Introduction
Myocardial infarction (MI) is one of the most
frequently encountered cardiovascular diseases that can result in
sudden cardiac death by inducing ventricular arrhythmias (VAs) or
heart failure (HF) (1). Despite
considerable advances in the treatment of MI, the 2-year mortality
rate is still high, with 49% for patients with type 2 MI and 26%
for those with type 1 MI (2).
Therefore, the development of more effective therapeutic strategies
for MI remains a major challenge faced by clinical
cardiologists.
Accumulating evidence reveals that cardiac
remodeling at the infarct border zone (IBZ) is the main contributor
for the occurrence of VAs or HF (3,4).
Post-MI remodeling in the IBZ is characterized by compensatory
cardiac hypertrophy with depressed contractile function (3,5,6);
extracellular matrix (ECM) rearrangement and cardiac fibrosis
(3,5); increased inflammatory response and
cardiomyocyte apoptosis (7,8) and
an insufficient angiogenic response (9,10).
Thus, targeting the genes involved in the aforementioned processes
of IBZ remodeling (11,12) may be a potential approach for the
treatment of MI and prevention of VAs or HF. This hypothesis has
been demonstrated by previous studies. For example, Wang et
al (13) directly injected
pro-angiogenic vascular endothelial growth factor (VEGF) into the
IBZ of MI rats and demonstrated that VEGF treatment could
significantly improve cardiac function by inducing myocardial
collateral vessel development and inhibiting myocardial apoptosis
by decreasing the expression levels of tumor necrosis factor
(TNF)-α and Bax. A decrease in ECM degradation enzymes (matrix
metalloproteinases), but an increase in gap junction protein
connexin 43 by artemisinin (14)
or doxycycline (15) can improve
myocardial IBZ contractility and decrease the vulnerability of VAs.
Asiatic acid was reported to inhibit cardiac hypertrophy, decrease
levels of inflammatory cytokines and decrease interstitial fibrosis
in the IBZ of MI model rats by blocking the activation of p38
mitogen-activated protein kinase (MAPK) and ERK1/2 pathways
(5). However, the molecular
mechanisms of cardiac remodeling at the IBZ following MI remain
unclear.
In addition to protein-coding genes, numerous
studies document that long non-coding RNAs (lncRNAs; sequences
>200 nucleotides in length) also play an important role in MI by
acting as competing endogenous RNAs (ceRNAs) to regulate microRNA
(miRNA)-mediated target repression (16,17)
or by directly controlling the transcription of target genes. For
example, Wu et al (18)
identified lncRNA ZFAS1 was significantly upregulated in the
myocardium IBZ of rats at 1–48 h post-MI. An RNA pull-down assay
indicated that ZFAS1 could directly interact with miR-150 to
regulate pro-inflammatory C-reactive protein. Knockdown of ZFAS1 or
overexpression of miR-150 effectively relieved MI in model rats.
Hao et al (19)
demonstrated that GAS5 expression was increased, while sema3a was
decreased in the IBZ of the MI group. Overexpression of GAS5 could
ameliorate cardiomyocyte apoptosis and decrease infarct size by
downregulating the protein expression of sema3a (19). By sequencing the IBZ regions of the
affected heart in porcine MI models, several novel lncRNAs
expressed in an antisense orientation to myocardial transcription
factors (including GATA-binding protein 4, GATA-binding protein 6
and Krüppel-like family transcription factor 6) were identified by
Kaikkonen et al (20);
while in the mouse MI model, Ounzain et al (21) screened hundreds of novel
heart-specific lncRNAs relevant to maladaptive remodeling, such as
Novlnc6, Novlnc15, Novlnc35 and Novlnc61. These lncRNAs may serve
as new targets for the treatment of MI. However, to the best of our
knowledge, there is a limited number of studies that focus on the
roles and mechanisms of lncRNAs in the IBZ of MI.
The aim of the present study was to further screen
crucial lncRNAs that are significantly differentially expressed in
the IBZ of MI mice compared with sham controls by constructing
lncRNA-miRNA-mRNA ceRNA and lncRNA-mRNA co-expression networks
using the high throughput data deposited in public databases. The
results of the present study may offer novel targets for the
treatment of MI.
Materials and methods
Data collection
GSE76592 (22) and
GSE52313 (21) datasets, which
investigated the miRNA and lncRNA/mRNA expression level profiles in
the border zone myocardium of MI model mice and sham controls, were
collected from the Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo). In
GSE76592 (22), the MI model was
established by surgical ligation of the left anterior descending
(LAD) coronary artery in mice aged 10 weeks. The ventricular septum
of the areas at risk of ischemia was sampled on post-operative day
28. The sham surgeries were performed by pericardiotomy through
left thoracotomy in mice aged 14 weeks to serve as the control and
then the area corresponding to the border zone for MI was acquired.
Total RNA was extracted from 38 MI and 11 control heart tissues and
subjected to microarray analysis using GeneChip® miRNA
3.0 Arrays (platform, GPL16384; Affymetrix; Thermo Fisher
Scientific, Inc.). In GSE52313 (21), the MI mouse model was also
constructed by ligation of the LAD artery in 12-week old mice, and
a sham operation in 12-week old mice was conducted by placing the
ligature in an identical location, but not tying it. A total of
four sham-operated and four infarcted heart samples were collected
2 weeks after surgery for RNA-sequencing with the Illumina
HiSeq2000 [platform, GPL9250; Illumina Genome Analyzer II (Mus
musculus)].
Differential expression analysis
The normalized data were downloaded from the GEO
database with the corresponding accession number. The
differentially expressed miRNAs (DEMs), genes (DEGs) and lncRNAs
(DELs) were identified using the Linear Models for Microarray data
software package (version 3.34.0; http://www.bioconductor.org/packages/release/bioc/html/limma.html)
(23) in R Bioconductor (version
3.4.1; http://www.bioconductor.org/). The
Benjamini-Hochberg method for multiple testing was used to adjust
P-values to false discovery rate (FDR) (24). P<0.05 and
|logFC(fold-change)|>0.5 were considered as the significant
cut-off in order to screen more targets for subsequent analysis.
Heatmaps were plotted using the pheatmap package (version: 1.0.8;
http://cran.r-project.org/web/packages/pheatmap)
based on the Euclidean distance measures.
Prediction of target encoding genes
for DEMs
Prediction of target mRNAs for DEMs was performed
using the miRWalk database (version 2.0; http://www.zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2)
(25), which provides 12
prediction algorithms [TargetScan (version 6.2), miRanda (version
1.0), DIANA-microT (version 4.0), miRBridge (version 1.0), miRDB
(version 4.0), miRMap (version 1.0), miRNAMap (version 1.0),
PicTar2 (version 2.0), PITA (version 6.0), RNA22 (version 2.0),
RNAhybrid (version 2.1) and miRWalk (version 1.0)]. Only target
mRNAs that were predicted by at least eight different algorithms
were selected. The intersections between the predicted target mRNAs
and DEGs were retained, among which the DEGs that were expressed in
the opposite direction to DEMs may represent the underlying
downstream targets of DEMs.
Construction of a protein-protein
interaction (PPI) network
In order to further screen crucial DEGs targeted by
DEMs, the interactions between these DEGs were predicted using the
Search Tool for the Retrieval of Interacting Genes (STRING; version
10.0; http://string db.org/) database
(26). Only interactions with a
combined score >0.4 were selected to construct the PPI network
using Cytoscape (version 3.4; www.cytoscape.org/) (27). Topological features were calculated
for each node (protein) in the PPI network using the CytoNCA plugin
in Cytoscape software (version 3.4; http://apps.cytoscape.org/apps/cytonca) (28) to screen hub genes, including degree
(DC), betweenness (BC), closeness (CC), eigenvector (EC) and
sub-gragh centrality (SC).
Construction of an lncRNA ceRNA
network
The DIANA- LncBase (version 2.0; http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted)
(29) database was used to predict
the interactions of lncRNAs with DEMs. The intersections between
the predicted target lncRNAs and DELs were retained, among which
the target DELs with expression in the opposite direction to DEMs
may represent the underlying sponge for DEMs. The lncRNA-miRNA
interaction pairs were then overlapped with the miRNA-mRNA
interaction pairs according to the common miRNAs in order to
generate the potential ceRNA axes, which were used to construct the
ceRNA network and visualized in Cytoscape.
Functional annotations for ceRNA
network genes
Functional analysis of the genes in the ceRNA
network was performed using the Database for Annotation,
Visualization and Integrated Discovery (DAVID) online tool (version
6.8; http://david.abcc.ncifcrf.gov)
(30). Only the enriched Gene
Ontology (GO) biological process terms and Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathways were highlighted to represent the
possible functions. P<0.05 was considered to indicate a
statistically significant difference.
Co-expression network between lncRNAs
and mRNAs
In order to further screen the crucial lncRNAs and
mRNAs, a co-expression network was also constructed based on the
correlation between DELs and DEGs. Pearson correlation coefficients
(PCC) were computed to evaluate the correlation using the WGCNA
(Weighted Gene Correlation Network Analysis; http://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/)
algorithm. Only the co-expressed pairs with |PCC≥0.900| and
P<0.001 were selected to establish the co-expression network
using Cytoscape. The lncRNAs and mRNAs in this co-expression
network that were overlapped with those in the ceRNA network may be
crucial for cardiac remodeling.
Results
Differential expression analysis
In GSE76592, 2.99% (23/769) miRNAs were found to be
differentially expressed in MI tissues compared with sham controls,
including 21 upregulated and two downregulated DEMs (Table I). In GSE52313, 2,563 of 9,205
mRNAs were identified to be differentially expressed between the MI
and sham control groups, including 1,705 upregulated and 858
downregulated DEGs (some are presented in Table I; all are presented in Table SI); while 91 upregulated and 77
downregulated DELs were yielded after differential analysis of 602
lncRNAs in two group samples (some are presented in Table I; all are presented in Table SII). The heatmaps indicated the MI
samples could be separated from the control samples according to
the expression levels of DEMs (Fig.
1A), DEGs (Fig. 1B) and DELs
(Fig. 1C).
 | Table I.Differentially expressed genes,
lncRNA and miRNAs between MI and sham control. |
Table I.
Differentially expressed genes,
lncRNA and miRNAs between MI and sham control.
| miRNA | logFC | P-value | FDR | lncRNA | logFC | P-value | FDR | mRNA | logFC | P-value | FDR |
|---|
| mmu-miR-382 | 0.74 |
1.90×10−13 |
1.46×10−10 | Dio3os | 2.70 |
2.39×10−7 |
9.83×10−5 | Col12a1 | 4.84 |
2.35×10−9 |
1.69×10−5 |
| mmu-miR-214 | 1.15 |
1.48×10−12 |
5.68×10−10 | Gm13749 | 3.32 |
3.26×10−7 |
9.83×10−5 | Lrp8 | 3.59 |
3.71×10−9 |
1.69×10−5 |
| mmu-miR-379 | 0.56 |
6.35×10−12 |
1.63×10−9 | Gm15867 | 2.48 |
2.50×10−6 |
5.02×10−4 | Nppa | 4.37 |
7.01×10−9 |
1.69×10−5 |
| mmu-miR-34c | 1.05 |
1.47×10−9 |
2.83×10−7 | A830039N20Rik | 2.88 |
4.18×10−6 |
6.29×10−4 | Serpina3n | 3.00 |
1.04×10−8 |
1.92×10−5 |
| mmu-miR-431 | 0.60 |
1.88×10−9 |
2.89×10−7 | Gm26512 | 2.60 |
1.60×10−5 |
1.93×10−3 | Vcan | 2.00 |
1.64×10−8 |
2.52×10−5 |
| mmu-miR-134 | 0.60 |
2.98×10−9 |
3.82×10−7 | Gm26740 | 1.11 |
3.35×10−5 |
3.01×10−3 | Crlf1 | 5.15 |
3.11×10−8 |
4.09×10−5 |
| mmu-miR-199a | 1.11 |
3.48×10−8 |
3.82×10−6 | 4930469K13Rik | 2.44 |
3.55×10−5 |
3.01×10−3 | Dkk3 | 3.96 |
4.02×10−8 |
4.25×10−5 |
| mmu-miR-21 | 0.90 |
9.74×10−8 |
8.33×10−6 | Pvt1 | 1.05 |
4.50×10−5 |
3.01×10−3 | Lox | 4.18 |
4.16×10−8 |
4.25×10−5 |
| mmu-miR-337 | 0.52 |
5.80×10−7 |
4.46×10−5 | 5330416C01Rik | 3.64 |
5.63×10−5 |
3.39×10−3 | Bst1 | 1.74 |
5.37×10−8 |
4.50×10−5 |
| mmu-miR-208b | 0.64 |
9.38×10−7 |
6.56×10−5 | 9430065F17Rik | 1.09 |
8.49×10−5 |
4.26×10−3 | Col8a1 | 3.09 |
6.01×10−8 |
4.50×10−5 |
| mmu-miR-411 | 0.51 |
1.66×10−6 |
9.80×10−5 | Gm15832 | 0.88 |
1.27×10−4 |
5.44×10−3 | Panx1 | 2.58 |
6.05×10−8 |
4.50×10−5 |
| mmu-miR-5130 | 0.59 |
2.64×10−6 |
1.45×10−4 | Has2os | 1.24 |
1.37×10−4 |
5.51×10−3 | Ptn | 3.31 |
6.35×10−8 |
4.50×10−5 |
| mmu-miR-125b-1 | 0.51 |
5.99×10−6 |
2.88×10−4 | A930029G22Rik | 0.92 |
2.36×10−4 |
6.77×10−3 | Dhrs9 | 3.82 |
7.10×10−8 |
4.57×10−5 |
| mmu-miR-762 | 1.53 |
9.06×10−6 |
4.10×10−4 | Gm6277 | 0.98 |
2.88×10−4 |
6.89×10−3 | Sprr1a | 5.92 |
7.80×10−8 |
4.57×10−5 |
| mmu-miR-5099 | 0.60 |
1.20×10−4 |
3.43×10−3 | A530020G20Rik | 2.61 |
3.27×10−4 |
6.89×10−3 | Chsy3 | 1.94 |
8.05×10−8 |
4.57×10−5 |
| mmu-miR-2861 | 0.52 |
2.89×10−4 |
7.40×10−3 | Gm15866 | 1.57 |
4.59×10−4 |
8.38×10−3 | Timp1 | 4.33 |
8.94×10−8 |
4.57×10−5 |
| mmu-miR-5126 | 0.52 |
1.24×10−3 |
2.03×10−2 | 1110046J04Rik | 1.10 |
4.93×10−4 |
8.74×10−3 | Met | 1.81 |
2.61×10−5 |
5.44×10−4 |
| mmu-miR-709 | 0.89 |
2.67×10−3 |
3.54×10−2 | A330023F24Rik | 0.76 |
1.57×10−3 |
1.69×10−2 | Itgam | 1.21 |
3.87×10−5 |
6.89×10−4 |
| mmu-miR-5121 | 0.67 |
7.26×10−3 |
6.85×10−2 | Gas5 | 0.63 |
3.77×10−3 |
2.70×10−2 | Egfr | 0.88 |
4.60×10−4 |
3.08×10−3 |
| mmu-miR-714 | 0.78 |
1.05×10−2 |
8.82×10−2 | Gm6634 | 0.78 |
3.78×10−3 |
2.70×10−2 | Esr1 | 0.84 |
7.35×10−3 |
2.19×10−2 |
| mmu-miR-3472 | 0.62 |
4.52×10−2 |
2.14×10−1 | Gm14636 | 1.03 |
7.80×10−3 |
4.27×10−2 | Tnf | 1.11 |
1.87×10−2 |
4.46×10−2 |
| mmu-miR-181a-2 |
−0.65 |
3.49×10−5 |
1.22×10−3 | Gm17281 |
−0.91 |
4.44×10−5 |
3.01×10−3 | Sod2 | −0.69 |
4.72×10−5 |
7.70×10−4 |
| mmu-miR-133a | −0.57 |
1.96×10−3 |
2.85×10−2 | 1810059H22Rik | −0.64 |
1.50×10−2 |
6.75×10−2 | Stat5a | −0.61 |
3.20×10−4 |
2.41×10−3 |
Screening of hub target mRNAs for
DEMs
A total of 299 downregulated DEGs were predicted to
be regulated by 21 upregulated DEMs (such as
mmu-miR-337-3p-Stat5a), while 184 upregulated DEGs were predicted
to be regulated by two downregulated DEMs. These 483 DEGs were
mapped onto the interaction pairs collected from the STRING
database. As a result, 394 DEGs (including 153 upregulated and 241
downregulated DEGs) were shown to interact with each other to
generate 1,013 interactions, which were used to construct the PPI
network (Fig. 2). Calculating the
topological features suggested Itgam (integrin α M), Sod2
(superoxide dismutase 2, mitochondrial), Met (met proto-oncogene),
TNF, Esr1 (estrogen receptor 1 α), Stat5a (signal transducer and
activator of transcription 5A) and Egfr (epidermal growth factor
receptor) may be hub genes as they ranked in the top 20 of all five
topological parameters (Table
II), indicating that their associated DEM interaction pairs may
be crucial for the development of MI.
 | Table II.Topological features of genes in the
protein-protein interaction network. |
Table II.
Topological features of genes in the
protein-protein interaction network.
| ID | DC | ID | BC | ID | CC | ID | EC | ID | SC |
|---|
| Tnf | 39 | Egfr | 19043.5 | Tnf | 0.0388 | Tnf | 0.356 | Tnf | 103778.8 |
| Egfr | 34 | Tnf | 14244.54 | Egfr | 0.0388 | Tlr4 | 0.288 | Itgam | 68163.56 |
| Itgam | 31 | Itgam | 12814.24 | Itgam | 0.0386 | Icam1 | 0.271 | Egfr | 60407.79 |
| Tlr4 | 23 | Ccnb1 | 9462.62 | Esr1 | 0.0385 | Cs | 0.254 | Tlr4 | 53020.38 |
| Cs | 23 | Ryr2 | 9017.83 | Sod2 | 0.0384 | Il10ra | 0.228 | Icam1 | 42583.11 |
| Esr1 | 22 | Eef1a2 | 8801.29 | Tlr4 | 0.0384 | Itgam | 0.206 | Stat5a | 34944.84 |
| Icam1 | 19 | Met | 8346.65 | Eef1a2 | 0.0384 | Ctla4 | 0.199 | Ctla4 | 32420.85 |
| Sod2 | 19 | Esr1 | 7894.01 | Dut | 0.0384 | Stat5a | 0.198 | Syk | 32246.88 |
| Stat5a | 18 | Kcna1 | 7429.03 | Icam1 | 0.0384 | Asb11 | 0.187 | Esr1 | 28649.09 |
| Syk | 18 | Sod2 | 7335.73 | Stat5a | 0.0383 | Klhl21 | 0.172 | Tnfsf11 | 24358.66 |
| Aco2 | 18 | Lamc1 | 6575.40 | Met | 0.03833 | Ube2cbp | 0.155 | Dut | 19740.2 |
| Met | 17 | Cnr1 | 6527.07 | Cnr1 | 0.0382 | Spn | 0.152 | Cd24a | 18906.82 |
| Ccnb1 | 17 | Eno3 | 5832.21 | Bcl2l11 | 0.0382 | Il4ra | 0.151 | Il10ra | 18840.3 |
| Ctla4 | 16 | Lpl | 5572.44 | Cd24a | 0.0382 | Uqcrc2 | 0.149 | Itga2 | 18214.35 |
| Itga2 | 15 | Stat5a | 5350.68 | Itga2 | 0.0382 | Syk | 0.146 | Jak1 | 17525 |
| Pdha1 | 15 | Vcan | 5071.53 | Rnase1 | 0.0382 | Jak1 | 0.138 | Sod2 | 16612.51 |
| Eno3 | 15 | Aco2 | 5031.69 | Ctla4 | 0.0381 | Wsb1 | 0.136 | Fcgr1 | 15776.24 |
| Gng7 | 15 | Gnai3 | 4899.65 | Eno3 | 0.0381 | Tnfsf11 | 0.132 | Spn | 14402.02 |
| Ryr2 | 15 | Tcap | 4856.46 | Tnfsf11 | 0.0381 | Uqcrc1 | 0.131 | Met | 14210.76 |
| Hadha | 14 | Arrb1 | 4844.48 | Flt1 | 0.0381 | Cd24a | 0.122 | Bcl2l11 | 12157.7 |
Construction of the ceRNA network
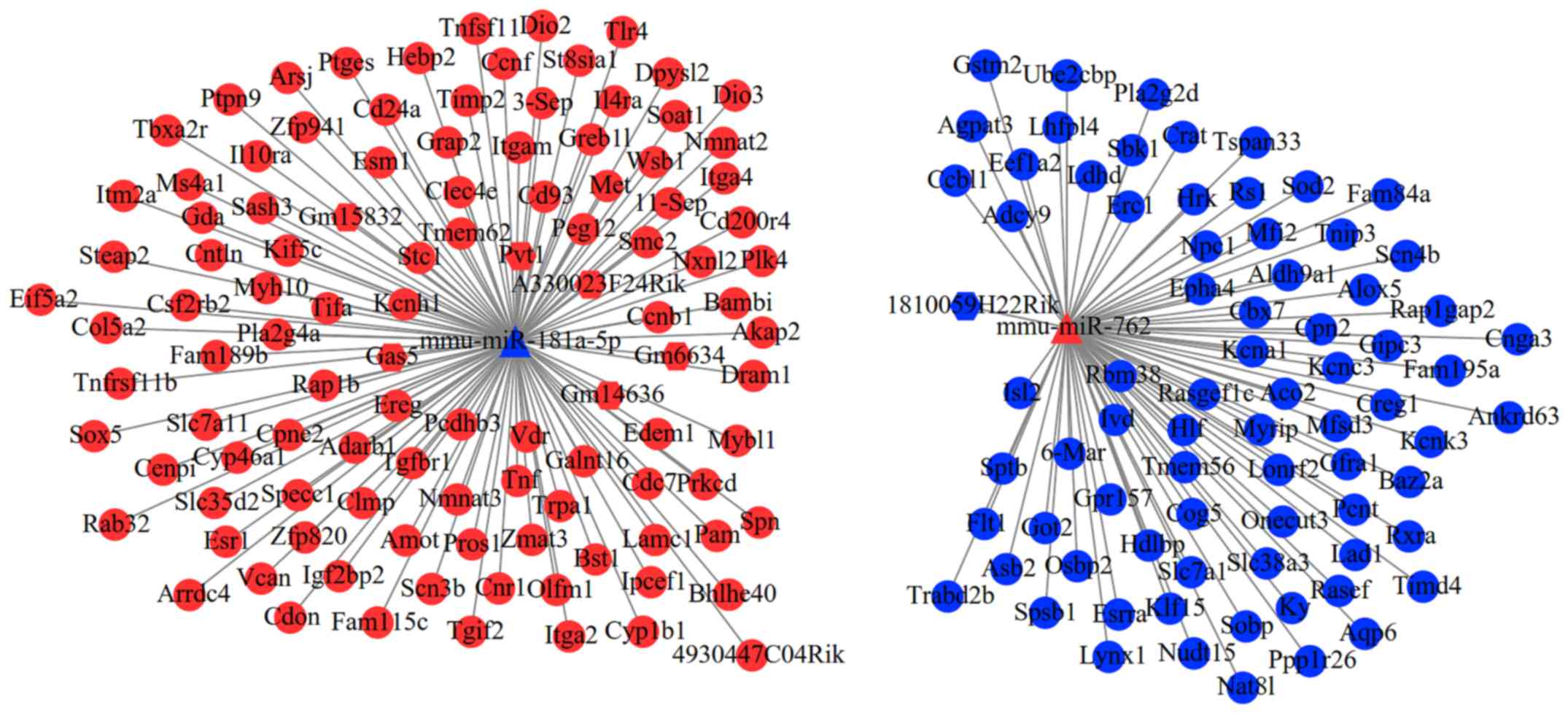
A total of 11 DEMs were predicted to interact with
32 lncRNAs, but only seven lncRNAs belonged to DELs. After further
excluding the DELs with consistent expression trend to DEMs, only
seven interaction pairs between two DEMs (mmu-miR-181a-5p and
mmu-miR-762) and seven DELs (Gm15832, Gas5, 1810059H22Rik, Gm6634,
Pvt1, Gm14636 and A330023F24Rik) remained. After overlapping the
aforementioned DEMs that regulated DEGs, a total of 693
lncRNA-miRNA-mRNA interaction pairs were yielded, which included
two DEMs, seven DELs and 178 DEGs (such as Met, Esr1, Sod2, TNF and
Itgam). These interaction pairs were used to construct the ceRNA
network (Fig. 3), in which hub
gene-associated ceRNA axes, such as lncRNA
Pvt1/Gm15832/A330023F24Rik/Gas5/Gm6634/Gm14636-
mmu-miR-181a-5p-Met/.
Esr1/TNF/Itgam and lncRNA
1810059H22Rik-mmu-miR-762-Sod2, may be important for cardiac
remodeling. In consideration of the fact that the FDR of Pvt1,
Gm15832, A330023F24Rik, Gas5, Gm6634 and Gm14636 were also less
than 0.05, ceRNA axes of these six lncRNAs may be inferred to be
particularly crucial for the post-MI cardiac remodeling at the
IBZ.
The 178 DEGs in the ceRNA network were uploaded into
the DAVID database to predict their potential functions. The
results indicated 12 significant KEGG pathways, such as
‘hematopoietic cell lineage (TNF and Itgam)’, ‘focal adhesion
(Met)’, ‘PI3K-Akt signaling pathway (Met)’, ‘cytokine-cytokine
receptor interaction (TNF)’ and ‘nicotinate and nicotinamide
metabolism’ [bst1 (bone marrow stromal cell antigen 1, also known
as CD157)] were enriched (Table
III and Fig. 4A and B).
Furthermore, the hub genes enriched in the KEGG pathways were also
included in 50 significant GO biological process terms, such as the
‘apoptotic signaling pathway (TNF)’, ‘positive regulation of MAP
kinase activity (TNF)’, ‘cell adhesion (Itgam)’, ‘positive
regulation of the inflammatory response (TNF)’, ‘leukocyte
cell-cell adhesion (Itgam)’, ‘negative regulation of cell
proliferation (TNF)’ and ‘positive regulation of mitotic nuclear
division (Met)’ (Table IV and
Fig. 4B).
 | Table III.KEGG pathways enriched for genes in
the ceRNA network. |
Table III.
KEGG pathways enriched for genes in
the ceRNA network.
| ID | Term | P-value | Genes |
|---|
| mmu04640 | Hematopoietic cell
lineage |
2.07×10−4 | TNF, IL4RA, MS4A1,
ITGA2, ITGA4, CD24A, ITGAM |
| mmu04611 | Platelet
activation |
1.04×10−2 | PLA2G4A, ADCY9,
TBXA2R, ITGA2, RAP1B, COL5A2 |
| mmu04510 | Focal adhesion |
1.82×10−2 | FLT1, MET, ITGA2,
RAP1B, LAMC1, ITGA4, COL5A2 |
| mmu04913 | Ovarian
steroidogenesis |
1.98×10−2 | PLA2G4A, CYP1B1,
ADCY9, ALOX5 |
| mmu05134 | Legionellosis |
1.98×10−2 | TNF, EEF1A2, TLR4,
ITGAM |
| mmu05145 | Toxoplasmosis |
2.16×10−2 | TNF, IL10RA, TLR4,
ALOX5, LAMC1 |
| mmu04151 | PI3K-Akt signaling
pathway |
2.52×10−2 | FLT1, RXRA, MET,
IL4RA, ITGA2, TLR4, LAMC1, ITGA4, COL5A2 |
| mmu05140 | Leishmaniasis |
2.69×10−2 | TNF, TLR4, ITGA4,
ITGAM |
| mmu05146 | Amoebiasis |
3.07×10−2 | TNF, TLR4, LAMC1,
COL5A2, ITGAM |
| mmu05152 | Tuberculosis |
3.27×10−2 | VDR, TNF, CLEC4E,
IL10RA, TLR4, ITGAM |
| mmu04060 | Cytokine-cytokine
receptor interaction |
3.57×10−2 | TNFRSF11B, TNFSF11,
TNF, CSF2RB2, TGFBR1, IL10RA, IL4RA |
| mmu00760 | Nicotinate and
nicotinamide metabolism |
4.38×10−2 | NMNAT3, NMNAT2,
BST1 |
 | Table IV.GO terms enriched for genes in the
ceRNA network. |
Table IV.
GO terms enriched for genes in the
ceRNA network.
| ID | Term | P-value | Genes |
|---|
| GO:0042493 | Response to
drug |
1.88×10−4 | CCNB1, PAM,
TNFRSF11B, NPC1, TRPA1, TBXA2R, ITGA2, DPYSL2, TIMP2, CBX7, KCNK3,
SOD2 |
| GO:0050966 | Detection of
mechanical stimulus involved in sensory perception of pain |
4.76×10−4 | TNF, KCNA1, TRPA1,
ITGA2 |
| GO:0097190 | Apoptotic signaling
pathway |
9.95×10−4 | VDR, TNFRSF11B,
TNF, CD24A, SPN |
| GO:0043406 | Positive regulation
of MAP kinase activity |
1.23×10−3 | FLT1, TNFSF11, TNF,
CD24A, PRKCD |
| GO:0032496 | Response to
lipopolysaccharide |
1.68×10−3 | TNFRSF11B, TNF,
PTGES, CNR1, IL10RA, TBXA2R, TLR4, SOD2 |
| GO:0043065 | Positive regulation
of apoptotic process |
2.66×10−3 | PLA2G4A, CYP1B1,
TNF, EEF1A2, CNR1, ZMAT3, RXRA, HRK, TLR4, PRKCD |
| GO:0007155 | Cell adhesion |
3.45×10−3 | EPHA4, CYP1B1,
CD93, CDON, ITGA2, VCAN, LAMC1, ITGA4, CD24A, ITGAM, MYH10,
RS1 |
| GO:0043627 | Response to
estrogen |
4.12×10−3 | TNFRSF11B, TGFBR1,
IL4RA, ESR1, CD24A |
| GO:0008202 | Steroid metabolic
process |
6.16×10−3 | SOAT1, HDLBP, NPC1,
CYP1B1, CYP46A1 |
| GO:0045429 | Positive regulation
of nitric oxide biosynthetic process |
6.98×10−3 | TNF, ESR1, TLR4,
SOD2 |
| GO:0034220 | Ion transmembrane
transport |
7.87×10−3 | KCNH1, AQP6, CNGA3,
KCNK3 |
| GO:0045670 | Regulation of
osteoclast differentiation |
9.35×10−3 | ESRRA, TNFSF11,
TNF |
| GO:0043401 | Steroid hormone
mediated signaling pathway |
1.10×10−2 | VDR, ESRRA, RXRA,
ESR1 |
| GO:0007420 | Brain
development |
1.15×10−2 | SLC38A3, KCNA1,
MET, DPYSL2, SLC7A11, KCNK3, MYH10 |
| GO:0043410 | Positive regulation
of MAPK cascade |
1.29×10−2 | TNFRSF11B, FLT1,
CDON, TIMP2, PRKCD |
| GO:0042404 | Thyroid hormone
catabolic process |
1.73×10−2 | DIO2, DIO3 |
| GO:0045994 | Positive regulation
of translational initiation by iron |
1.73×10−2 | TNF, RXRA |
| GO:0050729 | Positive regulation
of inflammatory response |
1.74×10−2 | PLA2G4A, TNF,
ITGA2, TLR4 |
| GO:0046697 |
Decidualization |
1.82×10−2 | VDR, PLA2G4A,
STC1 |
| GO:0007159 | Leukocyte cell-cell
adhesion |
1.82×10−2 | ITGA4, CD24A,
ITGAM |
| GO:0006629 | Lipid metabolic
process |
1.90×10−2 | SOAT1, PLA2G4A,
HDLBP, NPC1, CYP46A1, PTGES, ST8SIA1, CRAT, PLA2G2D, AGPAT3 |
| GO:0008285 | Negative regulation
of cell proliferation |
1.91×10−2 | VDR, ADARB1,
CYP1B1, TNF, EREG, PTGES, RXRA, TIMP2, SOD2 |
| GO:0006810 | Transport |
2.52×10−2 | KCNH1, SLC38A3,
HDLBP, KCNC3, MFSD3, SCN3B, ZMAT3, KCNA1, IGF2BP2, AQP6, GOT2,
SLC35D2, OSBP2, TRPA1, MFI2, CRAT, CNGA3, KCNK3, SLC7A11, COG5,
SLC7A1, SCN4B, STEAP2, ERC1, EIF5A2 |
| GO:0023041 | Neuronal signal
transduction |
2.58×10−2 | KCNA1, OLFM1 |
| GO:0060745 | Mammary gland
branching involved in pregnancy |
2.58×10−2 | VDR, ESR1 |
| GO:0034628 | ‘De novo’
NAD biosynthetic process from aspartate |
2.58×10−2 | NMNAT3, NMNAT2 |
| GO:0016477 | Cell migration |
2.58×10−2 | FLT1, GFRA1, LAMC1,
ITGA4, BAMBI, CD24A |
| GO:0045840 | Positive regulation
of mitotic nuclear division |
2.61×10−2 | TNF, EREG, MET |
| GO:0007507 | Heart
development |
2.66×10−2 | PAM, TGFBR1, RXRA,
VCAN, ITGA4, SOD2, MYH10 |
| GO:0071407 | Cellular response
to organic cyclic compound |
2.75×10−2 | CCNB1, CYP1B1, TNF,
RAP1B |
| GO:0019233 | Sensory perception
of pain |
2.94×10−2 | SCN3B, CNR1, TRPA1,
ALOX5 |
| GO:0045664 | Regulation of
neuron differentiation |
2.95×10−2 | CDON, DPYSL2,
TIMP2 |
| GO:0034765 | Regulation of ion
transmembrane transport |
3.02×10−2 | KCNH1, KCNC3,
SCN3B, KCNA1, SCN4B |
| GO:0006811 | Ion transport |
3.14×10−2 | KCNH1, SLC38A3,
KCNC3, SCN3B, MFI2, KCNA1, TRPA1, SCN4B, CNGA3, STEAP2, KCNK3 |
| GO:0055114 | Oxidation-reduction
process |
3.39×10−2 | PAM, CYP1B1,
CYP46A1, DIO2, DIO3, IVD, LDHD, CREG1, ALOX5, STEAP2, ALDH9A1,
SOD2 |
| GO:0045123 | Cellular
extravasation |
3.43×10−2 | TNF, ITGAM |
| GO:0051365 | Cellular response
to potassium ion starvation |
3.43×10−2 | SLC38A3, HRK |
| GO:0010718 | Positive regulation
of epithelial to mesenchymal transition |
3.51×10−2 | TGFBR1, BAMBI,
OLFM1 |
| GO:0048514 | Blood vessel
morphogenesis |
3.70×10−2 | FLT1, CYP1B1,
AMOT |
| GO:0019369 | Arachidonic acid
metabolic process |
3.89×10−2 | PLA2G4A, CYP1B1,
ALOX5 |
| GO:0009058 | Biosynthetic
process |
4.09×10−2 | GOT2, NMNAT3,
NMNAT2 |
| GO:0007411 | Axon guidance |
4.10×10−2 | EPHA4, DPYSL2,
LAMC1, CD24A, MYH10 |
| GO:0008203 | Cholesterol
metabolic process |
4.24×10−2 | SOAT1, HDLBP, NPC1,
CYP46A1 |
| GO:0033591 | Response to
L-ascorbic acid |
4.27×10−2 | ITGA2, SOD2 |
| GO:0071805 | Potassium ion
transmembrane transport |
4.36×10−2 | KCNH1, KCNC3,
KCNA1, KCNK3 |
| GO:0030199 | Collagen fibril
organization |
4.50×10−2 | CYP1B1, TGFBR1,
COL5A2 |
| GO:0001701 | In utero embryonic
development |
4.65×10−2 | CCNB1, PCNT,
TGFBR1, RXRA, SOX5, AMOT, MYH10 |
| GO:0009409 | Response to
cold |
4.71×10−2 | DIO2, TRPA1,
SOD2 |
| GO:0042127 | Regulation of cell
proliferation |
4.81×10−2 | KCNH1, PLA2G4A,
TNFRSF11B, ESRRA, TNF, IL4RA |
| GO:0008152 | Metabolic
process |
4.92×10−2 | GSTM2, PAM,
PLA2G4A, ACO2, IVD, ARSJ, EDEM1, AGPAT3, ALDH9A1 |
Construction of the co-expression
network
According to the threshold of |PCC≥0.900| and
P<0.001, 355 co-expression pairs between 44 DELs and 297 DEGs
were screened, which were used to construct the co-expression
network (Fig. 5).
In order to further screen the crucial lncRNAs and
mRNAs, the lncRNAs and mRNAs in co-expression and ceRNA networks
were compared. The comparison results demonstrated that three
lncRNAs (Pvt1, Gm15832 and A330023F24Rik) were shared between these
two networks. Subsequent comparisons demonstrated that Bst1 was the
common target of Pvt1, Gm15832 and A330023F24Rik, irrespective of
the co-expression or ceRNA network. Thus, Bst1-associated ceRNA
(Pvt1-mmu-miR-181a-5p-Bst1) and co-expression (Pvt1-Bst1) axes were
inferred to be pivotal for MI.
Discussion
From the PPI and ceRNA network analyses, the present
study identified six lncRNAs (Pvt1, Gm15832, A330023F24Rik, Gas5,
Gm6634 and Gm14636) that sponge mmu-miR-181a-5p to regulate the
three hub genes: TNF, Met and Itgam. These hub genes were involved
in ‘regulation of inflammatory response’ and ‘cell proliferation
(TNF)’, ‘mitotic nuclear division (Met)’ and ‘cell adhesion
(Itgam)’. Furthermore, based on the ceRNA and co-expression network
analyses, the present study demonstrated that Pvt1 may be important
for MI because it may serve a role in constituting the ceRNA axis
with mmu-miR-181a-5p-Bst1 or by directly co-expressing with Bst1 to
participate in nicotinate and nicotinamide metabolism.
There have been studies to prove the associations
between TNF, Met and Itgam with cardiac remodeling following MI.
For example, Jacobs et al (31) reported that TNF-α was detected in
the infarct, border and remote zones of rat hearts with MI. TNF-α
stimulation may lead to cardiac fibrosis by promoting the
proliferation of fibroblasts isolated from the infarct and
non-infarct-region hearts. Heba et al (32) observed that the expression of TNF-α
was increased at the border zone on days 1, 4, 11, 28 and 40 after
MI and the expression of TNF-α was parallel with the development of
HF after MI assessed by hemodynamic measurements. Artemisinin was
demonstrated to exert beneficial effects on ventricular remodeling
following MI by significantly decreasing the level of TNF-α at the
IBZ (14). Immunohistochemistry
analysis demonstrated that c-Met was intensely expressed in the
border zone myocardium of the infarcted and non-infarcted region in
patients with MI (33) and in
certain hypertrophic myocardial cells (34). Wang et al (35) reported that compound Longmaining
decoction may exert a protective effect on MI by decreasing the
expression level of Itgam. Furthermore, other integrin members
(such as α5β1 and αvβ3) were also demonstrated to be
upregulated to mediate abnormal vascular remodeling and
pro-inflammatory macrophage polarization in endothelial cells at
the IBZ following MI, further worsening cardiac hypoxia and
inflammation (36,37); while downregulation of integrin-β
by erythropoietin enhanced angiogenesis, decreased inflammation and
consequently improved myocardial functions (38). In line with these studies, the
present analysis of GSE52313 dataset (21) also found that TNF, Met and Itgam
were upregulated in the IBZ of MI model mice compared with sham
controls. These findings indicate upstream regulators may also
participate in cardiac remodeling by changing the expression levels
of these genes.
miRNAs are 21–25 nucleotide non coding RNA molecules
that bind to complementary sites in the 3′-untranslated regions of
target mRNAs to repress or silence their translation.
Theoretically, miRNAs (such as mmu-miR-181a-5p) that upregulate
TNF, Met and Itgam, may be downregulated in the IBZ of MI model
mice, notably analysis of the GSE76592 dataset (22) confirmed this in the present study.
Also, previous studies demonstrated that overexpression of
miR-181a, decreased cardiomyocyte cell size (i.e., hypertrophy) and
apoptosis induced by high glucose, which is a common risk factor
for MI (39); while miR-181a/b
deficiency mice exhibited an increase in infarct size (40). The negative regulatory associations
between miR-181a-5p and TNF (41,42)
as well as between miR-181a-5p and Met (43,44)
were also demonstrated to be present in immune cells and other
inflammatory diseases that are important mechanisms for MI
(31,32). Accordingly, the miR-181a-TNF/Met
interaction may be inferred to influence the development of post-MI
VAs.
lncRNAs can act as ceRNAs to regulate miRNA-mediated
target gene repression and thus they may also be important target
genes for the treatment of MI. The present study predicted that six
lncRNAs (Pvt1, Gm15832, A330023F24Rik, Gas5, Gm6634 and Gm14636)
could interact with miR-181a. Among these, Gm15832 and Gm6634 may
be new targets that, to the best of our knowledge, have not
previously been reported in any diseases. Gm14636 was previously
recorded to be upregulated to mediate the apoptotic effects of
etomidate on murine leukemia RAW264.7 cells in vitro
(45), which seemed to be in
accordance with its potential function in MI, as Gm14636 was also
observed to be upregulated in the present study. Vorinostat has
previously been implicated to possess anti-inflammatory effects by
increasing the expression of A330023F24Rik (46), which was not in line with the
expected pro-inflammatory roles of upregulated A330023F24Rik in the
IBZ in the present study. Further validation experiments should
therefore be performed. Similar to the results of the present
study, GAS5 had been shown to be highly expressed in the IBZ of MI
model animals, but its role is controversial. Hao et al
(19) suggested that
overexpression of GAS5 may exert a protective effect against MI by
decreasing cardiomyocyte apoptosis, while Du et al
demonstrated that silencing of GAS5 protected myocardial cells
against hypoxia-induced injury (47). Taking into consideration the roles
of downstream genes, the hypothesis of the present study was that
high expression levels of GAS5 may be detrimental for MI as
described by Du et al (47). This requires confirmation from
further studies. A previous study has demonstrated that Pvt1 was
upregulated in the myocardial tissues of sepsis rats, which
inhibited cardiac function and promoted the secretion of
inflammatory factors via activation of the MAPK/NF-κB pathway
(48). Furthermore, Pvt1 has also
previously been demonstrated to be upregulated in cardiac
hypertrophic model mice when compared with the sham group; while
knockout of Pvt1 by siRNA significantly reduced the cardiomyocyte
size (49). Consistent with these
two studies, the present study also demonstrated Pvt1 was
upregulated and may be involved in inflammation and cardiac
hypertrophy by regulating TNF, Met and Itgam. However, to the best
of our knowledge, there are a limited number of reports on the
mechanisms of Pvt1 in MI and the results of the present study
require additional verification.
In addition to miR-181a-5p-TNF/Met/Itgam ceRNA axes,
the present study also demonstrated that Pvt1 may function in MI by
acting as a ceRNA for mmu-miR-181a-5p-Bst1 or by directly
co-expressing with Bst1. CD157/Bst1 encodes a protein that exhibits
ADP-ribosyl cyclase (ADPRC) activity (50). ADPRCs can catalyze the conversion
of nicotinamide adenine dinucleotide to cyclic adenosine
diphosphoribose, which is a second messenger for regulating Ca(2+)
mobilization from intracellular stores. cADPR may exert
arrhythmogenic activity via its interaction with type 2 ryanodine
receptors in the heart; while inhibition of cardiac ADPRC was
previously reported to prevent Ca2+ overload-induced
ventricular fibrillation (51).
Thus, upregulation of Bst1 in the IBZ may cause VAs, which was
demonstrated in the present study. Taken together, these studies on
the roles of Pvt1 and miR-181a in MI along with the present study
suggested these Bst-associated mechanisms may also be verifiable
through further studies.
Certain limitations are present in the present
study. The datasets used in the present study present limitations,
such as the small sample size of the lncRNA-mRNA dataset, different
platforms of miRNA and lncRNA-mRNA datasets and different sample
collection time, which may bias the results. In addition,
lncRNA-associated co-expression and ceRNA axes were obtained by
database prediction, which may result in false positives.
Therefore, wet experiments (such as quantitative PCR, luciferase
assay, knockdown or overexpression) are needed to validate their
interactions and roles in cardiac remodeling after MI both in
vitro and in vivo.
In conclusion, the present study demonstrated that
upregulated Pvt1 may be crucial for cardiac remodeling in the IBZ
following MI, and may function by acting as a ceRNA for miR-181a to
regulate TNF/Met/Itgam/Bst1 or by directly co-expressing with Bst1
to participate in cardiomyocyte inflammation, hypertrophy,
apoptosis and contractile processes. These genes in ceRNA or
co-expression axes may present targets for the treatment of MI.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the GEO database in NCBI (http://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
BL, YC and XC conceived and designed the original
study. BL performed the bioinformatic analysis. JT and LZ
contributed to the acquisition and interpretation of data. BL and
YJC drafted the manuscript. XC revised the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fahed J, Floyd KC and Tighe DA: Sudden
cardiac death in postmyocardial-infarction patients. Panminerva
Med. 57:2605–86. 2015.
|
|
2
|
Saaby L, Poulsen TS, Diederichsen AC,
Hosbond S, Larsen TB, Schmidt H, Gerke O, Hallas J, Thygesen K and
Mickley H: Mortality rate in type 2 myocardial infarction:
Observations from an unselected hospital cohort. Am J Med.
127:295–302. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma S, Ma J, Mai X, Zhao X, Guo L and Zhang
M: Danqi soft capsule prevents infarct border zone remodelling and
reduces susceptibility to ventricular arrhythmias in
post-myocardial infarction rats. J Cell Mol Med. 23:5454–5465.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen RH, Li YG, Jiao KL, Zhang PP, Sun Y,
Zhang LP, Fong XF, Li W and Yu Y: Overexpression of Sema3a in
myocardial infarction border zone decreases vulnerability of
ventricular tachycardia post-myocardial infarction in rats. J Cell
Mol Med. 17:608–616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huo L, Shi W, Chong L, Wang J, Zhang K and
Li Y: Asiatic acid inhibits left ventricular remodeling and
improves cardiac function in a rat model of myocardial infarction.
Exp Ther Med. 11:57–64. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wenk JF, Klepach D, Lee LC, Zhang Z, Ge L,
Tseng EE, Martin A, Kozerke S, Gorman JH III, Gorman RC and
Guccione JM: First evidence of depressed contractility in the
border zone of a human myocardial infarction. Ann Thorac Surg.
93:1188–1193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu X, Wan N, Zhang XJ, Zhao Y, Zhang Y,
Hu G, Wan F, Zhang R, Zhu X, Xia H and Li H: Vinexin-β exacerbates
cardiac dysfunction post-myocardial infarction via mediating
apoptotic and inflammatory responses. Clin Sci (Lond). 128:923–936.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takahashi T, Anzai T, Kaneko H, Mano Y,
Anzai A, Nagai T, Kohno T, Maekawa Y, Yoshikawa T, Fukuda K and
Ogawa S: Increased C-reactive protein expression exacerbates left
ventricular dysfunction and remodeling after myocardial infarction.
Am J Physiol Heart Circ Physiol. 299:H1795–H1804. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song G, Li X, Shen Y, Qian L, Kong X, Chen
M, Cao K and Zhang F: Transplantation of iPSc restores cardiac
function by promoting angiogenesis and ameliorating cardiac
remodeling in a post-infarcted swine model. Cell Biochem Biophys.
71:1463–1473. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang X, Zhu D, Wei L, Zhao Z, Qi X, Li Z
and Sun D: OSM enhances angiogenesis and improves cardiac function
after myocardial infarction. Biomed Res Int.
2015:3179052015.PubMed/NCBI
|
|
11
|
Yang L, Gregorich ZR, Cai W, Zhang P,
Young B, Gu Y, Zhang J and Ge Y: Quantitative proteomics and
immunohistochemistry reveal insights into cellular and molecular
processes in the infarct border zone one month after myocardial
infarction. J Proteome Res. 16:2101–2112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiang F, Shi Z, Guo X, Qiu Z and Chen X,
Huang F, Sha J and Chen X: Proteomic analysis of myocardial tissue
from the border zone during early stage post-infarct remodelling in
rats. Eur J Heart Fail. 13:254–263. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang C, Zhang B, Lin Y and Dong Y: Effects
of adenovirus-mediated VEGF165 gene therapy on myocardial
infarction. Ann Clin Lab Sci. 48:208–215. 2018.PubMed/NCBI
|
|
14
|
Gu Y, Wu G, Wang X, Wang X, Wang Y and
Huang C: Artemisinin prevents electric remodeling following
myocardial infarction possibly by upregulating the expression of
connexin 43. Mol Med Rep. 10:1851–1856. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Spaulding K, Takaba K, Collins A, Faraji
F, Wang G, Aguayo E, Ge L, Saloner D, Wallace AW, Baker AJ, et al:
Short term doxycycline treatment induces sustained improvement in
myocardial infarction border zone contractility. PLoS One.
13:e01927202018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun F, Zhuang Y, Zhu H, Wu H, Li D, Zhan
L, Yang W, Yuan Y, Xie Y, Yang S, et al: lncRNA PCFL promotes
cardiac fibrosis via miR-378/GRB2 pathway following myocardial
infarction. J Mol Cell Cardiol. 133:188–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu H, Wu J, Li D, Zhou J, Yu H and Ma L:
Knockdown of lncRNA MALAT1 attenuates acute myocardial infarction
through miR-320-Pten axis. Biomed Pharmacother. 106:738–746. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu T, Wu D, Wu Q, Zou B, Huang X, Cheng X,
Wu Y, Hong K, Li P, Yang R, et al: Knockdown of long non-coding
RNA-ZFAS1 protects cardiomyocytes against acute myocardial
infarction via anti-apoptosis by regulating miR-150/CRP. J Cell
Biochem. 118:3281–3289. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hao S, Liu X, Sui X, Pei Y, Liang Z and
Zhou N: Long non-coding RNA GAS5 reduces cardiomyocyte apoptosis
induced by MI through sema3a. Int J Biol Macromol. 120:371–377.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kaikkonen MU, Halonen P, Liu OH, Turunen
TA, Pajula J, Moreau P, Selvarajan I, Tuomainen T, Aavik E, Tavi P
and Ylä-Herttuala S: Genome-wide dynamics of nascent noncoding RNA
transcription in porcine heart after myocardial infarction. Circ
Cardiovasc Genet. 10:e0017022017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ounzain S, Micheletti R, Beckmann T,
Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A,
Johnson R, et al: Genome-wide profiling of the cardiac
transcriptome after myocardial infarction identifies novel
heart-specific long non-coding RNAs. Eur Heart J. 36:353–368a.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsuji M, Kawasaki T, Matsuda T, Arai T,
Gojo S and Takeuchi JK: Sexual dimorphisms of mRNA and miRNA in
human/murine heart disease. PLoS One. 12:e01779882017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statist Soc B. 57:289–300. 1995.
|
|
25
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43((Database Issue)): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM and Hatzigeorgiou
AG: DIANA-LncBase: Experimentally verified and computationally
predicted microRNA targets on long non-coding RNAs. Nucleic Acids
Res. 41((Database Issue)): D239–D245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jacobs M, Staufenberger S, Gergs U, Meuter
K, Brandstätter K, Hafner M, Ertl G and Schorb W: Tumor necrosis
factor-alpha at acute myocardial infarction in rats and effects on
cardiac fibroblasts. J Mol Cell Cardiol. 31:1949–1959. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heba G, Krzemiński T, Porc M, Grzyb J and
Dembińska-Kieć A: Relation between expression of TNF alpha, iNOS,
VEGF mRNA and development of heart failure after experimental
myocardial infarction in rats. J Physiol Pharmacol. 52:39–52.
2001.PubMed/NCBI
|
|
33
|
Sato T, Fujieda H, Murao S, Sato H,
Takeuchi T and Ohtsuki Y: Sequential changes of hepatocyte growth
factor in the serum and enhanced c-Met expression in the myocardium
in acute myocardial infarction. Jpn Circ J. 63:906–908. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sato T, Tani Y, Murao S, Fujieda H, Sato
H, Matsumoto M, Takeuchi T and Ohtsuki Y: Focal enhancement of
expression of c-Met/hepatocyte growth factor receptor in the
myocardium in human myocardial infarction. Cardiovasc Pathol.
10:235–240. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang C, Bai X, Liu S, Wang J, Su Z, Zhang
W, Bu D, Yan Y and Song X: RNA-seq based transcriptome analysis of
the protective effect of compound longmaining decoction on acute
myocardial infarction. J Pharm Biomed Anal. 158:339–345. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee SJ, Lee CK, Kang S, Park I, Kim YH,
Kim SK, Hong SP, Bae H, He Y, Kubota Y and Koh GY: Angiopoietin-2
exacerbates cardiac hypoxia and inflammation after myocardial
infarction. J Clin Invest. 128:5018–5033. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Menichetti L, Kusmic C, Panetta D, Arosio
D, Petroni D, Matteucci M, Salvadori PA, Casagrande C, L'Abbate A
and Manzoni L: MicroPET/CT imaging of αvβ3 integrin via a novel
63Ga-NOTA-RGD peptidomimetic conjugate in rat myocardial
infarction. Eur J Nucl Med Mol Imaging. 40:1265–1274. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Furlani D, Klopsch C, Gäbel R, Ugurlucan
M, Pittermann E, Klee D, Wagner K, Li W, Wang W, Ong LL, et al:
Intracardiac erythropoietin injection reveals antiinflammatory
potential and improved cardiac functions detected by forced swim
test. Transplant Proc. 40:962–966. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Raut SK, Singh GB, Rastogi B, Saikia UN,
Mittal A, Dogra N, Singh S, Prasad R and Khullar M: miR-30c and
miR-181a synergistically modulate p53-p21 pathway in diabetes
induced cardiac hypertrophy. Mol Cell Biochem. 417:191–203. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Das S, Kohr M, Dunkerly-Eyring B, Lee DI,
Bedja D, Kent OA, Leung AK, Henao-Mejia J, Flavell RA and
Steenbergen C: Divergent effects of miR-181 family members on
myocardial function through protective cytosolic and detrimental
mitochondrial microRNA targets. J Am Heart Assoc. 6:e0046942017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhu J, Wang FL, Wang HB, Dong N, Zhu XM,
Wu Y, Wang YT and Yao YM: TNF-α mRNA is negatively regulated by
microRNA-181a-5p in maturation of dendritic cells induced by high
mobility group box-1 protein. Sci Rep. 7:122392017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Corsetti PP, de Almeida LA, Gonçalves ANA,
Gomes MTR, Guimarães ES, Marques JT and Oliveira SC: miR-181a-5p
regulates TNF-α and miR-21a-5p influences gualynate-binding protein
5 and IL-10 expression in macrophages affecting host control of
Brucella abortus Infection. Front Immunol. 9:13312018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Korhan P, Erdal E and Atabey N:
miR-181a-5p is downregulated in hepatocellular carcinoma and
suppresses motility, invasion and branching-morphogenesis by
directly targeting c-Met. Biochem Biophys Res Commun.
450:1304–1312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dong N, Tang X and Xu B: miRNA-181a
inhibits the proliferation, migration, and epithelial-mesenchymal
transition of lens epithelial cells. Invest Ophthalmol Vis Sci.
56:993–1001. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wu RS, Wu KC, Yang JS, Chiou SM, Yu CS,
Chang SJ, Chueh FS and Chung JG: Etomidate induces cytotoxic
effects and gene expression in a murine leukemia macrophage cell
line (RAW264.7). Anticancer Res. 31:2203–2208. 2011.PubMed/NCBI
|
|
46
|
Ye Y, Zhao X, Lu Y, Long B and Zhang S:
Varinostat alters gene expression profiles in aortic tissues from
ApoE-/-mice. Hum Gene Ther Clin Dev. 29:214–225. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Du J, Yang ST, Liu J, Zhang KX and Leng
JY: Silence of lncRNA GAS5 protects cardiomyocytes H9c2 against
hypoxic injury via sponging miR-142-5p. Mol Cells. 42:397–405.
2019.PubMed/NCBI
|
|
48
|
Feng F, Qi Y, Dong C and Yang C: PVT1
regulates inflammation and cardiac function via the MAPK/NF-κB
pathway in a sepsis model. Exp Ther Med. 16:4471–4478.
2018.PubMed/NCBI
|
|
49
|
Yu YH, Hu ZY, Li MH, Li B, Wang ZM and
Chen SL: Cardiac hypertrophy is positively regulated by long
non-coding RNA PVT1. Int J Clin Exp Pathol. 8:2582–2589.
2015.PubMed/NCBI
|
|
50
|
Higashida H, Liang M, Yoshihara T, Akther
S, Fakhrul A, Stanislav C, Nam TS, Kim UH, Kasai S, Nishimura T, et
al: An immunohistochemical, enzymatic, and behavioral study of
CD157/BST-1 as a neuroregulator. BMC Neuroscience. 18:352017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kannt A, Sicka K, Kroll K, Kadereit D and
Gögelein H: Selective inhibitors of cardiac ADPR cyclase as novel
anti-arrhythmic compounds. Naunyn Schmiedebergs Arch Pharmacol.
385:717–727. 2012. View Article : Google Scholar : PubMed/NCBI
|