase reporter assay
A pGL3 plasmid was purchased from Promega
Corporation. The wild-type (WT) or mutated 3′-UTR of PIK3CA was
synthesized and cloned into pGL3 downstream of the firefly
luciferase reporter. PanC1 cells were seeded in 96-well plates
until the confluence reached 70%. Subsequently, the cells were
co-transfected with WT or mutant 3′-UTR reporter vector and
miR-negative control (NC) or miR-142-5p mimic using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to manufacturer's protocol. At 48 h
post-transfection, firefly and Renilla luciferase activities
were measured using the Dual-Luciferase Reporter assay system
(Promega Corporation). The Renilla luciferase activity was
used for endogenous normalization.
Cell transfection
miR-142-5p mimic (cat. no. miR10000433-1-5; sense,
5′-CGUGUUCACAGCGGACCUUGAU-3′ and anti-sense,
5′-AUCAAGGUCCGCUGUGAACACG-3′) and the corresponding NC (cat. no.
miR1N0000002-1-5; sense, 5′-UAGCCAUAUCGUCGAUACU-3′ and anti-sense,
5′-AGUAUCGACGAUAUGGCUA-3′) were purchased from Guangzhou RiboBio
Co., Ltd. pcDNA3.1 was obtained from Invitrogen; Thermo Fisher
Scientific, Inc. The coding region of PIK3CA was inserted into the
pcDNA3.1 vector and termed pcDNA3.1-PIK3CA. PanC1 cells were seeded
into six-well plates at a density of 4×105 cells/well 24
h prior to transfection. Subsequently, the cells were transfected
with 20 nM miR-124-5p mimic, miR-NC, pcDNA3.1 or pcDNA3.1-PIK3CA
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in serum-free DMEM, according to the
manufacturer's protocol. At 6 h post-transfection, the cells were
incubated with complete medium. After 48 h of transfection, the
cells were harvested for further study. Untransfected cells were
used as the control group.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from paired pancreatic
cancer tissues, corresponding normal tissues and PanC1 cells using
the TRIzol® reagent (Invitrogen, Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Next,
the total RNA was reverse transcribed to cDNA at 60°C for 30 min
using the ThermoScript RT-PCR system (Invitrogen; Thermo Fisher
Scientific, Inc.). Subsequently, qPCR was performed using the
All-in-One™ miRNA RT-qPCR detection kit (GeneCopoeia, Inc.) to
detect miR-142-5p expression and using SYBR Green PCR Mastermix
(Beijing Solarbio & Technology, Co., Ltd.) to detect the mRNA
expression. All reactions were performed on an ABI PRISM 7500 Fast
Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The qPCR amplification conditions consisted of 95°C for 10
min, followed by 40 cycles of 95°C for 10 sec, 60°C for 20 sec and
72°C for 15 sec. The primers sequences were as follows: miR-142-5p
F: 5′-AACTCCAGCTGGTCCTTAG-3′, R: 5′-TCTTGAACCCTCATCCTGT-3′; small
nuclear RNA U6 F: 5′-CTCGCTTCGGCAGCACA-3′, R:
5′-AACGCTTCACGAATTTGCGT-3′; PIK3CA F:
5′-AAATGAAAGCTCACTCTGGATTCC-3′, R: 5′-TTGTGCAATTCCTATGCAATCG-3′;
FAK F: 5′-GTGCTCTTGGTTCAAGCTGGAT-3′, R:
5′-ACTTGAGTGAAGTCAGCAAGATGTGT-3′; MMP9 F:
5′-GTGCTGGGCTGCTGCTTTGCTG-3′, R: 5′-GTCGCCCTCAAAGGTTTGGAAT-3′; AKT
F: 5′-AGGCATCCCTTCCTTACAGC-3′, R: 5′-CAGCCCGAAGTCCGTTATCT-3′; GAPDH
F: 5′-GAAGGTGAAGGTCGGAGTC-3′, R: 5′-GAAGATGGTGATGGGATTTC-3′. U6 and
GAPDH were used as the loading control for miR-142-5p and mRNA,
respectively. The relative expression was calculated using the
2−ΔΔCq method (17).
Western blotting
The harvested PanC1 cells were lysed using RIPA
buffer (Beyotime Institute of Biotechnology) on ice for 30 min. The
Bicinchoninic Acid Protein assay kit (Beyotime Institute of
Biotechnology) was used to detect the protein concentration.
Subsequently, 30 µg protein was separated by 10% SDS-PAGE and
transferred onto PVDF membranes (EMD Millipore). After blocking at
room temperature for 1 h with TBST (containing 0.05% Tween-20;
Beijing Solarbio Science & Technology Co., Ltd.) containing 5%
skim milk, the membranes were incubated with primary antibodies all
purchased from Abcam: Anti-PIK3CA (cat. no. ab40776; 1:5,000);
anti-FAK (cat. no. ab40794; 1:1,000); anti-MMP9 (cat. no. ab38898;
1:1,000); anti-AKT (cat. no. ab227100; 1:1,000);
anti-phosphorylated (p)-AKT (cat. no. ab81283; 1:5,000); anti-GAPDH
(cat. no. ab181602; 1:10,000) overnight at 4°C. The next day,
secondary antibody [goat anti-rabbit IgG H&L (horseradish
peroxidase); cat. no. ab6721; 1:10,000; Abcam] was incubated with
the membranes at room temperature for 1 h. Finally, the membranes
were incubated with BeyoECL Plus (Beyotime Institute of
Biotechnology) and the protein bands were visualized. The relative
expression was calculated using ImageQuant LAS 4010 Imaging system
(GE Healthcare Life Sciences). GAPDH was used as an internal
control.
Cell proliferation assay
For the cell proliferation assay, a CCK-8
(BiotechWell) assay was performed according to the manufacturer's
protocols. Briefly, 2×103 transfected PanC1 cells were
seeded into 96-well plates and cultured in an incubator for 0, 12,
24 and 48 h. At the indicated time points, 10 µl CCK-8 solution was
added to each well and the cells were further incubated for 1 h.
The absorbance was measured at 450 nm using a microplate reader
(Thermo Fisher Scientific, Inc.).
Cell migration assay
A wound healing assay was performed to analyze cell
migration ability. Transfected cells were seeded into 6-well plates
at a density of 2×106 cells/well. After incubation for
24 h, cell layers were scratched with a 10 µl-sterile plastic tip
and cell debris was rinsed with PBS. The cells were then incubated
in serum-free medium at 37°C with 5% CO2 for 24 h.
Finally, wound closure was imaged under a light microscope
(magnification, ×100; Olympus Corporation) at 0 and 24 h after
wounding, and the width was quantified using a standard
caliper.
Cell invasion assay
Cell invasion was assessed by Transwell assay using
24-well Matrigel invasion chambers (8-µm pores; EMD Millipore). A
total of 1×105 cells were added to the upper chambers of
Transwell plates filled with serum-free DMEM and complete DMEM
added to the bottom chambers. The cells were incubated at 37°C with
5% CO2 for 24 h and the non-invading cells were then
removed with sterile swabs. The invaded cells were fixed with 4%
paraformaldehyde for 10 min at room temperature, followed by
staining with 0.1% crystal violet for 15 min at room temperature.
The cells were then photographed and quantified in five random
fields using an optical light microscope (magnification, ×200;
Olympus Corporation).
Statistical analysis
Each independent experiment was repeated a minimum
of three times. Statistical analysis was performed using GraphPad
Prism software (version 6.0; GraphPad Software, Inc.). An unpaired
student's t-test was used to evaluate the significant differences
between two groups in all cases except a paired student's t-test
was used to compare differences between tumor tissues and adjacent
normal tissues. A one-way analysis of variance followed by
Newman-Keuls post hoc test was used to measure the significant
differences among multiple groups. Data are presented as the mean ±
standard error of mean. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-142-5p is downregulated in
pancreatic cancer tissues and cell lines
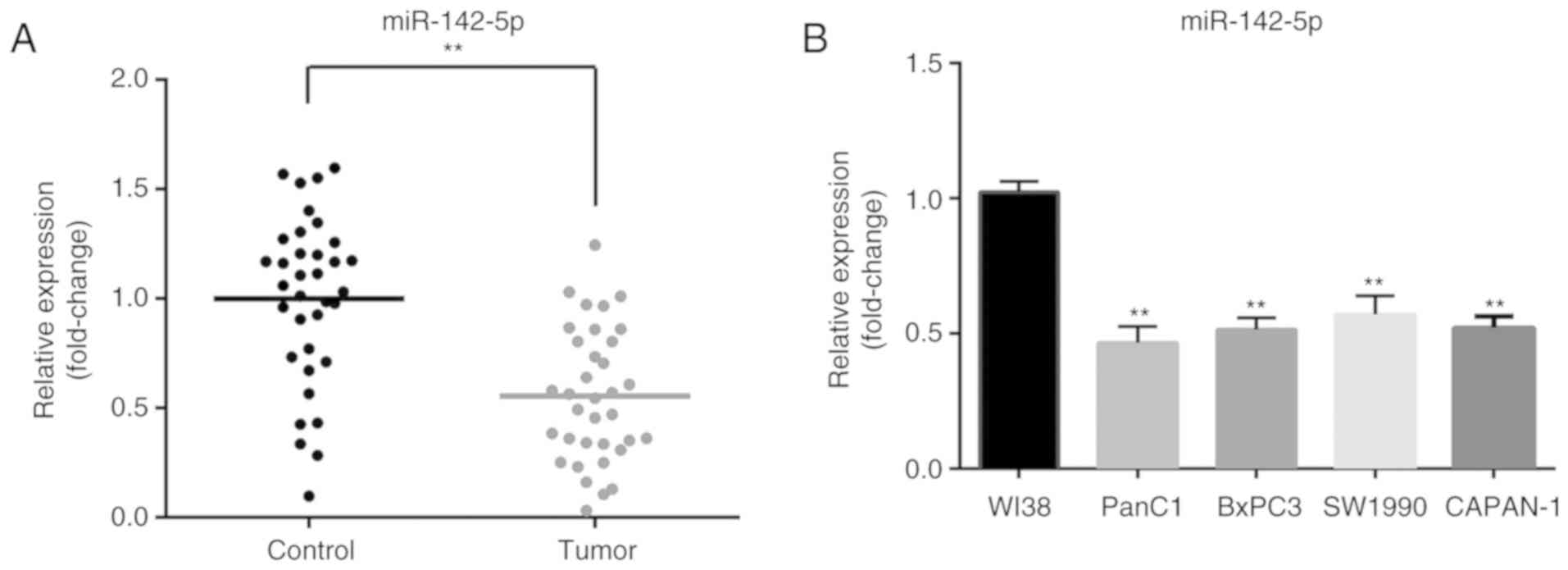
To determine the expression level of miR-142-5p,
RT-qPCR was performed to detect miR-142-5p expression in 35 pairs
of pancreatic cancer and matched adjacent normal tissues. Compared
with that in adjacent non-tumor tissues, the expression of
miR-142-5p was significantly decreased in pancreatic cancer tissues
(P<0.01; Fig. 1A). In addition,
miR-142-5p expression was significantly downregulated in four
pancreatic cancer cell lines (PanC1, BxPC3, SW1990 and CAPAN-1)
compared with normal human fibroblast WI-38 cells (P<0.01;
Fig. 1B). These results suggested
that low miR-142-5p expression may be closely associated with
pancreatic cancer progression.
PIK3CA is a target of miR-142-5p
To understand the molecular mechanism underlying
miR-142-5p, bioinformatics analysis was performed to identify the
potential targets of miR-142-5p. According to the TargetScan
database, miR-142-5p was identified to be able to bind to the
3′-UTR of PIK3CA at position 1,675-1,682 nt (Fig. 2A). To further confirm this
prediction, cells were co-transfected with WT or mutant 3′-UTR
reporter vector as well as miR-NC or miR-142-5p mimic to perform a
dual-luciferase reporter assay. As presented in Fig. 2B, overexpression of miR-142-5p
significantly decreased the luciferase activity of the plasmid
carrying the WT 3′-UTR of PIK3CA in PanC1 cells (P<0.01).
However, miR-142-5p did not affect the luciferase activity when the
3′-UTR of PIK3CA was mutated. Additionally, PanC1 cells were
transfected with miR-NC, miR-142-5p mimic, pcDNA3.1 and
pcDNA3.1-PIK3CA. miR-142-5p expression was significantly
upregulated when transfected with miR-142-5p mimic compared with
miR-NC (P<0.01) and PIK3CA mRNA and protein expression levels
were also significantly upregulated when transfected with
pcDNA3.1-PIK3CA compared with pcDNA3.1 (P<0.01; Fig. 2C-E). Furthermore, RT-qPCR and
western blotting results demonstrated that overexpression of
miR-142-5p resulted in a suppression of PIK3CA mRNA and protein
levels in PanC1 cells (P<0.01; Fig.
2F and G). These results indicated that PIK3CA expression is
negatively regulated by miR-142-5p. In summary, PIK3CA was
identified as a target of miR-142-5p.
PIK3CA expression is upregulated in
pancreatic cancer tissues
Subsequently, the expression of PIK3CA was examined
in clinical tissue samples. The mRNA and protein expression levels
of PIK3CA were increased in pancreatic cancer tissues compared with
matched adjacent normal tissues (P<0.01; Fig. 3A and B). Furthermore, an inverse
association was identified between miR-142-5p and PIK3CA expression
(P=0.0001; Fig. 3C). These results
suggested that the upregulation of PIK3CA is partly caused by a
downregulation of miR-142-5p and further supports the conclusion
that PIK3CA is a target of miR-142-5p.
miR-142-5p inhibits cell
proliferation, migration and invasion of pancreatic cancer in vitro
by targeting PIK3CA
To assess the role of miR-142-5p in pancreatic
cancer, CCK-8, wound healing and Transwell assays were performed to
measure the capability of cell proliferation, migration and
invasion, respectively. The results demonstrated that
overexpression of miR-142-5p significantly inhibited cell
proliferation after 48 h of transfection (P<0.01), while PIK3CA
reversed the inhibition induced by miR-142-5p (P<0.01; Fig. 4A). Furthermore,
miR-142-5p-overexpression led to slower wound closure in PanC1
cells compared with the miR-NC group (P<0.01), while
PIK3CA-overexpression significantly reversed the slow closure
compared with the miR-142-5p mimic group (P<0.01; Fig. 4B). Similarly, the invasion ability
of PanC1 cells was significantly inhibited by miR-142-5p mimic
(P<0.01), which was restored by PIK3CA (P<0.01; Fig. 4C). In summary, overexpression of
miR-142-5p inhibited the proliferation, migration and invasion in
pancreatic cancer cell by regulating PIK3CA.
miR-142-5p suppresses FAK and MMP9
expression, and the PI3K/AKT pathway by targeting PIK3CA
Finally, the expression levels of FAK, MMP9, AKT and
p-AKT were measured. As presented in Fig. 5A, B and D, overexpression of
miR-142-5p significantly suppressed the mRNA expression of FAK and
MMP9 (P<0.01), while PIK3CA significantly reversed the
suppression induced by miR-142-5p (P<0.01). Similar results were
observed for protein expression (P<0.01). In addition,
miR-142-5p-overexpression significantly reduced the protein level
of p-AKT (P<0.01), while PIK3CA reversed the inhibition
(P<0.01; Fig. 5D). However,
miR-142-5p and PIK3CA did not affect the expression of total AKT
(Fig. 5C and D). These results
suggested that miR-142-5p targets PIK3CA to suppress the expression
of FAK and MMP9, as well as inactivate the PI3K/AKT signaling
pathway.
 | Figure 5.miR-142-5p suppresses the expression
of FAK, MMP9 and inactivates the PI3K/AKT pathway through targeting
PIK3CA. The expression of (A) FAK, (B) MMP9 and (C) AKT was
detected by reverse transcription-quantitative PCR in transfected
PanC1 cells. (D) Protein expression of FAK, MMP9, AKT and p-AKT was
measured by western blotting. The relative protein expression,
normalized for GAPDH, was quantified and shown in the corresponding
histogram below. Data are presented as the mean ± standard error of
the mean. **P<0.01 vs. miR-NC mimics. FAK, focal adhesion
kinase; MMP, matrix metalloproteinase; PIK3CA,
phosphoinositide-3-kinase catalytic subunit α; AKT, protein kinase
B; p-, phosphorylated-; NC, negative control. |
Discussion
The present study revealed that miR-142-5p was
downregulated in pancreatic cancer tissues and cell lines. PIK3CA
was identified as a target of miR-142-5p. Notably, overexpression
of miR-142-5p inhibited cell proliferation, migration and invasion
of pancreatic cancer cells, suppressed the expression of FAK and
MMP9, and inhibited the PI3K/AKT pathway by targeting PIK3CA.
miR-142-5p has been reported to be a critical miRNA
during cancer progression (15).
miR-142-5p functions as an oncogene or a tumor suppressor in
different types of cancer. The expression of miR-142-5p is
upregulated in renal cell carcinoma cells and overexpression of
miR-142-5p promotes cell proliferation and migration (18). Similarly, miR-142-5p acts as an
oncogene in colorectal cancer by enhancing the capacities of cell
proliferation, colony formation and wound healing; however, it
inhibits cell apoptosis (19,20).
Furthermore, miR-142-5p-overexpression suppresses the proliferation
of non-small cell lung cancer (NSCLC) cells in vitro and
in vivo, while inhibition of miR-142-5p promotes cell growth
(21). miR-142-5p is also
downregulated in hepatocellular carcinoma, which inhibits tumor
cell viability, proliferation, migration and invasion, and induces
apoptosis (22,23). There are a limited number of
studies regarding the functional role of miR-142-5p in pancreatic
cancer cells. In the present study, miR-142-5p expression was
revealed to be upregulated in pancreatic cancer tissues and cells.
Furthermore, overexpression of miR-142-5p inhibited pancreatic
cancer cell proliferation, migration and invasion. These findings
indicated that miR-142-5p functions as a tumor suppressor in
pancreatic cancer. However, the underlying molecular mechanism
requires further investigation.
A previous study reported that PIK3CA is a direct
target of miR-142-5p in NSCLC (21). In the current study, bioinformatics
analysis predicted that PIK3CA is a target of miR-142-5p and a
dual-luciferase reporter assay confirmed this prediction. The
PIK3CA gene is involved in encoding the catalytic subunit of PI3K
P110a, which is activated by cell surface tyrosine kinase receptors
(24,25). PIK3CA often functions as an
oncogene in different types of human cancer, including prostate
cancer (26), lung cancer
(27,28), gastric cancer (29) and renal cell carcinoma (30). A previous study demonstrated that
PIK3CA is an oncogene in pancreatic cancer (31). However, the biological role of
PIK3CA remains unknown. In the present study, the expression of
PIK3CA was upregulated in pancreatic cancer tissues and was
negatively regulated by miR-142-5p expression in both tumor tissues
and cells, which further verifies that PIK3CA is a target of
miR-142-5p in pancreatic cancer. Furthermore, PIK3CA reversed the
miR-142-5p-induced inhibition of cell proliferation, migration and
invasion. These results demonstrated that miR-142-5p inhibits the
proliferation, migration and invasion of pancreatic cancer cells by
targeting PIK3CA.
FAK is a non-receptor tyrosine kinase that has been
demonstrated to be upregulated in cancer and is involved in the
progression of tumor aggressiveness (32). MMPs, including MMP9, have been
reported to serve a role in cancer initiation, tumor growth and
metastasis in pancreatic cancer (33). This study only focused on the
expression of MMP9 rather than activity of MMP9, just as other
miRNAs decrease cell migration and invasion through reducing MMP9
expression (34,35). Additionally, PIK3CA regulates the
activation of AKT and p-AKT activates various cellular processes
(36,37). In a previous study, knockdown of
PIK3CA reduced FAK, MMP9 and p-AKT levels in glioblastoma
multiforme cells (38). In the
present study, miR-142-5p suppressed the expression of FAK, MMP9
and p-AKT, but did not affect AKT expression, while PIK3CA reversed
the suppression. These findings suggested that miR-142-5p targets
PIK3CA to suppress the expression of FAK and MMP9, and inactivate
the PI3K/AKT signaling pathway.
In conclusion, the present study demonstrated that
miR-142-5p functions as a tumor suppressor in pancreatic cancer, as
it was identified to target PIK3CA and inhibit cell proliferation,
migration and invasion in vitro by suppressing FAK and MMP9
expression, and inactivating the PI3K/AKT signaling pathway. These
findings indicated that miR-142-5p may provide a novel target for
the therapeutic treatment of pancreatic cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JZ and YS contributed to study design. JZ, LZ, BW,
ZQ, JW and HH performed experiments and data analysis. JZ was a
major contributor in writing the manuscript. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The pancreatic cancer tissue and paired adjacent
normal tissue sample collection was approved by the Ethics
Committees of The Affiliated Huaian No. 1 People's Hospital of
Nanjing Medical University. All patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lin QJ, Yang F, Jin C and Fu DL: Current
status and progress of pancreatic cancer in China. World J
Gastroenterol. 21:7988–8003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goral V: Pancreatic cancer: Pathogenesis
and diagnosis. Asian Pac J Cancer Prev. 16:5619–5624. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carrato A, Falcone A, Ducreux M, Valle JW,
Parnaby A, Djazouli K, Alnwick-Allu K, Hutchings A, Palaska C and
Parthenaki I: A systematic review of the burden of pancreatic
cancer in Europe: Real-world impact on survival, quality of life
and costs. J Gastrointest Cancer. 46:201–211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Su D, Yamaguchi K and Tanaka M: The
characteristics of disseminated tumor cells in pancreatic cancer: A
black box needs to be explored. Pancreatology. 5:316–324. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li X, Li Y, Wan L, Chen R and Chen F:
miR-509-5p inhibits cellular proliferation and migration via
targeting MDM2 in pancreatic cancer cells. Onco Targets Ther.
10:4455–4464. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stathis A and Moore MJ: Advanced
pancreatic carcinoma: Current treatment and future challenges. Nat
Rev Clin Oncol. 7:163–172. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shah MY, Ferrajoli A, Sood AK,
Lopez-Berestein G and Calin GA: microRNA therapeutics in cancer-an
emerging concept. EBioMedicine. 12:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hata A and Lieberman J: Dysregulation of
microRNA biogenesis and gene silencing in cancer. Sci Signal.
8:re32015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Simonson B and Das S: MicroRNA
therapeutics: The next magic bullet? Mini Rev Med Chem. 15:467–474.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shrestha A, Mukhametshina RT, Taghizadeh
S, Vasquez-Pacheco E, Cabrera-Fuentes H, Rizvanov A, Mari B,
Carraro G and Bellusci S: MicroRNA-142 is a multifaceted regulator
in organogenesis, homeostasis, and disease. Dev Dynam. 246:285–290.
2017. View Article : Google Scholar
|
|
16
|
Jia L, Xi Q, Wang H, Zhang Z, Liu H, Cheng
Y, Guo X, Zhang J, Zhang Q, Zhang L, et al: miR-142-5p regulates
tumor cell PD-L1 expression and enhances anti-tumor immunity.
Biochem Biophys Res Commun. 488:425–431. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu L, Liu S, Duan Q, Chen L, Wu T, Qian
H, Yang S, Xin D, He Z and Guo Y: MicroRNA-142-5p promotes cell
growth and migration in renal cell carcinoma by targeting BTG3. Am
J Transl Res. 9:2394–2402. 2017.PubMed/NCBI
|
|
19
|
Islam F, Gopalan V, Vider J, Lu CT and Lam
AK: MiR-142-5p act as an oncogenic microRNA in colorectal cancer:
Clinicopathological and functional insights. Exp Mol Pathol.
104:98–107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu S, Xiao Z, Ai F, Liu F, Chen X, Cao K,
Ren W, Zhang X, Shu P and Zhang D: miR-142-5p promotes development
of colorectal cancer through targeting SDHB and facilitating
generation of aerobic glycolysis. Biomed Pharmacother.
92:1119–1127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Z, Liu Z, Fang X and Yang H:
MiR-142-5p suppresses tumorigenesis by targeting PIK3CA in
non-small cell lung cancer. Cell Physiol Biochem. 43:2505–2515.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsang FH, Au SL, Wei L, Fan DN, Lee JM,
Wong CC, Ng IO and Wong CM: MicroRNA-142-3p and microRNA-142-5p are
downregulated in hepatocellular carcinoma and exhibit synergistic
effects on cell motility. Front Med. 9:331–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lou K, Chen N, Li Z, Zhang B, Wang X, Chen
Y, Xu H, Wang D and Wang H: MicroRNA-142-5p overexpression inhibits
cell growth and induces apoptosis by regulating FOXO in
hepatocellular carcinoma cells. Oncol Res. 25:65–73. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Samuels Y and Waldman T: Oncogenic
mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol.
347:21–41. 2010.PubMed/NCBI
|
|
25
|
Vanhaesebroeck B, Stein RC and Waterfield
MD: The study of phosphoinositide 3-kinase function. Cancer Surv.
27:249–270. 1996.PubMed/NCBI
|
|
26
|
Zhang S, Cai J, Xie W, Luo H and Yang F:
miR-202 suppresses prostate cancer growth and metastasis by
targeting PIK3CA. Exp Ther Med. 16:1499–1504. 2018.PubMed/NCBI
|
|
27
|
Meng F and Zhang L: miR-183-5p functions
as a tumor suppressor in lung cancer through PIK3CA inhibition. Exp
Cell Res. 374:315–322. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu QQ, Wu H, Huang X, Shen H, Shu YQ,
Zhang B, Xiang CC, Yu SM, Guo RH and Chen L: MiR-1 targets PIK3CA
and inhibits tumorigenic properties of A549 cells. Biomed
Pharmacother. 68:155–161. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liang M, Shi B, Liu J, He L, Yi G, Zhou L,
Yu G and Zhou X: Downregulation of miR203 induces overexpression of
PIK3CA and predicts poor prognosis of gastric cancer patients. Drug
Des Devel Ther. 9:3607–3616. 2015.PubMed/NCBI
|
|
30
|
Chen K, Zeng J, Tang K, Xiao H, Hu J,
Huang C, Yao W, Yu G, Xiao W, Guan W, et al: miR-490-5p suppresses
tumour growth in renal cell carcinoma through targeting PIK3CA.
Biol Cell. 108:41–50. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Waddell N, Pajic M, Patch AM, Chang DK,
Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al:
Whole genomes redefine the mutational landscape of pancreatic
cancer. Nature. 518:495–501. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lv PC, Jiang AQ, Zhang WM and Zhu HL: FAK
inhibitors in Cancer, a patent review. Expert Opin Ther Pat.
28:139–145. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Knapinska AM, Estrada CA and Fields GB:
The roles of matrix metalloproteinases in pancreatic cancer. Prog
Mol Biol Transl Sci. 148:339–354. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu X, Gao F, Wang J, Tao L, Ye J, Ding L,
Ji W and Chen X: MiR-122-5p inhibits cell migration and invasion in
gastric cancer by down-regulating DUSP4. Cancer Biol Ther.
19:427–435. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zou Q, Yi W, Huang J, Fu F, Chen G and
Zhong D: MicroRNA-375 targets PAX6 and inhibits the viability,
migration and invasion of human breast cancer MCF-7 cells. Exp Ther
Med. 14:1198–1204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Osaki M, Oshimura M and Ito H: PI3K-Akt
pathway: Its functions and alterations in human cancer. Apoptosis.
9:667–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Weber GL, Parat MO, Binder ZA, Gallia GL
and Riggins GJ: Abrogation of PIK3CA or PIK3R1 reduces
proliferation, migration, and invasion in glioblastoma multiforme
cells. Oncotarget. 2:833–849. 2011. View Article : Google Scholar : PubMed/NCBI
|