Introduction
Previous epidemiological studies have shown that air
pollution is associated with increased morbidity and mortality
worldwide (1), and the total
global mortality caused by PM2.5 is ~7.6% (2). Outdoor air pollution has been
extensively studied, while little work has been performed to
evaluate the potential risk of exposure to the indoor environment
with regards to human health. Studies have indicated that
individuals spend ~90% of the time indoors (3). Therefore, it is important to evaluate
the potential effects of indoor air pollution exposure on human
health.
Indoor emission sources generate a variety of air
pollutants. For instance, indoor cooking can generate airborne
particulate matter (PM), carbon dioxide (CO2), black
carbon, polycyclic aromatic hydrocarbons (PAH), formaldehyde and
nitrogen oxide, all of which have the potential to cause
respiratory diseases (4–6). Moreover, cooking oil fumes enhance
the generation of secondary fine particulate matter
(PM2.5), which contain various organic compounds and
hazardous chemicals, including metals, PAHs, carbonyl compositions,
benzene and quinines, and PM and PM2.5 are present at
higher concentrations and durations indoors compared with outdoor
environmental fine particulate matter (7).
Currently, printers are widely and frequently used,
and the printing room has become a source of PM2.5
emission and represents a potential health hazard (8). Previous studies have shown that
toners used in printers contain metal carbon nanotubes made of iron
oxide, titanium dioxide and other metal oxides, which are released
into the air along with printer emitted particles (PEPs) (9). PEPs are potentially hazardous to
individuals who spend a lot of time in the printing room (10). As early as 1979, studies have been
performed, which indicated that long-term use of wet toner causes
eye itching and skin redness in post office staff (11). Moreover, serious diseases like
pneumoconiosis (12), thrombus
(13) and peritoneal deposition of
carbon nanoparticles (14) may
occur in individuals subjected to long-term particulate
exposure.
PM2.5, which are atmospheric PM with a
diameter <2.5 µm, are composed of biological components,
particles, PAHs, organic carbon, elemental carbon, inorganic
water-soluble ions and heavy metals (15). In recent years, due to its adverse
effects on the climate, environment and health, PM2.5
has attracted attention and extensive research examining its
effects have been performed (16).
Research has indicated that the PM2.5 is harmful to the
respiratory system, since it accumulates in lung tissue and
stimulates alveoli (17). In
addition, toxic substances in PM2.5 can harm the human
respiratory system and immune response, which both cause health
problems (17). The lung is the
main organ of the respiratory system, which is sensitive to the
harmful effects of PM2.5. Toxicological evidence has
indicated that PM2.5 exposure induced lung injury,
alveoli collapse and inflammatory response (18). Previous work in our lab revealed
differences between chemical components associated with indoor
PM2.5 derived from printer functioning and outdoor
PM2.5, and chemical components, such as water-soluble
particles, were increased indoors compared with the outdoors
(19,20). The main emission characteristics of
PM2.5 produced throughout the printing process are as
follows: i) The primary emission rate of PM2.5 under
working printer conditions is 1.70-2.62 mg/h; ii) working
conditions of some printers result in direct discharge of fine and
ultrafine particles (particle size is mainly distributed between 10
nm-1 µm) and iii) in addition to releasing primary particles
throughout printer functioning, volatile organic compounds are
released, which generate secondary organic aerosols in the printing
room (data not yet published). Therefore, to understand the effects
of exposure to PM2.5 collected from printing rooms,
pulmonary injury both in vitro and in vivo were
examined using molecular biology, histology, proteomics and
high-throughput sequencing techniques.
Materials and methods
Collection of PM2.5 from a
printing room
PM2.5 samples were collected from a
printing room in the School of Resources Environmental &
Chemical Engineering at Nanchang University (Nanchang, China).
Middle-volume samplers using a flow rate of 100 l/min (Wuhan
Tianhong Environmental Protection Industry Co., Ltd.; http://www.thyb.cn/index.html.) were used.
PM2.5 collected on a glass fiber filter (90 mm; Shanghai
Lanhua Scientific Instrument Co., Ltd.; http://www.instrument.com.cn/netshow/SH103948/)
were diluted with double distilled water, ultrasonically shaken,
filtered using 6–8 layers of gauze to remove debris and stored at
−80°C (Thermo Fisher Scientific, Inc.) for 48 h. Finally, a vacuum
freeze dryer was used to produce dry, powdered particles.
Cell types and culture conditions
Human bronchial epithelial (HBE) cells and human
umbilical vein endothelial cells (HUVECs) were provided by The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
HUVECs were cultured in DMEM (HyClone; Cytiva) supplemented with
10% FBS (HyClone; Cytiva). HBE cells were cultured in RPMI-1640
medium (Genaxxon Bioscience GmbH) supplemented with 10% FBS. Both
cell types were cultured in an incubator supplied with 5%
CO2 at 37°C. Four groups of cells were cultured to
compare and study the toxic effects of different concentrations of
PM2.5. These four groups were: i) Cells cultured with
100 µl PBS (C group); ii) cells cultured with 5 µg/ml
PM2.5 suspension (PRL group); iii) cells cultured with
10 µg/ml PM2.5 suspension (PRM group) and iv) cells
cultured with 15 µg/ml PM2.5 suspension (PRH group).
Cell treatment
When cells reached 70–80% confluence, different
concentrations of PM2.5 were co-cultured in HUVECs and
HBE cells. Subsequently, commercially available Cell Counting Kit-8
(cat. no. G021-1-1), superoxide dismutase (SOD; cat. no. A001-3-2)
and malondialdehyde (MDA; cat. no. A003-1-2) kits (all purchased
from Nanjing Jiancheng Bioengineering Institute and were used
according to the manufacturer's instructions) were used to evaluate
the effects of PM2.5 on cell viability, intracellular
SOD activity and MDA content.
To study the effects of the three different
concentrations of PM2.5 on cytokine production, an ELISA
kit (all purchased from Beijing 4A Biotech Co., Ltd.) was used to
measure levels of interleukin IL-6 (cat. no. CME0006; sensitivity
range: 7.8–500 pg/ml; concentrations used to create a calibration
curve: 500, 250, 125, 62.5, 31.2, 15.6, 7.8 and 0 pg/ml), IL-2
(cat. no. CME0001; sensitivity range: 15.6-1,000 pg/ml;
concentrations used to create a calibration curve: 1,000, 500, 250,
125, 62.5, 31.2, 15.6 and 0 pg/ml), tumor necrosis factor-α (TNF-α;
cat. no. CME0004; sensitivity range: 7.8–500 pg/ml; concentrations
used to create a calibration curve: 500, 250, 125, 62.5, 31.2,
15.6, 7.8 and 0 pg/ml) and IL-1β (cat. no. CME0015; sensitivity
range: 31.25-2,000 pg/ml; concentrations used to create a
calibration curve: 2000, 1000, 500, 250, 125, 62.5, 31.2 and 0
pg/ml) in accordance with the manufacturer's instructions.
Western blot analysis
Samples were lysed with RIPA buffer (Beijing
Solarbio Science & Technology Co., Ltd.) and protease
inhibitors, and the supernatant of each lysate was obtained via
centrifugation at 12,000 × g for 10 min at 4°C after fragmented on
ice. The protein concentration was determined with the BCA kit
(cat. no. 23225; Thermo Fisher Scientific, Inc.). Proteins were
separated via 10% SDS-PAGE, transferred to nitrocellulose membranes
(the mass of protein loaded per lane was 30 µg), and blocked using
8% BSA (Beijing Solarbio Science & Technology Co., Ltd.) in
Tris-buffered saline-Tween-20 (1% TBS-T) for 1.5 h at room
temperature. Subsequently, membranes and primary antibodies were
incubated overnight at 4°C, washed with TBS-T and incubated with
appropriate quantities of horseradish peroxidase-conjugated
secondary antibodies (1:2,000; cat. no. 7076; Cell Signaling
Technology, Inc.) for another 1.5 h at room temperature.
Subsequently, DAB (cat. no. 34577; Thermo Fisher Scientific, Inc.)
was then used to detect the proteins and the band density was
determined using ImageJ (version 2.1.0; National Institutes of
Health) software. The expression levels were normalized against the
housekeeping gene of actin. Antibodies used included β-actin
(1:10,000; cat. no. 60008-1-lg; ProteinTech Group, Inc.), Bax
(1:2,500; cat. no. 50599-2-lg; ProteinTech Group, Inc.), Bcl-2
(1:1,000; cat. no. 12789-1-AP; ProteinTech Group, Inc.),
transforming growth factor-β1 (TGF-β1; 1:2,000; cat. no.
21898-4-AP; ProteinTech Group, Inc.), ERK (1:2,000; cat. no.
16443-1-AP; ProteinTech Group, Inc.), phosphorylated (p)-ERK
(1:1,000; cat. no. AF1015; Affinity Biosciences), cyclooxygenase-2
(COX2; 1:1,000; cat. no. 12375-1-AP; ProteinTech Group, Inc.),
NF-κB (p65; 1:2,000; cat. no. 10745-1-AP; ProteinTech Group, Inc.)
and p-NF-κB (p-p65; 1:1,000; cat. no. bs0982R; BIOSS).
Animals and treatments
In total, 48 male C57BL/6 mice (age, 8 weeks;
weight, 25–30 g) were purchased from Hunan SJA Laboratory Animal
Co., Ltd. (http://hnslk.mfqyw.com/) and housed
in cages in the Institute of Translational Medicine of Nanchang
University using standard conditions (humidity 51±13%, temperature
23±3°C, 12/12-h light-dark cycle). Each mouse was distinguished via
ear piercing (a loophole in the front, middle and rear of the
margin of the left ear represented numbers 1, 2 and 3,
respectively, and a loophole in the front, middle and rear of the
margin of the right ear represented numbers 4, 5 and 6,
respectively). Subsequently, mice were randomly divided into four
groups as follows: The first group, mice treated with PBS (C group,
n=12); The second group, mice given a 5 µg/g PM2.5
suspension on days 1 and 3 via tracheal perfusion (PRL group,
n=12); The third group, mice given a 10 µg/g PM2.5
suspension on days 1 and 3 via tracheal perfusion (PRM group, n=12)
and the fourth group, mice given a 15 µg/g PM2.5
suspension on days 1 and 3 via tracheal perfusion (PRH group,
n=12). On day 4, 100 µl serum was collected via tail vein blood
collection after mice were anesthetized with 1% pentobarbital
sodium solution (40 mg/kg), then mice were sacrificed and the lung
tissue were collected and stored at −80°C for use in subsequent
experiments. The concentrations of PM2.5 used in the
present study were determined via referencing previous studies
(21,22) and the present pre-experimental
results (date not shown). The present study was approved by the
Ethical Committee of the Second Affiliated Hospital of Nanchang
University, and all experiments were performed in accordance with
approved guidelines (The Guide for Care and Use of Laboratory
Animals; National Institutes of Health publication no. 85–23)
(23).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from lung tissue using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), and RNA purity was determined using a NanoDrop™ 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). cDNA was
reversed transcribed from RNA (cat. no. 4374967; Applied
Biosystems; Thermo Fisher Scientific, Inc.) and qPCR was performed
using a 7900HT Fast Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and 2X SYBRGreen master mix
(Bio-Rad Laboratories, Inc.). Samples with appropriate primers were
incubated at 95°C for 30 sec, and 40 cycles of 60°C for 30 sec, and
72°C for 30 sec were performed after a 1 min incubation step at
95°C to activate the reaction. Expression levels of IL-6 (forward
primer 5′-GAAATCGTGGAAvATGAG-3′, reverse primer
5′-GCTTAGGCATAACGCACT-3′), TNF-α (forward primer
5′-GTGGAACTGGCAGAAGAGGCA-3′, reverse primer
5′-AGAGGGAGGCCATTTGGGAAC-3′), IL-1β (forward primer
5′-GTGTCTTTCCCGTGGACCTTC-3′, reverse primer
5′-TCATCTCGGAGCCTGTAGTGC-3′), IL-2 (forward primer
5′-GCCAAGAGCTGACCAACTTC-3′, reverse primer
5′-ATCGCCCACACTAAGAGCAT-3′) and GAPDH (forward primer
5′-CTCGTGGAGTCTACTGGTGT-3′, reverse primer
5′-GTCATCATACTTGGCAGGTT-3′) were analyzed using the
2−ΔΔCq method (24).
Hematoxylin and eosin (HE) staining
and immunohistochemistry (IHC)
Samples containing tissues from each group were
fixed in 10% buffered formalin for 24 h at 4°C, embedded into
paraffin and sliced into 4–5 µm-thick sections, The wax sheets were
smoothed on polylysine-treated slides in the spreading machine tank
(45°C). The surrounding water was drained with absorbent paper and
tissues were place in an oven at 60°C for 4 h. Subsequently,
H&E staining was performed (hematoxylin staining for 5–10 min;
0.6% ammonia reflux blue; eosin staining for 3–5 min; treatment
with 70% ethanol and 80% ethanol for 10–20 sec, 95% ethanol for 3–5
min, 100% ethanol for 3–5 min, and transparent xylene for 3–5 min.
All the operations were performed at room temperature) and tissues
were observed using a light microscope (magnification, ×200;
Olympus Corporation) to compare the pathological features of
samples. IHC was performed using caspase-3 (1:250; cat. no.
19677-1-AP; ProteinTech Group, Inc.), caspase-8 (1:250; cat. no.
13426-1-P; ProteinTech Group, Inc.), Bax (1:2,500; cat. no.
50599-2-lg; ProteinTech Group, Inc.) and Bcl-2 (1:1,000; cat. no.
12789-1-AP; ProteinTech Group, Inc.) antibodies (incubated at 4°C
overnight; reheated at room temperature for 45 min), and secondary
antibody (1:1,000; Servicebio, Inc.; cat. no. GB24303; 37°C, 1 h)
antibodies.
High-throughput sequencing
analyses
Lung tissues from groups C (n=6), PRL (n=6), PRM
(n=6) and PRH (n=6) were collected, and total genomic DNA was
extracted from samples using a genomic DNA extraction kit (cat. no.
69506; Qiagen GmbH) and a bead beating method. Subsequently, 515F
(5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACVSGGGTATCTAAT-3′)
primers were used to amplify the V4 region of the 16S ribosomal DNA
gene for high-throughput sequencing, and PCR products were
sequenced using an IlluminaHiSeq 2000 platform (Illumina, Inc.;
sequencing read archive (SRA) accession no. SRP217605]. The
extraction quality of DNA was determined using 0.8% agar-gel
electrophoresis, and the DNA was quantified using
Uv-spectrophotometer (Thermo Fisher Scientific, Inc.). The Quant-iT
PicoGreen dsDNA Assay kit (cat. no. P11496; Thermo Fisher
Scientific, Inc.) was used to quantify DNA in the Promega
QuantiFluor fluorescence quantitative system. The cut adapt and
UCHIME algorithm, from the UPARSE software package (UPARSE;
http://drive5.com/uparse/ and Supplementary
Software; version 7.0.100), Qiime software (Quantitative insights
Into Microbial Ecology; versions 1.8.0 and 1.9.1; http://qiime.org/index.html; used to compare the
richness and diversity indexes, such as Shannon index and Simpson
index), and SIMCA-P software (Umetrics; version 11.5) were used to
evaluate α (intra-sample) and β (inter-sample) diversity.
The Kyoto Encyclopedia of Genes and Genomes (KEGG)
database (http://www.genome.jp/kegg/pathway.html) was used to
predict the correlation between microbiota changes with
environmental adaptation, immune responses and energy metabolism
(25). The Operational taxonomic
units (OTUs) and species classification were analyzed based on
effective data (excluded errors and questionable data), and then
the abundance and diversity indexes of OTUs were analyzed.
Principal co-ordinates analysis (PCoA) was used to reflect the
degree of similarity between community samples. KEGG was also used
to assess xenobiotic biodegradation, endocrine system and
infectious diseases. The high-throughput sequencing experiment was
performed by Shanghai Personal Biotechnology Co., Ltd.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software version 7.0 (GraphPad Software, Inc.). All data were
presented as the mean ± SD. Statistical significance was assessed
via one-way ANOVA and Tukey's multiple comparison tests. P<0.05
was considered to indicate a statistically significant
difference.
Results
Effects of different concentrations of
PM2.5 on cell viability, oxidative stress and cellular
cytokines
To assess the effects of PM2.5 exposure
on HBE and HUVEC cells, PM2.5 and cells were
co-cultured. Cell proliferation experiments revealed that
PM2.5 exposure significantly inhibited growth of both
HBE (survival rates were 92.34, 50.56 and 24.67%, respectively)
cells and HUVECs (survival rates were 94.34, 64.54 and 32.12%,
respectively) in PRL, PRM and PRH groups in a dose-dependent manner
(Fig. 1A). PM2.5
treatment in PRL, PRM and PRH groups also significantly enhanced
MDA activity (63.7, 175.21, and 420.99 U/mg protein, respectively),
and significantly reduced SOD activity (69.81, 45.72, and 27.36
U/mg protein, respectively) in HBS cells and HUVECs (Fig. 1B), in a dose-dependent manner. The
effect of PM2.5 on expression of cellular cytokines was
then assessed. It was found that PM2.5 exposure in PRL,
PRM and PRH groups significantly increased the accumulation of
pro-inflammatory factors including IL-1β (111.63, 144.15 and 182.11
pg/ml, respectively), TNF-α (181.5, 200.63 and 231.79 pg/ml,
respectively) and IL-6 (57.53, 75.21 and 86.15 pg/ml,
respectively), and significantly reduced accumulation of the
anti-inflammatory factor, IL-2 (122.76, 84.87 and 46.63 pg/ml,
respectively) in both HBE and HUVEC cells (Fig. 1C and D), in a dose-dependent
manner.
 | Figure 1.Oxidative damage resulting from
printing room-derived PM2.5 exposure in human HBE and
HUVEC cells. (A) Determination of HBE and HUVEC cell viability. (B)
MDA and SOD activity in HBE and HUVEC cells. Levels of IL-1β, IL-6,
TNF-α and IL-2 expression in (C) HBE cells and (D) HUVECs. Data are
presented as the mean ± standard deviation. n=12/group. *P<0.05
and **P<0.01. PM2.5, fine particulate matter; C,
cells treated with PBS; PRL, cells treated with 5 µg/ml
PM2.5; PRM, cells treated with 10 µg/ml
PM2.5; PRH, cells treated with 15 µg/ml
PM2.5; HBE, human bronchial epithelial cells; HUVEC,
human umbilical vein endothelial cells; MDA, malondialdehyde; SOD,
superoxide dismutase; IL, interleukin; TNF, tumor necrosis factor;
OD, optical density. |
Effects of exposure to different
concentrations of PM2.5 on inflammatory response and
apoptosis in vitro
The expression levels of key proteins involved in
inflammatory and apoptosis pathways were studied using western
blotting. The results indicated that exposure to high levels of
PM2.5 significantly increased the accumulation of
inflammatory proteins COX2 (HUVECs, 0.99 vs. 0.61; HBE cells, 1.07
vs. 0.45 for PRH and group C, respectively) and p-p65/p65 (HUVECs,
1.21 vs. 0.67; HBE cells, 1.08 vs. 0.60 for PRH and group C,
respectively) compared with controls (Fig. 2D and A). Overexpression of key
proteins in inflammatory pathways significantly increased the
expression of apoptotic factors Bax/Bcl-2 (HUVECs, 1.87 vs. 0.58;
HBE cells, 2.05 vs. 0.87 for PRH and group C, respectively)
(Fig. 2F and C) and TGF-β1
(HUVECs, 0.93 vs. 0.43; HBE cells, 1.03 vs. 0.63 for PRH and group
C, respectively) (Fig. 2E and
B).
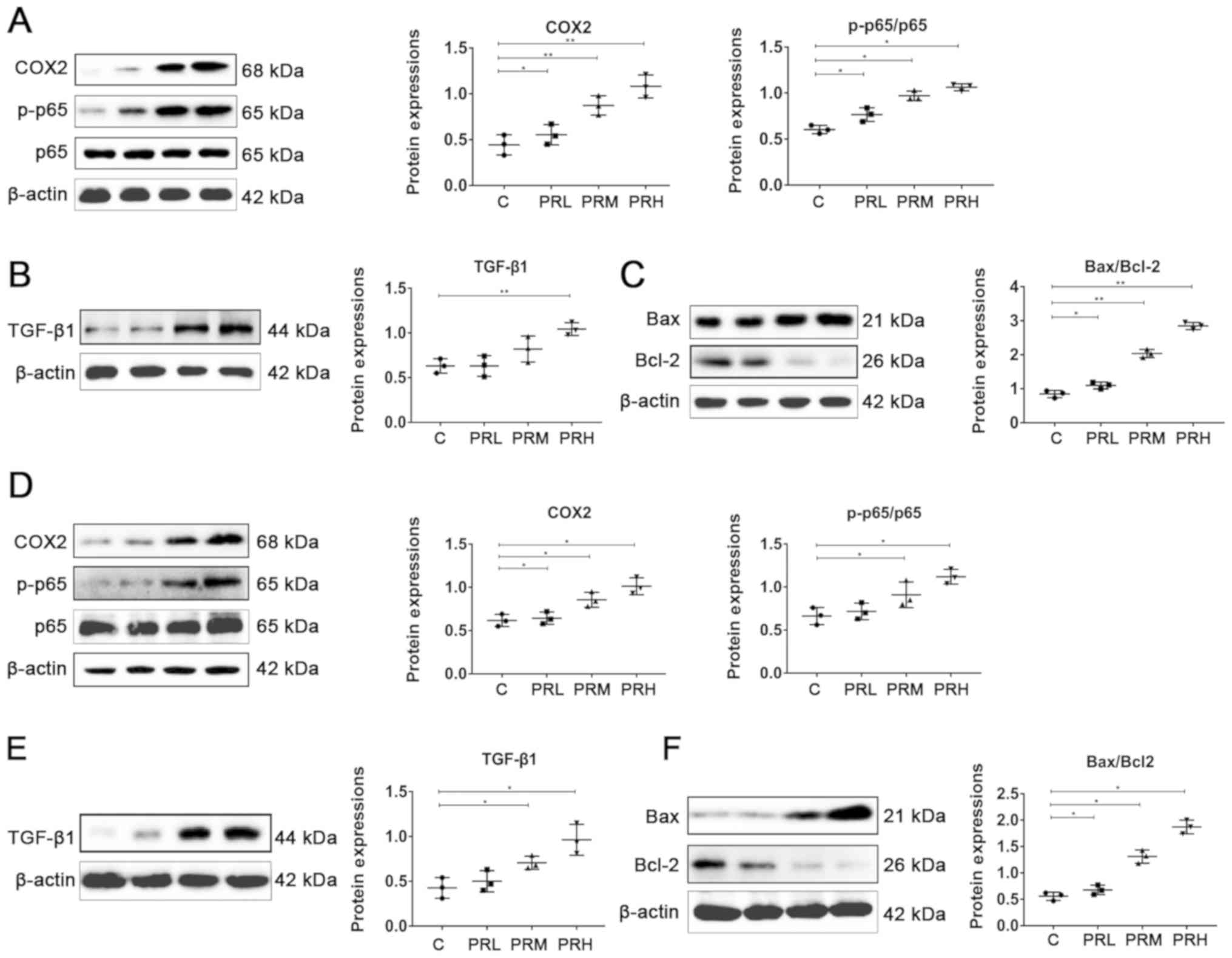 | Figure 2.Effects of printing room-derived
PM2.5 exposure on protein expression in human HBE and
HUVEC cells. Levels of (A) COX2, p-p65 and p65, (B) TGF-β1 and (C)
Bcl-2 and Bax protein in HBE cells. Levels of (D) COX2, p-p65, p65,
(E) TGF-β1 and (F) Bcl-2 and Bax protein in HUVECs. Data are
presented as the mean ± standard deviation. n=12/group. *P<0.05
and **P<0.01. PM2.5, fine particulate matter; C,
cells treated with PBS; PRL, cells treated with 5 µg/ml
PM2.5; PRM, cells treated with 10 µg/ml
PM2.5; PRH, cells treated with 15 µg/ml
PM2.5; HBE, human bronchial epithelial cells; HUVEC,
human umbilical vein endothelial cells; COX2, cyclooxygenase 2; p,
phosphorylated; TGF, transforming growth factor. |
Effects of different printing
room-derived PM2.5 concentrations on lung injury in
mice
To further evaluate whether harmful effects of
PM2.5 occur in lung tissues, a mouse model was
established and the H&E staining was used to assess the
histopathological effects of lung tissue exposure. As shown in
Fig. 3A, HE staining results
indicated that there was little infiltration by eosinophil-based
inflammatory cells within lung tissue in the PRL group. However,
eosinophil-based inflammatory cells infiltrated PRM, and especially
PRH groups, and airway epithelial cells displayed shedding and
necrosis, increased mucosal folds, mucous glands hyperplasia and
mucous wall thickening.
 | Figure 3.Inflammatory damage in lung tissues
occurs post-PM2.5 exposure in male C57BL/6 mice. (A)
Hematoxylin and eosin staining images (magnification, ×200). (B)
MDA and (C) SOD activity. (D) Levels of IL-1β, IL-6, TNF-α and
IL-2. (E) Gene expression of IL-1β, IL-6, TNF-α and IL-2. Data are
presented as the mean ± standard deviation. n=12/group. *P<0.05
and **P<0.01. Scale bar, 100 µm. PM2.5, fine
particulate matter; C, cells treated with PBS; PRL, cells treated
with 5 µg/ml PM2.5; PRM, cells treated with 10 µg/ml
PM2.5; PRH, cells treated with 15 µg/ml
PM2.5; MDA, malondialdehyde; SOD, superoxide dismutase;
IL, interleukin; TNF, tumor necrosis factor; OD, optical
density. |
Subsequently, SOD and MDA activity was evaluated,
and it was determined whether expression levels of the inflammatory
factors were altered in lung tissue homogenates post-exposure. The
results indicated that PM2.5 significantly increased MDA
activity in in PRL, PRM and PRH groups (28.18, 56.73 and 102.55
U/mg, respectively) and significantly decreased SOD activity in
PRL, PRM and PRH groups (33.91, 25.07 and 21.64) compared with
controls (Fig. 3B and C).
Moreover, PM2.5 significantly promoted the accumulation
of pro-inflammatory factors IL-6 (protein expression, 96.11 and
42.16 pg/ml for PRH and group C, respectively; gene expression,
sample/control ratios were 2.98 and 0.88 for PRH and group C,
respectively), IL-1β (protein expression, 436.5 and 330.5 pg/ml for
PRH and group C, respectively; gene expression, sample/control
ratios were 4.17 and 1.03 for PRH and group C, respectively), TNF-α
(protein expression: 48.33 and 32.35 pg/ml for PRH and group C,
respectively; gene expression, sample/control ratios were 4.55 and
0.98 for PRH and group C, respectively), and significantly reduced
the expression of inflammatory factor IL-2 (protein expression,
32.58 and 55.33 pg/ml for PRH and group C; gene expression,
sample/control ratios were 0.67 and 3.15 for PRH and group C,
respectively) (Fig. 3D and E).
Effects of different printing
room-derived PM2.5 concentrations on the expression of
key proteins in exposed lung tissue
Similar to in vitro findings, printing
room-derived PM2.5 exposure significantly increased the
expression of mitogen-activated protein kinase (MAPK) pathway genes
COX2 (1.59 vs. 0.71 for PRH and group C, respectively), p-p65/p65
(1.25 vs. 0.83 for PRH and group C, respectively; Fig. 4A) and p-ERK/ERK (1.21 vs. 0.79 for
PRH and group C, respectively; Fig.
4B). Furthermore, PM2.5 significantly increased the
expression of apoptosis pathway genes TGF-β1 (0.87 vs. 0.56 for PRH
and group C, respectively; Fig.
4C), and Bax/Bcl-2 (1.22 vs. 0.71 for PRH and group C,
respectively; Fig. 4D). IHC
staining was performed to further evaluate lung tissue-related
effects of printing room-derived PM2.5 exposure, and the
results showed that PM2.5 exposure markedly enhanced
production of caspase-3, caspase-8 and Bax, and inhibited Bcl-2
accumulation in all groups assessed, particularly in the PRH group
(Fig. 5).
 | Figure 4.Effects of printing room-derived
PM2.5 exposure on signaling pathways in vivo.
Protein levels of (A) COX2 and p-p65/p65 (B) p-ERK/ERK (C) TGF-βand
(D) Bcl-2 and Bax in mouse lungs are shown. Data are presented as
the mean ± standard deviation. n=12/group. *P<0.05.
PM2.5, fine particulate matter; C, cells treated with
PBS; PRL, cells treated with 5 µg/ml PM2.5; PRM, cells
treated with 10 µg/m PM2.5; PRH, cells treated with 15
µg/ml PM2.5; HBE, human bronchial epithelial cells;
HUVEC, human umbilical vein endothelial cells; COX2, cyclooxygenase
2; p, phosphorylated; TGF, transforming growth factor. |
Effects of different concentrations of
printing room-derived PM2.5 on microbial diversity of
the lungs
Subsequently, the effects of PM2.5
exposure on microbial diversity within lung cells were evaluated
using high-throughput sequencing. The results showed that low
concentrations of PM2.5 did not influence α diversity
(Shannon index and Simpson index), while high PM2.5
concentration markedly reduced Shannon and Simpson index values
compared with control groups (Fig. 6A
and B). Assessment using Venn diagrams revealed 541 OTUs
commonly identified for all groups, which represented 29.84%
(541/1813), 29.56% (541/1830), 31.09% (541/1740) and 32.97%
(541/1641) of the total for C, PRL, PRM and PRH groups,
respectively (Fig. 6C). Moreover,
PCoA showed that dots corresponding to PRH scattered away from the
other groups, which indicated that PM2.5 exposure
markedly altered the microbial composition of the group (Fig. 6D).
A further comparison of microbial diversity at the
order (Fig. 6E) and class
(Fig. 6F) levels was then
performed, and bacteria most associated with infection were
assessed. The results indicated that in PRH, PM2.5
exposure significantly increased the richness of pathogenic
bacteria of the genus Burkholderiales (0.44 vs. 0.17 for PRH
and group C, respectively), Coriobacteriia (0.024 vs. 0.0017
for PRH and group C, respectively), and Betaproteobacteria
(0.43 vs. 0.17 for PRH and group C, respectively). Exposure also
resulted in microbial changes including reduced potential to adapt
to the environment (0.0011 vs. 0.0015 for PRH and group C,
respectively), immune system activity (0.00055 vs. 0.00087 for PRH
and group C, respectively), and altered energy metabolism (0.05 vs.
0.055 for PRH and group C, respectively). PM2.5 exposure
notably enhanced the appearance of biodegradation and metabolism of
xenobiotics (0.057 and 0.036 for PRH and group C, respectively),
endocrine system activity (0.0061 and 0.0041 for PRH and group C,
respectively) and infectious diseases (0.0041 and 0.0032 for PRH
and group C, respectively) that occurred in PRH compared with
control cells (Fig. 7). A
graphical summary of the study is presented in Fig. 8.
 | Figure 7.Effects of printing room-derived
PM2.5 exposure on metabolic pathways. The relative
abundance of (A) Burkholderiales, (B) Coriobacteriia
and (C) Betaproteobacteria were assessed. Furthermore, (D)
environmental adaptations, (E) immunity, (F) energy metabolism, (G)
xenobiotic biodegradation and metabolism, (H) endocrine system and
(I) infectious disease of groups were compared. *P<0.05.
PM2.5, fine particulate matter; C, cells treated with
PBS; PRL, cells treated with 5 µg/ml PM2.5; PRM, cells
treated with 10 µg/m PM2.5; PRH, cells treated with 15
µg/ml PM2.5. |
Discussion
Exposure to carbon dust and volatile organic
compounds affects the respiratory, immune and nervous systems, and
staff working in offices may have symptoms of cough and throat
discomfort that are caused by PM present in the air (26). Since the importance of indoor
particulate matter on health has been neglected, the effects of
PM2.5 (collected from the printing room) exposure on
lung injury, as well as its potential mechanisms, was studied both
in vitro and in vivo.
First, the effects of exposure to different
concentrations of PM2.5 on HBE and HUVEC cells was
evaluated. The results indicated that PM2.5 exposure
significantly reduced cell viability in a dose-dependent manner,
enhanced the lipid oxidation index of MDA and decreased the
anti-oxidative stress index of SOD, which was in line with
previously published reports (27,28).
Moreover, PM2.5 addition to cell culture media
significantly increased the accumulation of pro-inflammatory
factors IL-1β, IL-6 and TNF-α, inhibited the production of
inflammatory factor IL-2, activated the inflammatory response and
promoted lung injury and fibrosis. These present results are
consistent with previously published studies (29,30).
Therefore, PM2.5 activated oxidative stress and
inflammatory responses in lung cells, which has the potential to
cause cellular injury and even death.
To further study the potential mechanisms by which
PM2.5 alters lung cell function, key proteins of
inflammatory, fibrosis and apoptosis pathways were analyzed.
PM2.5 significantly increased the levels of COX2 and
p-p65/p65. COX2 is an important pro-inflammatory gene, and studies
have shown that overexpression of COX2 promotes inflammation and
adversely affects fibrosis (31).
p65 belongs to the NF-κB family of proteins, and these proteins
exist either as homologous or heterologous dimers formed between
family members under normal conditions (31). When cells were stimulated by
different external factors, such as stress, lipopolysaccharide,
viruses and free oxygen radicals, NF-κB immediately disassociates
and translocates to the nucleus to enhance transcription of
inflammatory genes (32). p65 has
also been shown to be a transcription factor involved in the NF-KB
pathway via regulation of pro-inflammatory cytokines TNF-α and IL-6
(33). As PM2.5
concentration increased, the expression of COX2 and p-p65/p65 were
enhanced in a dose-dependent manner. This finding indicated that as
PM2.5 concentrations increase, the degree of pulmonary
inflammation and fibrosis become increasingly severe as a result of
enhanced expression of COX2 and p-p65/p65.
Overexpression of COX2 and p-p65/p65 greatly
enhanced pulmonary fibrosis via enhancing the expression of
pro-fibrotic cytokine TGF-β1 (34)
and apoptosis markers, including Bax/Bcl-2 (35). TGF-β1 is critical for regulatory T
cell function and T-helper (Th) 9 and Th17 differentiation, which
play important roles in the regulation of tissue repair,
embryogenesis, cartilage homeostasis, cellular growth and
proliferation and cancer, and are associated with the development
of autoimmune disorders, chronic inflammatory conditions and
allergic diseases (36). Bcl-2 is
an anti-apoptotic marker, and downregulation of Bcl-2 may promote
the production of pro-apoptotic proteins (37). For example, increased Bax levels
can lead to apoptotic cell death (38). Of TGF-β1 and Bax expression levels
were upregulated in groups exposed to PM2.5.
Simultaneously, Bcl-2 expression was downregulated. Taken together,
these results indicated that exposure to increased concentrations
of PM2.5 enhanced pulmonary injury as a result of
enhanced pulmonary inflammation and fibrosis via downregulation of
Bcl-2 and upregulation of TGF-β1.
A mouse model was established and the results
indicated that PM2.5 exposure lead to inflammatory cell
infiltration, airway epithelial cell shedding and necrosis.
Consistent with cell culture findings, exposure to PM2.5
significantly increased the accumulation of pro-inflammatory
factors IL-1β, IL-6 and TNF-α, and reduced production of the
inflammatory factor, IL-2. This finding indicated that
PM2.5 induced inflammation in the lung via increasing
the production of pro-inflammatory factors in lung tissue. Western
blotting results indicated that protein expression of COX2,
p-p65/p65, TGF-β1 and Bax/Bcl-2 were enhanced post-exposure to
PM2.5, which indicated that PM2.5 induced
fibrosis and apoptosis in the lung via increasing the production of
pro-fibrotic cytokines and apoptosis markers. In addition, the
expression of p-ERK/ERK was evaluated, and it was found that
increased concentrations of PM2.5 resulted in MAPK
pathway activation, whereby increased levels of p-ERK relative to
ERK were observed. The MAPK pathway involves p38, JNK and ERK1/2,
which play key roles in acute lung injury, and the ERK pathway
exerts a critical function in the management of levels of various
cytokines, including TNF-α, IL-1β and IL-6 (39). Furthermore, ERK is involved in the
regulation of numerous cellular processes, including stress
response, inflammatory pathways, proliferation, differentiation,
apoptosis and survival (34).
Elevated p-ERK/ERK ratios in the MAPK pathway indicated that
increased concentrations of PM2.5 can lead to lung
inflammation. Additionally, IHC results indicated that
PM2.5 upregulated the expression of the pro-apoptotic
protein, Bax, and downregulated the accumulation of the
anti-apoptotic protein, Bcl-2, which eventually lead to enhanced
expression of apoptosis proteins caspase-8 and caspase-3 (40).
The lungs have previously been considered a sterile
environment, and assessment of pulmonary disease has been examined
from a bacterial pathology perspective (41). Recently, 16s ribosomal RNA studies
and metagenomics revealed the presence of a flexible microbiota in
the upper and lower respiratory tract, blood, placenta and amniotic
fluid (42). Increasing evidence
has shown that bacteria are present in the lungs and serve key
roles in fatal pneumonia, which can be both beneficial and harmful
(43,44). The present high-throughput
sequencing results showed that PM2.5 altered microbial
diversity in the lung by increasing abundance of
Burkholderiales (at the class level),
Betaproteobacteria (at the order level) and
Coriobacteriia (at the order level). Both
Betaproteobacteria and Burkholderiales can cause lung
inflammation associated with lung disease, and as abundance of
Betaproteobacteria and Burkholderiales increase, the
degree of lung damage is expected to correspondingly increase
(45). Therefore, PM2.5
exposure has the ability to damage human health through increasing
pathogen abundance in the lung. This can lead to increased lung
inflammation and damage. Environmental PM2.5 is also
capable of causing lung damage and energy metabolism disorders,
such as diarrhea (46). Multiple
lines of evidence have shown that PM2.5 exposure is
associated with metabolic disorders, especially in children and the
elderly or genetically susceptible experimental animal models
(47). Environmental adaptation,
immune response and energy metabolism in PRH groups markedly
decreased compared group C. Furthermore, infectious disease
occurrence, endocrine activity and the xenobiotic biodegradation
and metabolism may have an association with PM2.5
exposure.
In the present study, the effect of printing room
PM2.5 exposure on the lungs was assessed. The results
indicated that PM2.5 caused cellular and tissue injury
as a result of increased oxidative stress, inflammation, fibrosis
and apoptosis. Additionally, PM2.5 exposure enhanced
pathogen abundance in lung tissues. Therefore, printing room
PM2.5 may negatively affect individuals exposed for long
periods. However, considering limitations of the present acute lung
injury model, more work will be needed to clarify the health risks
associated with printing room PM2.5 exposure. In
conclusion, the present study evaluated mechanisms of lung injury
caused by printing room-derived PM2.5, and found that
they are closely related the inflammation, apoptosis and fibrosis
caused, and provided a theoretical foundation for further
research.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 41765009), the Excellent
Youth Foundation of the Jiangxi Scientific Committee (grant no.
20171BCB23028), the Science and Technology Plan of the Jiangxi
Health Planning Committee (grant no. 20175526) and the Science and
Technology Project of Jiangxi (grant nos. 20181BBG70028 and
20181BCB24003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TC and HH made substantial contributions to
conception and design. CZ, HY, LC and XC performed the analysis and
interpretation of data. CZ, TC, HY, LC and XC drafted the
manuscript and revised the important intellectual content. All
authors read and approved the final manuscript, and TC gave the
final approval of the version to be published.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of the Second Affiliated Hospital of Nanchang University,
and all experiments were performed in accordance with approved
guidelines (The Guide for Care and Use of Laboratory Animals;
National Institutes of Health publication 85–23).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Holzer M, Bihari P, Praetner M, Uhl B,
Reichel C, Fent J, Vippola M, Lakatos S and Krombach F:
Carbon-based nanomaterials accelerate arteriolar thrombus formation
in the murine microcirculation independently of their shape. J Appl
Toxicol. 34:3213–1176. 2014. View
Article : Google Scholar
|
|
2
|
Cohen AJ, Brauer M, Burnett R, Anderson
HR, Frostad J, Estep K, Balakrishnan K, Brunekreef B, Dandona L,
Dandona R, et al: Estimates and 25-year trends of the global burden
of disease attributable to ambient air pollution: An analysis of
data from the global burden of diseases study 2015. Lancet.
389:1907–1918. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Urlaub S, Grün G, Foldbjerg P and
Sedlbauer K: Ventilation and health - a review. Proc AVIC Conf.
2015.
|
|
4
|
Chen Y, Du W, Shen G, Zhuo S, Zhu X, Shen
H, Huang Y, Su S, Lin N, Pei L, et al: Household air pollution and
personal exposure to nitrated and oxygenated polycyclic aromatics
(PAHs) in rural households: Influence of household cooking
energies. Indoor Air. 27:169–178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Du B, Gao J, Chen J, Stevanovic S,
Ristovski Z and Wang L and Wang L: Particle exposure level and
potential health risks of domestic Chinese cooking. Build Environ.
123:564–574. 2017. View Article : Google Scholar
|
|
6
|
Liu T, Liu Q, Li Z, Huo L, Chan M, Li X,
Zhou Z and Chan CK: Emission of volatile organic compounds and
production of secondary organic aerosol from stir-frying spices.
Sci Total Environ. 599-600:1614–1621. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Y, Chen YY, Cao JY, Tao FB, Zhu XX,
Yao CJ, Chen DJ, Che Z, Zhao QH and Wen LP: Oxidative stress,
apoptosis, and cell cycle arrest are induced in primary fetal
alveolar type II epithelial cells exposed to fine particulate
matter from cooking oil fumes. Environ Sci Pollut Res Int.
22:9728–9741. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khatri M, Bello D, Pal AK, Woskie S,
Gassert TH, Demokritou P and Gaines P: Toxicological effects of
PM0.25-2.0 particles collected from a photocopy center in three
human cell lines. Inhal Toxicol. 25:621–632. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pirela SV, Bhattacharya K, Wang Y, Zhanga
Y, Wanga G, Christophic CA, Godleskia J, Thomasd T, Qiane Y,
Orandle MS, et al: A 21-day sub-acute, whole-body inhalation
exposure to printer-emitted engineered nanoparticles in rats:
Exploring pulmonary and systemic effects. NanoImpact.
15:1001762019. View Article : Google Scholar
|
|
10
|
Pirela S, Molina R, Watson C, Cohen JM,
Bello D, Demokritou P and Brain J: Effects of copy center particles
on the lungs: A toxicological characterization using a Balb/c mouse
model. Inhal Toxicol. 25:498–508. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jensen M and Rold-Petersen J: Itching
erythema among post office workers caused by a photocopying
illachine with wet toner. Contact Dermatitis. 5:389–391. 1979.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Galiardo M, Romero P, Sánchez-Quevedo MC
and López-Caballero JJ: Siderosilicosis due to photocopier toner
dust. Lancet. 344:412–413. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grifka J: Use of drugs in renal
impairment. Internist (Berl). 49:1262008.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Theegarten D, Boukercha S, Philippou S and
Anhenn O: Submesothelial deposition of carbon nanoparticles after
toner exposition: Case report. Diagn Pathol. 5:772010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ya P, Xu H, Ma Y, Fang M, Yan X, Zhou J
and Li F: Liver injury induced in Balb/c mice by PM2.5 exposure and
its alleviation by compound essential oils. Biomed Pharmacother.
105:590–598. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tavera Busso I, Vera A, Mateos AC,
Amarillo AC and Carreras H: Histological changes in lung tissues
related with sub-chronic exposure to ambient urban levels of PM2.5
in Córdoba, Argentina. Atmos Environ. 167:616–624. 2017. View Article : Google Scholar
|
|
17
|
Yang B, Guo J and Xiao C: Effect of PM2.5
environmental pollution on rat lung. Environ Sci Pollut Res Int.
25:36136–36146. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H, Zhao Q, Liu R, Yang L, Chen H and
Cui X: Protective effect and potential mechanism of simvastatin on
myocardial injury induced by diabetes with hypoglycemia. Exp Clin
Endocrinol Diabetes. 126:148–161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang H, Zou C, Cao J and Tsang P:
Carbonaceous aerosol characteristics in outdoor and indoor
environments of Nanchang, China, during summer 2009. J Air Waste
Manag Assoc. 61:1262–1272. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang H, Zou C, Cao J, Tsang P, Zhu F, Yu
C and Xue S: Water-soluble Ions in PM2.5 on the Qianhu Campus of
Nanchang University, Nanchang City: Indoor-outdoor distribution and
source implications. Aerosol Air Qual Res. 12:435–443. 2012.
View Article : Google Scholar
|
|
21
|
Wang H, Song L, Ju W, Wang X, Dong L,
Zhang Y, Ya P, Yang C and Li F: The acute airway inflammation
induced by PM2.5 exposure and the treatment of essential oils in
Balb/c mice. Sci Rep. 7:442562017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang X, Zhong W, Meng Q, Lin Q, Fang C,
Huang X, Li C, Huang Y and Tan J: Ambient PM2.5 exposure
exacerbates severity of allergic asthma in previously sensitized
mice. J Asthma. 52:785–794. 2015.PubMed/NCBI
|
|
23
|
Tian P, Xu D, Huang Z, Meng F, Fu J, Wei H
and Chen T: Evaluation of truncated G protein delivered by live
attenuated Salmonella as a vaccine against respiratory syncytial
virus. Microb Pathog. 115:299–303. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hammery, Harper DA and Ryan PD: PAST:
Paleontological statistics software package for education and data
analysis. Palaeontol Electron. 4:1–9. 2001.
|
|
26
|
Tang T, Hurraß J, Gminski R and
Mersch-Sundermann V: Fine and ultrafine particles emitted from
laser printers as indoor air contaminants in German offices.
Environ Sci Pollut Res Int. 19:3840–3849. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kohl H, Orth R, Riebartsch O, Galeitzke M
and Cap JP: Support of innovation networks in manufacturing
industries through identification of sustainable collaboration
potential and best-practice transfer. Procedia CIRP. 26:185–189.
2015. View Article : Google Scholar
|
|
28
|
Zhang ZQ, Zhang CZ, Shao B, Pang DH, Han
GZ and Lin L: Effects of abnormal expression of fusion and fission
genes on the morphology and function of lung macrophage
mitochondria in SiO2-induced silicosis fibrosis in rats in vivo.
Toxicol Lett. 312:181–187. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Smith RE, Strieter RM, Phan SH, Lukacs N
and Kunkel SL: TNF and IL-6 mediate MIP-1alpha expression in
bleomycin-induced lung injury. J Leukoc Biol. 64:528–536. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cavarra E, Carraro F, Fineschi S, Naldini
A, Bartalesi B, Pucci A and Lungarella G: Early response to
bleomycin is characterized by different cytokine and cytokine
receptor profiles in lungs. Am J Physiol Lung Cell Mol Physiol.
287:L1186–L1192. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dai P, Shen D, Shen J, Tang Q, Xi M, Li Y
and Li C: The roles of Nrf2 and autophagy in modulating
inflammation mediated by TLR4-NFκB in A549 cell exposed to layer
house particulate matter 2.5 (PM2.5). Chemosphere. 235:1134–1145.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hayden MS and Ghosh S: NF-κB in
immunobiology. Cell Res. 21:223–244. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Barker HE, Paget JT, Khan AA and
Harrington KJ: The tumour microenvironment after radiotherapy:
Mechanisms of resistance and recurrence. Nat Rev Cancer.
15:409–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lazzara F, Fidilio A, Platania CBM,
Giurdanella G, Salomone S, Leggio GM, Tarallo V, Cicatiello V, De
Falco S, Eandi CM, et al: Aflibercept regulates retinal
inflammation elicited by high glucose via the PlGF/ERK pathway.
Biochem Pharmacol. 168:341–351. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kuroki M, Noguchi Y, Shimono M, Tomono K,
Tashiro T, Obata Y, Nakayama E and Kohno S: Repression of
bleomycin-induced pneumopathy by TNF. J Immunol. 170:567–574. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ndaw VS, Abebayehu D, Spence AJ, Paez PA,
Kolawole EM, Taruselli MT, Caslin HL, Chumanevich AP, Paranjape A,
Baker B, et al: TGF-β1 suppresses IL-33-induced mast cell function.
J Immunol. 199:866–873. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ashkenazi A, Fairbrother WJ, Leverson JD
and Souers AJ: From basic apoptosis discoveries to advanced
selective BCL-2 family inhibitors. Nat Rev Drug Discov. 16:273–284.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hii LW, Lim SE, Leong CO, Chin SY, Tan NP,
Lai KS and Mai CW: The synergism of Clinacanthus nutans
Lindau extracts with gemcitabine: Downregulation of anti-apoptotic
markers in squamous pancreatic ductal adenocarcinoma. BMC
Complement Altern Med. 19:2572019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang C, Zhong R, Zhang J, Wang X, Ding G,
Xiao W and Ma S: Reduning injection ameliorates paraquat-induced
acute lung injury by regulating AMPK/MAPK/NF-κB signaling. J Cell
Biochem. 120:12713–12723. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Che Z, Liu Y, Chen Y, Cao J, Liang C, Wang
L and Ding R: The apoptotic pathways effect of fine particulate
from cooking oil fumes in primary fetal alveolar type II epithelial
cells. Mutat Res Genet Toxicol Environ Mutagen. 761:35–43. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Chen J, Tang B, Zhang X and Hua
ZC: Systemic administration of attenuated salmonella typhimurium in
combination with interleukin-21 for cancer therapy. Mol Clin Oncol.
1:461–465. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu K, Rodriguez MD, Paul Z, Gordon E, Rice
K, Triplett EW, Keller-Wood M and Wood CE: Proof of principle:
Physiological transfer of small numbers of bacteria from mother to
fetus in late-gestation pregnant sheep. PLoS One. 14:e02172112019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kaimala S, Al-Sbiei A, Cabral-Marques O,
Fernandez-Cabezudo MJ and Al-Ramadi BK: Attenuated bacteria as
immunotherapeutic tools for cancer treatment. Front Oncol.
8:1362018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao C, He J, Cheng H, Zhu Z and Xu H:
Enhanced therapeutic effect of an antiangiogenesis peptide on lung
cancer in vivo combined with salmonella VNP20009 carrying a Sox2
shRNA construct. J Exp Clin Cancer Res. 35:1072016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pacello F, D'Orazio M and Battistoni A: An
ERp57-mediated disulphide exchange promotes the interaction between
Burkholderia cenocepacia and epithelial respiratory cells. Sci Rep.
6:211402016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ning X, Ji X, Li G and Sang N: Ambient
PM2.5 causes lung injuries and coupled energy metabolic disorder.
Ecotoxicol Environ Saf. 170:620–626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pan K, Jiang S, Du X, Zeng X, Zhang J,
Song L, Zhou J, Kan H, Sun Q, Xie Y and Zhao J: AMPK activation
attenuates inflammatory response to reduce ambient PM2.5-induced
metabolic disorders in healthy and diabetic mice. Ecotoxicol
Environ Saf. 179:290–300. 2019. View Article : Google Scholar : PubMed/NCBI
|






















