Introduction
Thyroid cancer is the most common subtype of
endocrine cancer worldwide, with an increasing incidence level
(1). Thyroid cancer can be divided
into multiple subtypes, the majority of which originate from
follicular cells. Medullary thyroid cancer (MTC), which accounts
for 5–10% of all cases of thyroid cancer, is the only type that
originates from parafollicular C cells (2,3).
Patients with MTC with an obvious thyroid nodule frequently present
with cervical metastases and ~13% present with distant metastases
(4,5). The 10-year MTC-specific mortality
rate ranges from 13.5–38% worldwide (6). The clinical stage at the time of
diagnosis and the probability of complete surgical removal of the
tumor are the two most important determinants for the successful
treatment of MTC (7). The
prognostic factors of patients with MTC who undergo surgical
resection include tumor volume, metastases and location, age,
calcitonin level and carcinoembryonic antigen doubling times
(8). In the last decade,
advancements have been made in the diagnosis and therapeutic
treatment of human non-MTC; however, the physiopathology of MTC is
not completely understood (9,10).
Therefore, exploring molecular targets that may improve the
accuracy of MTC diagnosis is important.
MicroRNAs (miRNA) are small single-stranded
non-coding RNAs, ~22 nucleotides in length, that widely exist in
mammalian cells (11,12). Due to the limitations of research
technologies, miRNAs were initially considered to be noise of
transcription without biological functions (13). Progression in next-generation
sequencing has made it possible to identify the expression profiles
of miRNAs, thereby allowing their functions during the pathogenesis
of various human diseases to be investigated (14). At present, the regulatory role of
miRNAs on gene expression has been well demonstrated, with
increasing evidence indicating that miRNAs can degrade target mRNAs
or repress translation (15).
miRNA dysregulation is a critical event during the initiation and
progression of tumors (16,17),
and recently, it has also been reported that miRNAs are implicated
during the tumorigenesis of MTC (18); however, the roles of the majority
of miRNAs in MTC are not completely understood.
To identify unique miRNAs associated with the
tumorigenesis of MTC, differentially expressed miRNAs in the
GSE40807 dataset were assessed using the Gene Expression Omnibus
(GEO)2R method. miR-592 expression was increased in MTC samples
compared with normal samples, which indicated that miR-592 may
serve a role during the tumorigenesis of MTC. It has been
previously reported that miR-592 affects the development of various
human tumors, including glioma and acute myeloid leukemia, as well
as gastric and breast cancer (19–22);
however, the role and mechanism of miR-592 during MTC is not
completely understood. Therefore, the present study aimed to
investigate the physiological functions and potential mechanisms
underlying miR-592 during MTC tumorigenesis.
Materials and methods
Microarray dataset
The MTC-associated miRNA GEO dataset GSE40807,
consisting of 80 samples, was downloaded from the GEO database
(www.ncbi.nlm.nih.gov/geo) (23). Differentially expressed miRNAs in
GSE40807 were identified using GEO2R.
MTC tumor samples
A total of 20 paired cancer and normal tissue
specimens (distance from tumor margin, 5 cm) were collected from
patients with MTC (mean age, 66.53±12.48 years; 8 female patients
and 12 male patients) who were diagnosed at the Affiliated Wuhan
Central Hospital of Tongji Medical College between March 2015 and
October 2019. Tissue samples were stored in liquid nitrogen until
further analysis. The present study was approved by the Ethics
Committee of The Affiliated Wuhan Central Hospital of Tongji
Medical College, Huazhong University of Science and Technology.
Written informed consent was obtained from each participant. The
basic clinicopathological features of the 20 patients with MTC are
presented in Table I. Moreover,
the number of patients with different pathological grade, tumor
size, T stage, N stage, M stage and TNM stage was determined.
 | Table I.Basic clinicopathological features of
patients with MTC. |
Table I.
Basic clinicopathological features of
patients with MTC.
| Parameters | Patients with MTC
(n=20) |
|---|
| Age, years | 66.53±12.48 |
| Sex,
male/female | 12/8 |
| Pathological
grade |
|
| Well
differentiation | 4 (20%) |
|
Moderate differentiation | 13 (65%) |
| Poor
differentiation | 3 (15%) |
| Tumor size, cm |
|
|
<5 | 11 (55%) |
| ≥5 | 9 (45%) |
| T stage, n (%) |
|
| T1 | 4 (20) |
| T2 | 2 (10) |
| T3 | 5 (25) |
| T4 | 9 (45) |
| N stage, n (%) |
|
| N0 | 12 (60) |
|
N1a | 3 (15) |
|
N1b | 2 (10) |
|
N1c | 1 (5) |
|
N2a | 1 (5) |
|
N2b | 1 (5) |
| M stage, n (%) |
|
| M0 | 19 (95) |
|
M1a | 1 (5) |
| TNM stage, n
(%) |
|
| II | 3 (15) |
|
IIA | 8 (40) |
|
IIIB | 5 (25) |
|
IIIC | 3 (15) |
|
IVA | 1 (5) |
Cell lines
The immortalized normal thyroid follicular
NThy-ori-3.1 cell line, and the MTC TT and MZ-CRC-1 cell lines were
purchased from the American Type Culture Collection. Cells were
cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.)
containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C and 5% CO2.
RNA transfection
miR-592 mimic (5′-UGUAGUAGCGUAUAACUGUGUU-3′),
scramble mimic (5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′)
and the CDK8 plasmid were designed and obtained from Shanghai
GenePharma Co., Ltd. TT and MZ-CRC-1 cells were seeded
(1×105 cells/well) into 6-well plates and cultured at
37°C for 8 h. Subsequently, cells were transfected with 200 µl
mimic (100 nM) or scramble (100 nM) or 1 µg plasmid using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. At 48
h post-transfection, cells were used for subsequent
experiments.
RNA extraction and quantitative
real-time PCR (RT-qPCR) assay
Total RNA was extracted from MTC tissue samples and
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). RNA quality was determined using a NanoDrop
2000c spectrophotometer (Thermo Fisher Scientific, Inc.). Total RNA
(3 µg) was reverse transcribed into cDNA using the Bestar qPCR RT
kit (DBI Bioscience; cat. no. DBI-2220). The temperature protocol
used for reverse transcription was 37°C for 15 min and 98°C for 5
min. Subsequently, qPCR was performed using the Bestar qPCR
MasterMix (DBI Bioscience) and an ABI 7500 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The following thermocycling conditions
were used for qPCR: 95°C for 2 min; 95°C for 10 sec, 60°C for 34
sec, 72°C for 30 sec; and the solubility curve was obtained at
98°C. The primers used for qPCR are presented in Table II. miRNA and mRNA expression
levels were quantified using the 2−∆∆Cq method (24) normalized to the internal reference
genes U6 and GAPDH, respectively.
 | Table II.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table II.
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5′-3′) |
|---|
| GAPDH | F:
TGTTCGTCATGGGTGTGAAC |
|
| R:
ATGGCATGGACTGTGGTCAT |
| miR-592 | F:
CCATGACATTGTGTCAATATGCGA |
|
| R:
CGTCATGATGTTGCGTCACC |
| SCGB2A2 | F:
GAACACCGACAGCAGCA |
|
| R:
TCTCCAATAAGGGGCAGCC |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| LINC00632 | F:
CACGCCTGTTATCCC |
|
| R:
CAACCTCCGCCTCTT |
| CREB3L3 | F:
CAGTCAGCTCAAGAAAGCAGG |
|
| R:
TGGTTCTGGGCAGTACACG |
| OR4F4 | F:
ATAGCCATGGGCTTTGACAG |
|
| R:
TGGGACCACAGAAGGGTAAG |
| CCDC149 | F:
CTCTCCAAGGAGCTGGACAC |
|
| R:
TCCAAGCCTTTGCTGAAGTT |
| CDK8 | F:
GCCGGTTGTCAAATCCCTTAC |
|
| R:
TGTGACTGCTGTCTTGATTCCCT |
Data analysis
The data used for the analysis of miR-592 or CDK8
expression and the overall survival of patients with high and low
miR-592 or CDK8 expression were downloaded from starBase (version
3; starbase.sysu.edu.cn/panCancer.php). Kaplan-Meier
plots (www.kmplot.com) was applied to analyze
the association between overall survival and miR-592 or CDK8
expression in patients with MTC. Kalpan-Meier plots were compared
using the log-rank test.
Western blotting
Total protein was extracted from transfected TT and
MZ-CRC-1 cells using RIPA buffer (cat. no. R0278; Sigma-Aldrich;
Merck KGaA). Total protein was quantified using a BCA kit (Pierce;
Thermo Fisher Scientific, Inc.). Subsequently, proteins (30 µg)
were separated via 10% SDS-PAGE and transferred to nitrocellulose
membranes (EMD Millipore). Following blocking with 5% skim milk at
room temperature for 1.5 h, the membranes were incubated at 4°C
overnight with anti-CDK8 (1:1,000; cat. no. ab224828; Abcam) and
anti-GAPDH (1:2,000; cat. no. ab8245; Abcam) primary antibodies.
Following washing with PBS, the membranes were incubated with
anti-mouse or anti-rabbit horseradish peroxidase-conjugated
secondary antibodies (cat. nos. SC-2005 and SC-2004, respectively)
at room temperature for 1 h. Protein bands were visualized using
ECL Plus reagent (Beyotime Institute of Biotechnology). GAPDH was
used as the loading control. Protein expression levels were
quantified using Quantity One software (version 4.62; Bio-Rad
Laboratories, Inc.).
MTT assay
TT and MZ-CRC-1 cell viability was determined using
the MTT assay at 12, 24, 36 and 72 h post-transfection. Briefly,
MTC cells in the exponential growth phase were collected and seeded
(3×104 cells/well) into 96-well plates. Following
incubation at 37°C for 8 h, 20 µl MTT solution was added to each
well and incubated at 37°C for 4 h. Subsequently, 200 µl DMSO (cat.
no. D4540; Sigma-Aldrich; Merck KGaA) was added to each well at
room temperature for 15 min. The absorbance of each well was
determined at a wavelength of 490 nm using a microplate reader.
Colony formation assay
For the cell colony formation assay, at 24 h
post-transfection, TT and MZ-CRC-1 cells were seeded
(3×103 cells/dish) into 35 mm culture dishes containing
RPMI-1640 and cultured for two weeks at 37°C with 5% CO2
and 95% O2. The visible colonies were fixed in 4%
paraformaldehyde at room temperature for 10 min and stained with
10% Giemsa solution (cat. no. G4507; Sigma-Aldrich; Merck KGaA) at
room temperature for 10 min. Subsequently, the number of colonies
was manually counted using a light microscope (magnification,
×10).
Cell cycle analysis
TT and MZ-CRC-1 cells in the exponential growth
phase were collected and fixed using ethanol (75%) overnight at
4°C. Cells were washed with pre-cooled PBS to remove the excessive
ethanol and stained with 200 µl PI at 37°C for 30 min.
Subsequently, cells were stained with 100 µl RNaseA staining buffer
(BD Pharmingen) at room temperature for 20 min. Cell cycle was
detected by flow cytometry using a FACSCalibur flow cytometer (BD
Biosciences). The number of cells in G0/G1, S
and G2/M phases was calculated using ModFit software
(version 4.1; Verity Software House, Inc.).
Functional analysis
The functional roles of miR-592 target genes were
analyzed using the Database for Annotation, Visualization and the
Integrated Discovery (DAVID; version 6.8; david.ncifcrf.gov/home.jsp). In addition, integration
of the Gene Ontology (GO) (24,25)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) (26–28)
databases was performed.
Dual-luciferase reporter assay
The target genes of miR-592 were identified by
TargetScan (version 7.1; www.targetscan.org/vert_71) and the interaction
between miR-592 and CDK8 was verified using StarBase (version 2.0;
starbase.sysu.edu.cn). The interaction between miR-592 and CDK8 in
MTC cells was examined using a dual-luciferase reporter assay. The
wild-type (WT) and mutant (Mut) CDK8 3′-untranslated region (UTR)
containing miR-592 complementary sequences were cloned into the
pGL3 vector (Promega Corporation) to form CDK8-WT and CDK8-Mut
luciferase plasmids, respectively. TT and MZ-CRC-1 cells
(1×105 cells/well) were plated in a 6-well plate and
cultured for 8 h. Subsequently, cells were co-transfected with 100
ng CDK8-WT or 100 ng CDK8-Mut and 20 nM miR-592 mimic or 20 nM
scramble mimic using Lipofectamine® 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Following incubation at 37°C for 48 h, luciferase
activities were detected using the Dual-Luciferase Assay System
(Promega Corporation), according to the manufacturer's protocol.
Firefly luciferase activity was normalized to Renilla
luciferase activity.
Gene regulatory network
Based on previous studies (29–31),
the gene regulatory network of CDK8 was analyzed and identified
using GENEVESTIGATOR® (genevestigator.com/gv).
Statistical analysis
Data are presented as the mean ± SD. Statistical
analyses were performed using GraphPad Prism software (version 7;
GraphPad Software, Inc.). Differences between two groups were
analyzed using the paired (comparisons between paired and normal
tissue samples) or unpaired (other data) Student's t-test.
Differences among multiple groups were analyzed using one-way ANOVA
with Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-592 expression is increased during
MTC
To identify miRNAs that may contribute to the
tumorigenesis of MTC, differentially expressed miRNAs in the
GSE40807 dataset were analyzed using GEO2R. The relative expression
of miR-592 was significantly increased in MTC samples compared with
normal samples (P<0.05; Fig.
1A). To further investigate the expression of miR-592 during
MTC, miR-592 expression levels were examined in 20 paired MTC and
corresponding normal samples using RT-qPCR. miR-592 expression
levels were significantly increased in MTC samples compared with
normal samples (P<0.001; Fig.
1B). Moreover, miR-592 expression levels were significantly
increased in metastatic MTC samples compared with non-metastatic
samples (P<0.001; Fig. 1C). In
addition, the expression levels of miR-592 in MTC cell lines were
assessed. The results indicated that miR-592 expression was also
significantly increased in TT and MZ-CRC-1 cells compared with
NThy-ori 3.1 cells (P<0.001; Fig.
1D). Furthermore, the starBase analysis suggested that miR-592
expression levels were increased in MTC samples compared with
normal samples (Fig. 1E). The
starBase analysis results also indicated that overall survival was
not significantly different between patients with MTC with high and
low miR-592 expression (Fig. 1F).
However, the Kaplan-Meier survival curves demonstrated that
patients with high miR-592 expression exhibited a significantly
less favorable prognosis compared with patients with low miR-592
expression (P=0.034; Fig. 1G).
Collectively, the results indicated that miR-592 may serve a role
during MTC tumorigenesis.
miR-592 overexpression facilitates MTC
cell proliferation
To investigate the precise functions of miR-592
during MTC tumor development, TT and MZ-CRC-1 cells were
transfected with miR-592 mimic. Subsequently, cell proliferation,
viability and cell cycle distribution were assessed using MTT,
colony formation and flow cytometry assays. The miR-592 mimic group
exhibited significantly increased miR-592 expression levels
compared with the scramble mimic group in both TT and MZ-CRC-2
cells, which indicated that miR-592 mimic transfection was
successful (P<0.05; Fig. 2A and
B). miR-592 overexpression significantly increased TT and
MZ-CRC-1 cell viability compared with the scramble group
(P<0.05; Fig. 2C and D).
miR-592 overexpression also significantly increased the colony
formation rate compared with the scramble group in both TT and
MZ-CRC-1 cells (P<0.05; Fig. 2E and
F). Moreover, miR-592 overexpression significantly decreased
the number of cells in the G0/G1 phase and
significantly increased the number of cells in the S phase compared
with the scramble group in both TT and MZ-CRC-1 cells (P<0.05;
Fig. 2G and H). The results
suggested that miR-592 overexpression promoted MTC cell
proliferation.
Significantly enriched GO terms and
KEGG pathways of miR-592
The target genes of miR-592 were identified by
TargetScan and categorized into biological process (BP), cellular
component (CC) and molecular function (MF) GO categories via DAVID
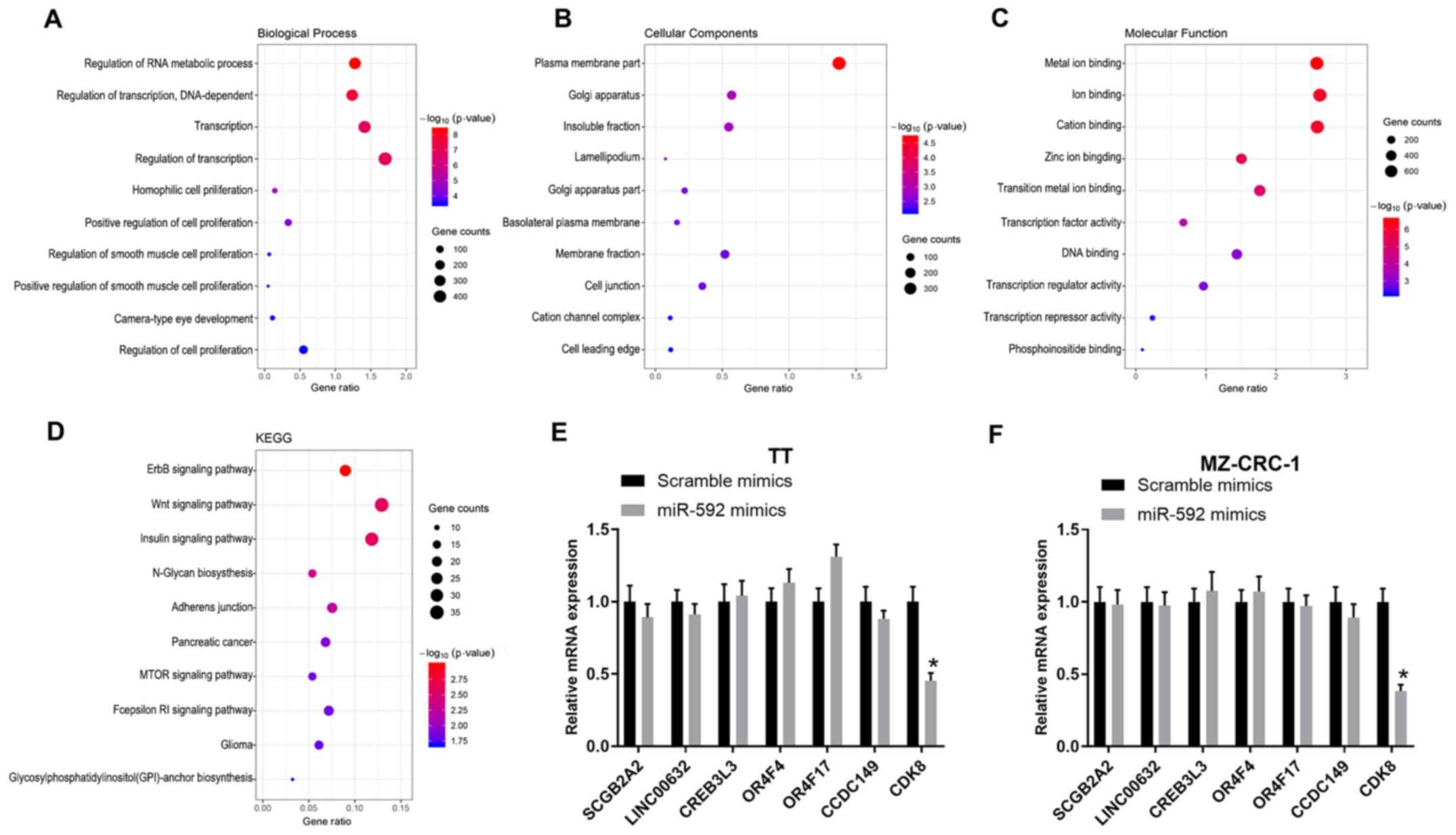
analysis (Fig. 3A-C). Moreover,
the target genes were functionally assessed by KEGG analysis, which
indicated that the target genes were associated with several
pathways, including the ‘Wnt signaling pathway’, ‘ErbB signaling
pathway’, ‘insulin signaling pathway’, ‘N-Glycan biosynthesis’ and
‘Adherens junction’ (Fig. 3D).
Moreover, the mRNA expression levels of multiple miR-592 target
genes in miR-592 mimic-transfected TT and MZ-CRC-1 cells were
assessed by RT-qPCR. miR-592 overexpression significantly decreased
CDK8 expression levels compared with the scramble mimic group in
both TT and MZ-CRC-1 cells (P<0.05; Fig. 3E and F); therefore, CDK8 was
selected for further analysis.
miR-592 binds to and negatively
regulates CDK8 in MTC cells
To determine whether CDK8 was regulated by miR-592
in MTC cells, the mRNA expression levels of CDK8 in miR-592
mimic-transfected TT and MZ-CRC-1 cells were measured by RT-qPCR.
Compared with the scramble mimic group, the expression levels of
CDK8 were significantly decreased in miR-592 mimic-transfected TT
and MZ-CRC-1 cells (Fig. 4A).
Moreover, it was predicted that CDK8 3′-UTR possessed an miR-592
binding site (Fig. 4B).
Subsequently, a dual-luciferase reporter assay was performed in TT
and MZ-CRC-1 cells to verify the interaction between miR-592 and
CDK8. The luciferase activities of TT and MZ-CRC-1 cells
co-transfected with miR-592 mimic and CDK8-WT were significantly
decreased compared with TT and MZ-CRC-1 cells co-transfected with
scramble mimic and CDK8-WT. The luciferase activities of TT and
MZ-CRC-1 cells co-transfected with miR-592 mimic and CDK8-Mut were
not significantly different compared with TT and MZ-CRC-1 cells
co-transfected with scramble mimic and CDK8-Mut (P<0.05;
Fig. 4C and D). In addition, CDK8
expression levels in 510 MTC samples and 58 normal samples obtained
from the starBase database were assessed. CDK8 expression was
downregulated in MTC samples compared with normal samples (Fig. 4E). There was no significant
difference in the survival rate of patients with MTC with high and
low CDK8 expression (Fig. 4F and
G). Furthermore, a negative correlation between the expression
levels of miR-592 and CDK8 in MTC was identified using starBase
(Fig. 4H).
CDK8 overexpression reverses
miR-592-mediated MTC cell proliferation
Subsequently, whether CDK8 was associated with
miR-592-mediated MTC cell proliferation was investigated by
co-transfecting TT and MZ-CRC-1 cells with miR-592 mimic and a CDK8
overexpression plasmid. Co-transfection of TT and MZ-CRC-1 cells
with miR-592 mimic and the CDK8 overexpression plasmid reversed
miR-592 mimic-mediated downregulation of CDK8 expression
(P<0.05; Fig. 5A-D). The
results of the colony formation assay indicated that miR-592 mimic
and CDK8 overexpression plasmid co-transfection significantly
decreased miR-592 mimic-induced colony formation of TT and MZ-CRC-1
cells (P<0.05; Fig. 5E and F).
miR-592 overexpression decreased the number of cells in the
G0/G1 phase and increased the number of cells
in the S phase in both TT and MZ-CRC-1 cells; however,
co-transfection of miR-592 mimic and CDK8 overexpression plasmid
reversed miR-592 overexpression-induced effects on cell cycle
distribution (P<0.05; Fig. 5G and
H). The results indicated that CDK8 overexpression reversed the
miR-592-mediated effects on MTC cell proliferation.
CDK8 interaction network and the
mechanism of miR-592/CDK8 axis in MTC
To understand the regulatory network underlying CDK8
during MTC, CDK8-associated genes were identified using
GENEVESTIGATOR and the top 30 CDK-8-associated genes are presented
in Fig. 6A. Collectively, the
results suggested that miR-592 overexpression promoted MTC
tumorigenesis by downregulating CDK8 expression (Fig. 6B).
Discussion
During the past few decades, the expression profiles
of miRNAs in different subtypes of thyroid cancer have been
analyzed (32). A large number of
mammalian miRNAs are upregulated in thyroid tumors, such as miR-650
(33), miR-340-5p (34), miR-424-5p (35) and miR-155 (36); while a number of miRNAs have been
reported to be downregulated, such as miR-26a (37), miR-215 (38), miR-34a (39) and miR-206 (40). In addition, a previous study
indicated that the expression levels of numerous miRNAs exhibit
significant differences in MTC (41).
At present, MTC accounts for <10% of all cases of
thyroid cancer worldwide, and only a small number of studies have
been conducted to examine the expression profiles and biological
functions of miRNAs during MTC tumorigenesis (42). The first MTC-associated microarray
analysis of miRNA expression was conducted by Nikiforova et
al (26) in 2008 using two MTC
specimens. Hereditary MTC (hMTC) and sporadic MTC (sMTC) are the
two subtypes of MTC that account for 25 and 75% of MTC cases,
respectively. A subsequent study demonstrated that the expression
levels of miR-183 and miR-375 were increased, while miR-9
expression levels were decreased in sMTC samples compared with hMTC
samples using a miRNA microarray analysis in 19 patients with MTC,
including 7 hMTC and 12 sMTC cases (43). Moreover, it was also demonstrated
that miR-183 and miR-375 upregulation were closely associated with
lateral lymph node metastases, representing two promising
biomarkers for MTC prognosis (43). Recently, a large miRNA microarray
profiling study was conducted to identify differentially expressed
miRNAs in MTC, and the results also validated that miR-375 and
miR-10a were upregulated, while miR-455 was downregulated in MTC
samples compared with normal samples (44). To further investigate the
regulatory network and functions of miRNAs during MTC, an miRNA
microarray was performed in the present study to analyze
differentially expressed miRNAs between MTC and normal samples in
the GSE40807 dataset, which was downloaded from the GEO database.
Among the differentially expressed miRNAs, miR-592 exhibited a high
fold-change; therefore, it was selected for subsequent functional
analysis. In the last few years, miR-592 has been reported to be
involved in the development of several different types of human
cancer (20,45). For example, high expression of
miR-592 was demonstrated to be associated with colorectal cancer
tumorigenesis and poor prognosis (46,47),
miR-592 exhibits an oncogenic effect on prostate cancer cells by
repressing Forkhead box O3A (48)
and miR-592 facilitates the proliferation, migration and invasion
of gastric cancer (49).
Therefore, it was speculated that miR-592 may serve as a
carcinogenic marker for different types of cancer. Nevertheless,
the role of miR-592 in MTC has not been previously reported. In the
present study, the RT-qPCR results indicated that miR-592
expression was upregulated in MTC tissue samples and cell lines
compared with normal tissue samples and cell lines, and miR-592
overexpression promoted TT and MZ-CRC-1 cell proliferation. To the
best of our knowledge, the present study was the first study to
suggest that miR-592 may serve an oncogenic role during MTC.
To further explore the mechanism underlying miR-592
in MTC, the target genes of miR-592 were predicted by
bioinformatics followed by functional analysis. CDK8 was identified
as a target gene of miR-592, which was negatively regulated by
miR-592. Numerous studies have demonstrated that CDK8 serves as a
critical oncogenic molecule in various types of human cancer,
including colorectal, breast and prostate cancer (50–52).
Although the majority of studies support the oncogenic role of
CDK8, a number of studies have indicated that CDK8 exerts
repressive functions in different types of cancer, including colon
(53), breast (54,55),
pancreatic (56) and non-small
cell lung (57) cancer. The
context-specific roles of CDK8 in distinct types of human cancer
have received increasing interest and there is considerable
controversy regarding the development of CDK8-based therapeutics.
In the present study, CDK8 overexpression abolished
miR-592-mediated effects on MTC cell proliferation, which implied
that CDK8 may serve as a tumor suppressor during MTC.
The present study indicated that miR-592 may serve
as an oncogene during MTC by decreasing CDK8 expression, providing
a novel therapeutic target for MTC treatment. However, the present
study had a number of limitations. The effects of miR-592
inhibitors on the functions of MTC, including cell apoptosis,
autophagy, migration, invasion and tumor growth in vivo
require further investigation. Additionally, the detailed
mechanisms underlying the miR-592/CDK8 axis also require further
investigation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TL, JM and YZ designed the study, performed the
bioinformatics analysis and the experiments. TL and YZ performed
the statistical analysis. YZ wrote the manuscript. TL and JM
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Affiliated Wuhan Central Hospital of Tongji
Medical College, Huazhong University of Science and Technology.
Written informed consent was obtained from each participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cabanillas ME, McFadden DG and Durante C:
Thyroid cancer. Lancet. 388:3316–2795. 2016. View Article : Google Scholar
|
|
2
|
Kushchayev SV, Kushchayeva YS, Tella SH,
Glushko T, Pacak K and Teytelboym OM: Medullary thyroid carcinoma:
An update on imaging. J Thyroid Res. 2019:18930472019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hao WJ, Zhang H, Yu Y, Zhao J, Ge ZJ, Ding
PX, Sun XX, Liu H, Wen SY and You J: Clinical significance and
cost-benefit analysis of serum calcitonin assay in diagnosis and
treatment of medullary thyroid carcinoma. Zhonghua Er Bi Yan Hou
Tou Jing Wai Ke Za Zhi. 54:506–509. 2019.(In Chinese). PubMed/NCBI
|
|
4
|
Kebebew E, Greenspan FS, Clark OH, Woeber
KA and Grunwell J: Extent of disease and practice patterns for
medullary thyroid cancer. J Am Coll Surg. 200:890–896. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roman S, Lin R and Sosa JA: Prognosis of
medullary thyroid carcinoma: Demographic, clinical, and pathologic
predictors of survival in 1252 cases. Cancer. 107:2134–2142. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kuo EJ, Sho S, Li N, Zanocco KA, Yeh MW
and Livhits MJ: Risk factors associated with reoperation and
disease-specific mortality in patients with medullary thyroid
carcinoma. JAMA Surg. 153:52–59. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ceolin L, Duval M, Benini AF, Ferreira CV
and Maia AL: Medullary thyroid carcinoma beyond surgery: Advances,
challenges, and perspectives. Endocr Relat Cancer. 26:R499–R518.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meijer JA, le Cessie S, van den Hout WB,
Kievit J, Schoones JW, Romijn JA and Smit JWA: Calcitonin and
carcinoembryonic antigen doubling times as prognostic factors in
medullary thyroid carcinoma: A structured meta-analysis. Clin
Endocrinol (Oxf). 72:534–542. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Y, Zhong Q, Chen X, Fang J and Huang
Z: Diagnostic value of microRNAs in discriminating malignant
thyroid nodules from benign ones on fine-needle aspiration samples.
Tumour Biol. 35:9343–9353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nishino M: Molecular cytopathology for
thyroid nodules: A review of methodology and test performance.
Cancer Cytopathol. 124:14–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mohr AM and Mott JL: Overview of microRNA
biology. Semin Liver Dis. 35:3–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu X, Jiao Z, Chen H and Wang L: A
correlational study on miR-34s and cervical lesions. Eur J Gynaecol
Oncol. 39:786–789. 2018.
|
|
13
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian T, Wang J and Zhou X: A review:
microRNA detection methods. Org Biomol Chem. 13:2226–2238. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Catalanotto C, Cogoni C and Zardo G:
microRNA in control of gene expression: An overview of nuclear
functions. Int J Mol Sci. 17:17122016. View Article : Google Scholar
|
|
16
|
Yeh M, Oh CS, Yoo JY, Kaur B and Lee TJ:
Pivotal role of microRNA-138 in human cancers. Am J Cancer Res.
9:1118–1126. 2019.PubMed/NCBI
|
|
17
|
Humphries B, Wang Z and Yang C: microRNA
regulation of epigenetic modifiers in breast cancer. Cancers
(Basel). 11:8972019. View Article : Google Scholar
|
|
18
|
Chu YH, Hardin H, Schneider DF, Chen H and
Lloyd RV: microRNA-21 and long non-coding RNA MALAT1 are
overexpressed markers in medullary thyroid carcinoma. Exp Mol
Pathol. 103:229–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou W, Zhang H, Bai X, Liu X, Yu Y, Song L
and Du Y: Suppressive role of miR-592 in breast cancer by
repressing TGF-β2. Oncol Rep. 38:3447–3454. 2017.PubMed/NCBI
|
|
20
|
Slattery ML, Mullany LE, Sakoda LC, Wolff
RK, Samowitz WS and Herrick JS: Dysregulated genes and miRNAs in
the apoptosis pathway in colorectal cancer patients. Apoptosis.
23:237–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu Y, Li K, Wang SB and Yang SG: Mir-592
functions as a tumor suppressor in acute myeloid leukemia by
targeting ROCK1 and predicts patients' prognosis. Eur Rev Med
Pharmacol Sci. 23:1610–1619. 2019.PubMed/NCBI
|
|
22
|
Gao S, Chen J, Wang Y, Zhong Y, Dai Q,
Wang Q and Tu J: MiR-592 suppresses the development of glioma by
regulating rho-associated protein kinase. Neuroreport.
29:1391–1399. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Clough E and Barrett T: The gene
expression omnibus database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
The Gene Ontology Consortium, . The gene
ontology resource: 20 years and still going strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nikiforova M N, Tseng G C, Steward D, et
al: MicroRNA Expression Profiling of Thyroid Tumors: Biological
Significance and Diagnostic Utility. Journal of Clinical
Endocrinology & Metabolism. 93:1600–1608. 2008. View Article : Google Scholar
|
|
27
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanehisa M: Toward understanding the
origin and evolution of cellular organisms. Protein Sci.
28:1947–1951. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Besso MJ, Rosso M, Lapyckyj L, Moiola CP,
Matos ML, Mercogliano MF, Schillaci R, Reventos J, Colas E,
Gil-Moreno A, et al: FXYD5/dysadherin, a biomarker of endometrial
cancer myometrial invasion and aggressiveness: Its relationship
with TGF-β1 and NF-κB pathways. Front Oncol. 9:13062019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cari L, Nocentini G, Migliorati G and
Riccardi C: Potential effect of tumor-specific treg-targeted
antibodies in the treatment of human cancers: A bioinformatics
analysis. Oncoimmunology. 7:e13877052018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li C, Zhou D, Jiang X, Liu M, Tang H and
Mei Z: Identifying hepatocellular carcinoma-related hub genes by
bioinformatics analysis and CYP2C8 is a potential prognostic
biomarker. Gene. 698:9–18. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu G, Xie L and Miller D: Expression of
microRNAs in thyroid carcinoma. Methods Mol Biol. 1617:261–280.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Orlandella FM, Mariniello RM, Iervolino
PLC, Imperlini E, Mandola A, Verde A, De Stefano AE, Pane K,
Franzese M, Esposito S, et al: miR-650 promotes motility of
anaplastic thyroid cancer cells by targeting PPP2CA. Endocrine.
65:582–594. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao P, Ma W, Hu Z, Zhang Y, Zhang S and
Wang Y: Up-regulation of miR-340-5p promotes progression of thyroid
cancer by inhibiting BMP4. J Endocrinol Invest. 41:1165–1172. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu X, Fu Y, Zhang G, Zhang D, Liang N, Li
F, Li C, Sui C, Jiang J, Lu H, et al: Mir-424-5p promotes anoikis
resistance and lung metastasis by inactivating hippo signaling in
thyroid cancer. Mol Ther Oncolytics. 15:248–260. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang W, Ji W and Zhao X: Mir-155 promotes
anaplastic thyroid cancer progression by directly targeting socs1.
BMC Cancer. 19:10932019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gong Y, Wu W, Zou X, Liu F, Wei T and Zhu
J: Mir-26a inhibits thyroid cancer cell proliferation by targeting
arpp19. Am J Cancer Res. 8:1030–1039. 2018.PubMed/NCBI
|
|
38
|
Han J, Zhang M, Nie C, Jia J, Wang F, Yu
J, Bi W, Liu B, Sheng R, He G, et al: Mir-215 suppresses papillary
thyroid cancer proliferation, migration, and invasion through the
AKT/GSK-3β/Snail signaling by targeting ARFGEF1. Cell Death Dis.
10:1952019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu H, Deng H, Zhao Y, Li C and Liang Y:
Lncrna xist/miR-34a axis modulates the cell proliferation and tumor
growth of thyroid cancer through met-Pi3K-Akt signaling. J Exp Clin
Cancer Res. 37:2792018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang P, Gu J, Wang K, Shang J and Wang W:
miR-206 inhibits thyroid cancer proliferation and invasion by
targeting RAP1B. J Cell Biochem. 120:18927–18936. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Saiselet M, Pita JM, Augenlicht A, Dom G,
Tarabichi M, Fimereli D, Dumont JE, Detours V and Maenhaut C: miRNA
expression and function in thyroid carcinomas: A comparative and
critical analysis and a model for other cancers. Oncotarget.
7:52475–52492. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chu YH and Lloyd RV: Medullary thyroid
carcinoma: Recent advances including microRNA expression. Endocr
Pathol. 27:312–324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Abraham D, Jackson N, Gundara JS, Zhao JT,
Gill AJ, Delbridge L, Robinson BG and Sidhu SB: microRNA profiling
of sporadic and hereditary medullary thyroid cancer identifies
predictors of nodal metastasis, prognosis, and potential
therapeutic targets. Clin Cancer Res. 17:4772–4781. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hudson J, Duncavage E, Tamburrino A,
Salerno P, Xi L, Raffeld M, Moley J and Chernock RD: Overexpression
of miR-10a and miR-375 and downregulation of YAP1 in medullary
thyroid carcinoma. Exp Mol Pathol. 95:62–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yerukala Sathipati S and Ho SY:
Identifying a miRNA signature for predicting the stage of breast
cancer. Sci Rep. 8:161382018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu M, Zhi Q, Wang W, Zhang Q, Fang T and
Ma Q: Up-regulation of miR-592 correlates with tumor progression
and poor prognosis in patients with colorectal cancer. Biomed
Pharmacother. 69:214–220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fu Q, Du Y, Yang C, Zhang D, Zhang N, Liu
X, Cho WC and Yang Y: An oncogenic role of miR-592 in tumorigenesis
of human colorectal cancer by targeting forkhead box o3a (FoxO3A).
Expert Opin Ther Targets. 20:771–782. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lv Z, Rao P and Li W: MiR-592 represses
FOXO3 expression and promotes the proliferation of prostate cancer
cells. Int J Clin Exp Med. 8:15246–15257. 2015.PubMed/NCBI
|
|
49
|
He Y, Ge Y, Jiang M, Zhou J, Luo D, Fan H,
Shi L, Lin L and Yang L: MiR-592 promotes gastric cancer
proliferation, migration, and invasion through the Pi3K/Akt and
MAPK/ERK signaling pathways by targeting spry2. Cell Physiol
Biochem. 47:1465–1481. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bragelmann J, Klümper N, Offermann A, von
Mässenhausen A, Böhm D, Deng M, Queisser A, Sanders C, Syring I,
Merseburger AS, et al: Pan-cancer analysis of the mediator complex
transcriptome identifies CDK19 and CDK8 as therapeutic targets in
advanced prostate cancer. Clin Cancer Res. 23:1829–1840. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
McDermott MS, Chumanevich AA, Lim CU,
Liang J, Chen M, Altilia S, Oliver D, Rae JM, Shtutman M, Kiaris H,
et al: Inhibition of CDK8 mediator kinase suppresses estrogen
dependent transcription and the growth of estrogen receptor
positive breast cancer. Oncotarget. 8:12558–12575. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liang J, Chen M, Broude EV and Roninson
IB: Role of transcription-regulating kinase CDK8 in colon cancer
metastasis. Oncotarget. 10:622–623. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Philip S, Kumarasiri M, Teo T, Yu M and
Wang S: Cyclin-dependent kinase 8: A new hope in targeted cancer
therapy? J Med Chem. 61:5073–5092. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Crown J: CDK8: A new breast cancer target.
Oncotarget. 8:14269–14270. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Song W, Wu S, Wu Q, Zhou L, Yu L, Zhu B
and Gong X: The microRNA-141-3p/CDK8 pathway regulates the
chemosensitivity of breast cancer cells to trastuzumab. J Cell
Biochem. 120:14095–14106. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wei R, Kong L, Xiao Y, Yuan H, Song Y,
Wang J, Yu H, Mao S and Xu W: CDK8 regulates the angiogenesis of
pancreatic cancer cells in part via the CDK8-β-catenin-klf2 signal
axis. Exp Cell Res. 369:304–315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Xing S, Xu Q, Fan X, Wu S and Tian F:
Downregulation of miR-138-5p promotes non-small cell lung cancer
progression by regulating CDK8. Mol Med Rep. 20:5272–5278.
2019.PubMed/NCBI
|