Introduction
Ovarian cancer (OC) is a frequently occurring
malignant tumor worldwide (1). At
the stage of diagnosis, the majority of patients with OC present
with cancer cell metastases (2).
Despite significant advances in the treatment of OC, the
therapeutic efficiency and prognosis of the disease remain
unsatisfactory (3,4). Therefore, although the pathogenesis
of OC has been previously reported (5), the exact regulatory mechanism
underlying OC requires further investigation.
Long non-coding RNAs (lncRNAs) are composed of
>200 nucleotides and originate from the multitudinous class of
genome transcripts (6). Increasing
evidence has indicated that lncRNAs serve a vital role in diverse
cellular functions, such as cell proliferation, apoptosis,
migration and autophagy (7–9).
Over the past few decades, it has been revealed that the aberrant
expression of lncRNAs is associated with the progression and
initiation of various tumors, including human OC (10–12).
In particular, metastasis-associated lung adenocarcinoma transcript
1 is associated with poor prognosis and may serve as a biomarker in
colorectal cancer (13). High
expression levels of HOX transcript antisense RNA are related to
poor prognosis and tumor metastasis in human OC (14). Moreover, lncRNA nuclear paraspeckle
assembly transcript 1 (NEAT1), which is highly expressed in
multiple types of cancer, is related to OC prognosis (15). In addition, it has also been
reported that the aberrant expression of NEAT1 promotes
tumorigenesis, metastasis and paclitaxel resistance in human OC
(16,17). The aforementioned studies indicate
that NEAT1 is highly associated with OC progression, therefore, the
precise function of NEAT1 in the molecular mechanisms underlying OC
requires further investigation.
MicroRNAs (miRNAs/miRs) are a class of small
non-coding RNAs (~22 nucleotides in length) that can suppress
target protein expression by repressing translation or promoting
mRNA degradation (18). Previous
studies have demonstrated that miRNA dysfunction participates in
cancer progression (19–22). The role and sequences of miR-4500
have been previously reported (23,24),
and the inhibitory effects of miR-4500 on cancer progression have
been partially identified (25,26).
For example, miR-4500 limits tumor development by targeting high
mobility group AT-hook 2 in colorectal cancer (26). By contrast, the function of
miR-4500 in OC is not completely understood; therefore, the present
study aimed to explore the effects of miR-4500 on OC cell behaviors
in vitro.
Basic leucine zipper and W2 domain-containing
protein 1 (BZW1; also known as BZAP45) is expressed in a diverse
number of human tissues and cell lines (27). BZW1 promotes salivary
mucoepidermoid carcinoma development (28), whereas its influence during OC
pathogenesis is not completely understood.
In the present study, the expression levels and
roles of NEAT1, miR-4500 and BZW1 during the progression and
initiation of OC were investigated.
Materials and methods
Patient samples and cell culture
The present study was approved by the Ethical
Committee of Jingzhou Hospital of Traditional Chinese Medicine, The
Third Clinical Medical College of Yangtze University. Written
informed consent was obtained from all participants. Tumor tissues
and adjacent noncancerous tissues (>5 cm from the tumor margin)
were obtained from 30 patients with OC (aged 37–68 years) who
received resection treatment at Jingzhou Hospital of Traditional
Chinese Medicine, The Third Clinical Medical College of Yangtze
University between June 2017 and June 2018. OC was validated by
histological examination according to World Health Organization
criteria (29). All patients had
not received adjuvant chemotherapy prior to surgery.
The OC cell lines (CAOV3 and ES-2) were purchased
from ATCC and normal control cells (IOSE80) were purchased from
BeNa Culture Collection, Beijing Beina Chunglian Biotechnology
Research Institute. CAOV3 and IOSE80 cells were cultured in DMEM
(Corning Inc.), and ES-2 cells were cultured in McCoy's 5a
(modified) Medium (Sigma-Aldrich; Merck KGaA). Both culture mediums
were supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.), 100 units/ml penicillin and 100 mg/ml streptomycin. Cells
were cultured at 37°C with 5% CO2.
Transient transfection
A small interfering RNA (siRNA) targeted against
NEAT1 (si-NEAT1, 5′-GCCAUCAGCUUUGAAUAAAUU-3′) and its negative
control (NC; si-NC, 5′-UUCUCCGAACGUGUCACUTT-3′) were designed by
Guangzhou RiboBio Co., Ltd. BZW1 sequences were inserted into the
pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.) to
construct a BZW1 overexpression vector. An empty pcDNA3.1 vector
serves as the control (pcDNA). miR-4500 mimic (miR-4500,
5′-UGAGGUAGUAGUUUCUUGCGCC-3′), inhibitor (anti-miR-4500,
5′-GGCGCAAGAAACUACUACCUCA-3′) and their relative controls (miR-NC,
5′-GAAAUGUACUUGAGCGUGGAGAC-3′; and anti-miR-NC,
5′-CUAAAACCGGCCGUACGGCGUU-3′) were designed and synthesized by
Guangzhou RiboBio Co., Ltd. BZW1 overexpression vector or pcDNA
(0.2 µg) was transfected into CAOV3 and ES-2 cells
(5×104 cells) with 0.5 µl Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Moreover,
si-NEAT1, si-NC, miR-4500 mimic, anti-miR-4500, miR-NC or
anti-miR-NC (0.5 µg) was transfected into cells using 0.6 µl
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to manufacturer's protocol. After 48 h transfection, the
transfected cells were used for subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from patient samples and
cell lines (CAOV3 and ES-2) using RNAiso Plus (Takara Biotechnology
Co., Ltd.). Total RNA was reverse transcribed into cDNA using
PrimeScript RT Master Mix (Takara Biotechnology Co., Ltd.) at 50°C
for 15 min and 85°C for 5 sec. Subsequently, qPCR was performed
using SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd) and an
AB7900 Real-Time PCR System machine (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The amplification parameters were as
follows: Denaturation at 95°C for 10 min; followed by 40 cycles of
denaturation at 95°C for 30 sec and annealing at 60°C for 30 sec;
and final extension at 72°C for 1 min. The following primers
(designed and synthesized by Guangzhou RiboBio Co., Ltd.) were used
for qPCR: SOX4 forward, 5′-GGCCTGTTTCGCTGTCGGGT-3′ and reverse,
5′-GCCTGCATGCAACAGACTGGC-3′; HOTAIR forward,
5′-TAGGCAAATGTCAGAGGGTT-3′ and reverse, 5′-CTTAAATTGGGCTGGGTCT-3′;
plasmacytoma variant translocation 1 forward,
5′-TGAGAACTGTCCTTACGTGACC-3′ and reverse,
5′-AGAGCACCAAGACTGGCTCT-3′; growth arrest-specific transcript 5
forward, 5′-CCCAAGGAAGGATGAG-3′ and reverse,
5′-ACCAGGAGCAGAACCA-3′; oryzacystatin-1 forward,
5′-TTCCAGCGCATGTCGGCGCTC-3′ and reverse,
5′-GGTACTAGTCCGTGGTTCTTC-3′; human ovarian cancer specific
transcript 2 forward, 5′-GGACAGGTCCCTTGTTTCAA-3′ and reverse,
5′-CTGGTCTTTCCTTGCCTCTG-3′; NEAT1 forward,
5′-TGGCTAGCTCAGGGCTTCAG-3′ and reverse,
5′-TCTCCTTGCCAAGCTTCCTTC-3′; miR-4500 forward,
5′-CGTCGCACTGTGAGGTAGTAG-3′ and reverse,
5′-TATGCTTGTTCTCGTCTCTGTGTC-3′; BZW1 forward,
5′-TACCGTCGATATGCAGAAACAC-3′ and reverse,
5′-CCATTAGCCAGAAGAACACCAG-3′; GAPDH forward,
5′-TGCACCACCAACTGCTTAGC-3′ and reverse,
5′-GGCATGCACTGTGGTCATGAG-3′; and U6 forward,
5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse,
5′-GGAACGCTTCACGAATTTG-3′. miRNA and mRNA expression levels were
quantified using the 2−∆∆Cq method (30) and normalized to the internal
reference genes U6 and GAPDH, respectively.
Cell Counting Kit-8 (CCK-8) assay for
cell proliferation
CAOV3 and ES-2 cells were seeded (5×104
cells/well) into a 96-well plate. At 70% confluence, cells were
transfected according to the aforementioned protocol. After
incubation for 24, 48 or 72 h at 37°C, CCK-8 reagent (Beyotime
Institute of Biotechnology) was added to each well according to
manufacturer's protocol. Following incubation for 4 h at 37°C, the
absorbance of each well was measured using a microplate reader.
Colony formation
Transfected CAOV3 and ES-2 cells (5×102
cells/well) were seeded into 6-well plates and cultured for 2 weeks
at 37°C, and the culture medium was replaced with fresh medium
every 4 days. Then, cells were washed using PBS, fixed with 4%
paraformaldehyde for 15 min at room temperature, and stained with
0.5% crystal violet (Sigma-Aldrich; Merck KGaA) for 10 min at room
temperature. The number of colonies (containing >50 cells) was
observed and counted using an optical microscope.
Flow cytometry assay to detect cell
apoptosis
Cell apoptosis was detected using the Annexin
V-FITC/PI detection kit (Nanjing KeyGen Biotech Co., Ltd.),
according to the manufacturer's protocol. Briefly, transfected
CAOV3 and ES-2 cells (5×104) were incubated with 5 µl
Annexin V-FITC and 5 µl propidium iodide in the dark for 15 min at
4°C. Apoptotic cells were detected by flow cytometry using a
FACScan flow cytometer (BD Biosciences) and analyzed using Cell
Quest software version 3.3 (BD Bioscience). The rate of apoptosis
was presented as the percentage of early apoptosis OC cells in the
total number of OC cells.
Transwell assay
At 60–80% confluence, CAOV3 and ES-2 were
transfected according to the aforementioned protocol. At 24 h
post-transfection, cells were collected and washed with Hanks'
balanced salt solution (Invitrogen; Thermo Fisher Scientific,
Inc.). Cell migration and invasion were assessed using a 24-well
Transwell system (pore size, 8 µm; Costar; Corning Inc.). To assess
cell invasion, the upper chambers were precoated with
Matrigel® at 4°C for 12 h. Transfected cells
(5×104 cells/well) were seeded into the upper chamber
with 200 µl serum-free medium. Complete medium with 10% FBS was
seeded into the lower chamber. Following incubation for 24 h at
37°C, migratory/invading cells were stained with 0.5% crystal
violet (Sigma-Aldrich; Merck KGaA) at room temperature for 20 min.
Stained cells were observed and photographed using an inverted
microscope.
Glycolysis analysis
The extracellular acidification rate (ECAR) of OC
cells was evaluated using an XF96 metabolic flux analyzer (Seahorse
Biosciences; Agilent Technologies, Inc.), according to the
manufacturer's protocol. Briefly, 10 mM glucose, 1 mM oligomycin
and 80 mM 2-deoxyglucose were added at the indicated time points.
Seahorse XF-96 Wave software (Seahorse Bioscience; Agilent
Technologies, Inc.) was used to analyze the ECAR of OC cells.
Dual-luciferase reporter assay
StarBase (http://starbase.sysu.edu.cn/starbase2/) was used to
screen the putative target. The complementary sequences between
miR-4500 and wild-type (WT) or mutant (Mut) NEAT1 were amplified
and cloned into the pGL3 basic vector (Promega Corporation) to
construct luciferase reporter vectors (NEAT1-WT and NEAT1-Mut,
respectively). Similarly, the complementary sequences between
miR-4500 and WT or Mut BZW1 were cloned into the pGL3 basic vector
to construct luciferase reporter vectors (BZW1-WT and BZW-Mut,
respectively). Subsequently, CAOV3 and ES-1 cells (5×104
cells) were co-transfected with 400 ng of luciferase reporter
(NEAT1-WT, NEAT1-Mut, BZW-WT or BZW-Mut) and miR-4500 mimic (50 nM)
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.). After incubation for 48 h at 37°C, luciferase activities
were detected using the Dual-Luciferase Reporter Assay System
(Promega Corporation), according to the manufacturer's protocol.
Firefly luciferase activities were normalized to Renilla
luciferase activities.
RNA binding protein
immunoprecipitation (RIP)
The RIP assay was performed using the Magna RIP
RNA-Binding Protein Immunoprecipitation kit (EMD Millipore).
Western blotting was performed to detect the expression levels of
Ago2 (cat. no. ab32381; 1:1,000; Abcam) and RT-qPCR was performed
to measure the expression levels of NEAT1 and miR-4500 in the
precipitates. IgG served as the control (cat. no. ab150077;
1:1,000; Abcam).
Western blotting
Western blotting was performed as previously
described (31). In brief, total
protein was extracted from tissues and cell lines using RIPA lysis
buffer (EMD Millipore). Total protein concentration was measured
using a BCA Protein assay kit (Nanjing KeyGen Biotech Co., Ltd.),
according to the manufacturer's protocol. Equal amounts of protein
(~30 µg) were separated via SDS-PAGE on a 12% gel, and subsequently
transferred onto PVDF membranes (EMD Millipore). After blocking
with 5% skimmed milk at room temperature for 2 h, the membranes
were incubated overnight at 4°C with the following primary
antibodies: BZW1 (cat. no. ab85090; 1:800; Abcam) and GAPDH (cat.
no. ab8245; 1:8,000; Abcam). Following primary incubation, the
membranes were incubated with horseradish peroxidase
(HRP)-conjugated goat anti-rabbit IgG H&L (cat. no. ab205718;
1:2,000; Abcam) and HRP-conjugated goat anti-mouse IgG H&L
(cat. no. ab205719; 1:2,000; Abcam) at room temperature for 40 min.
Protein bands were visualized using the Enhanced Chemiluminescence
Plus kit (EMD Millipore) and semi-quantified by densitometric
analysis of protein signals using ImageJ version 1.49 (National
Institutes of Health).
Statistical analysis
All experiments were repeated at least three times.
Statistical analyses were performed using GraphPad software
(version 5.0; GraphPad Software, Inc.). Data are expressed as the
mean ± SD. Differences between paired tumor tissues and adjacent
noncancerous tissues were analyzed using the paired Student's
t-test. Differences between two groups were analyzed using the
unpaired Student's t-test. Differences among multiple groups were
analyzed using one-way ANOVA followed by Tukey's post hoc test.
Spearman's correlation coefficient was used to analyze the
relationship between miR-4500 expression and NEAT1 expression.
P<0.05 was considered to indicate a statistically significant
difference.
Results
NEAT1 is upregulated in OC tissues and
cell lines
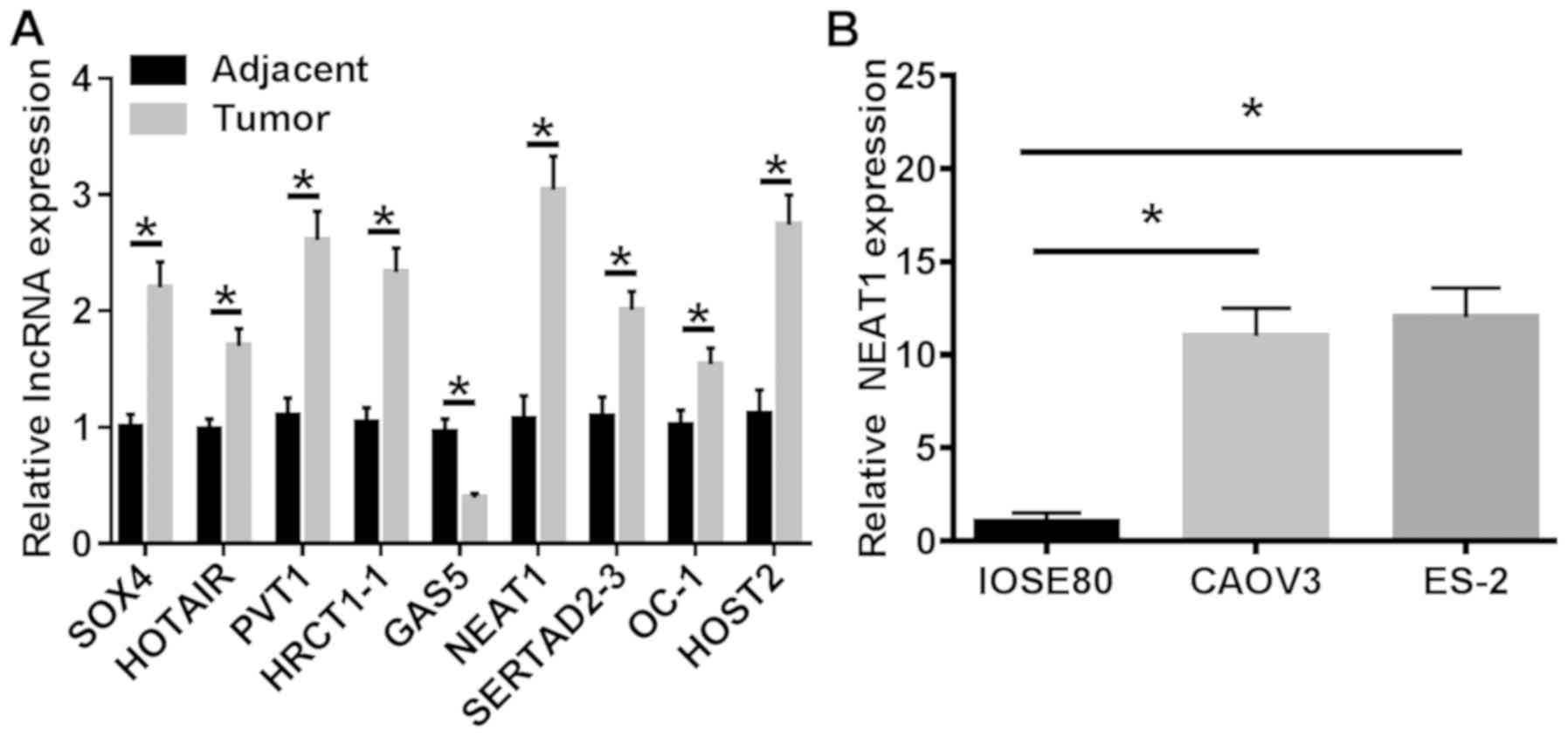
RT-qPCR was performed to assess the expression of
multiple lncRNAs in 30 paired OC and adjacent noncancerous tissue
samples. NEAT1 was chosen for subsequent experiments as it
displayed the highest expression levels in tumor samples compared
with matched normal tissue samples (Fig. 1A). The expression levels of NEAT1
in OC cell lines (CAOV3 and ES-2) were also detected via RT-qPCR.
The results indicated that NEAT1 expression was significantly
higher in OC cell lines compared with normal ovarian cells
(Fig. 1B). Collectively, the
results demonstrated that the aberrant expression of NEAT1 may be
associated with the progression of human OC.
NEAT1 knockdown increases cell
apoptosis, and decreases cell proliferation, colony formation,
migration, invasion and glycolysis in vitro
To investigate whether NEAT1 served a critical role
in ovarian carcinogenesis, NEAT1 knockdown was performed in CAOV3
and ES-2 cells. si-NEAT1 significantly reduced NEAT1 expression
levels compared with si-NC (Fig.
2A). CAOV3 and ES-2 cell proliferation was assessed by the
CCK-8 assay, which indicated that NEAT1 knockdown significantly
decreased cell proliferation in both cell lines compared with si-NC
(Fig. 2B and C). In addition, the
effect of si-NEAT1 on colony formation was assessed. Colony
formation was inhibited by NEAT1 knockdown in OC cell lines
(Fig. 2D). By contrast, cell
apoptosis was significantly increased following transfection with
si-NEAT1 compared with si-NC (Fig.
2E). Furthermore, the role of NEAT1 in cell migration and
invasion was analyzed using Transwell assays. The results suggested
that CAOV3 and ES-2 cell migration and invasion were significantly
inhibited by si-NEAT1 compared with si-NC (Fig. 2F and G). Moreover, si-NEAT1
significantly inhibited OC cell glycolysis compared with si-NC
(Fig. 2H and I). The
aforementioned results indicated that NEAT1 knockdown suppressed OC
cell progression in vitro.
 | Figure 2.NEAT1 knockdown increases cell
apoptosis, and inhibits cell proliferation, colony formation,
migration, invasion and glycolysis in vitro. CAOV3 and ES-2
cells were transfected with si-NEAT1 or si-NC. (A) Transfection
efficiency of NEAT1 knockdown. The CCK-8 assay was performed to
assess (B) CAOV3 and (C) ES-2 cell proliferation at 72 h. (D) The
colony formation assay was performed to assess colony formation.
(E) Cell apoptosis was assessed via flow cytometry. (F) CAOV3 and
(G) ES-2 cell migration and invasion were determined using
Transwell assays (magnification, ×100). ECAR in (H) CAOV3 and (I)
ES-2 cells was detected using an XF96 metabolic flux analyzer.
*P<0.05, as indicated. NEAT1, nuclear paraspeckle assembly
transcript 1; si, small interfering RNA; NC, negative control;
CCK-8, Cell Counting Kit-8; ECAR, extracellular acidification rate;
PI, propidium iodide; OM, oligomycin; 2-DG, 2-deoxyglucose. |
NEAT1 is a sponge of miR-4500
The starBase database was used to predict potential
targets of NEAT1, and RT-qPCR was performed to detect the
expression levels of the potential targets in si-NC and
si-NEAT1-transfected ES-2 cells. Following NEAT1 knockdown, the
expression levels of miR-4500 were significantly increased in ES-2
cells compared with si-NC-transfected ES-2 cells (Fig. 3A). Furthermore, the binding sites
between miR-4500 and NEAT1 were identified (Fig. 3B). The transfection efficiency of
miR-4500 mimic is presented in Fig.
3C. Subsequently, CAOV3 and ES-2 cells were transfected with
luciferase reporters, including NEAT1-WT and NEAT1-Mut. miR-4500
mimic significantly inhibited NEAT-WT luciferase activities in
CAOV3 and ES-2 cells compared with miR-NC. By contrast, miR-4500
mimic displayed no effect on the luciferase activities of NEAT1-Mut
(Fig. 3D and E). The binding
between miR-4500 and NEAT1 was also verified using the RIP assay
(Fig. 3F and G). In addition,
NEAT1 knockdown significantly upregulated miR-4500 expression in
CAOV3 cells compared with si-NC-transfected cells (Fig. 3H). miR-4500 expression levels were
significantly decreased in OC tissue samples and cell lines
compared with the corresponding controls (Fig. 3I and J). The results also indicated
that miR-4500 expression was negatively correlated with NEAT1
expression (Fig. 3K). Therefore,
the results indicated that miR-4500 was targeted by NEAT1.
miR-4500 inhibitor abolishes NEAT1
knockdown-mediated effects on cell behaviors
Based on the interaction between NEAT1 and miR-4500,
CAOV3 and ES-2 cells were transfected with si-NC, si-NEAT1,
si-NEAT1 + anti-miR-NC or si-NEAT1 + anti-miR-4500. miR-4500
knockdown significantly decreased miR-4500 expression levels
compared with the anti-miR-NC group (Fig. 4A). miR-4500 knockdown also reversed
NEAT1 knockdown-mediated effects on miR-4500 expression (Fig. 4B). Furthermore, NEAT1
knockdown-mediated inhibition of cell proliferation was decreased
by miR-4500 knockdown (Fig. 4C and
D). Similarly, the inhibitory effects of NEAT1 knockdown on
colony formation were reversed by miR-4500 knockdown (Figs. 4E and S1A). miR-4500 knockdown also decreased
the rate of NEAT1 knockdown-induced cell apoptosis (Figs. 4F and S1B). Cell migration and invasion were
assessed using the Transwell assay, which indicated that si-NEAT1
significantly decreased cell migration and invasion compared with
si-NC. However, following transfection with miR-4500 inhibitor,
NEAT1 knockdown-mediated effects on cell migration and invasion
were reversed (Figs. 4G, H,
S1C and D). Moreover, miR-4500
knockdown reversed the inhibitory effect of NEAT1 knockdown on OC
cell glycolysis (Fig. 4I and J).
Taken together, the results indicated that NEAT1 knockdown-mediated
effects on cell proliferation, colony formation, apoptosis,
migration, invasion and glycolysis were reversed by miR-4500
knockdown.
 | Figure 4.miR-4500 inhibitor reverses NEAT1
knockdown-mediated effects on cell behavior. (A) Transfection
efficiency of anti-miR-4500. (B) miR-4500 expression levels in
CAOV3 and ES-2 cells transfected with si-NC, si-NEAT1, si-NEAT1 +
anti-miR-NC or si-NEAT1 + anti-miR-4500. The CCK-8 assay was
performed to assess (C) CAOV3 and (D) ES-2 cell proliferation at 72
h. (E) The effects of NEAT1 knockdown and miR-4500 inhibitor on
colony formation. (F) Cell apoptosis was assessed via flow
cytometry. Transwell assays were conducted to investigate the
effect of NEAT1 knockdown and miR-4500 inhibitor on (G) CAOV3 and
(H) ES-2 cell migration and invasion. ECAR in (I) CAOV3 and (J)
ES-2 cells determined using an XF96 metabolic flux analyzer.
*P<0.05, as indicated. miR, microRNA; NEAT1, nuclear paraspeckle
assembly transcript 1; si, small interfering RNA; NC, negative
control; CCK-8, Cell Counting Kit-8; ECAR, extracellular
acidification rate; OM, oligomycin; 2-DG, 2-deoxyglucose. |
BZW1 is a target gene of miR-4500
Subsequently, the mechanism underlying the tumor
suppressor role of miR-4500 in OC carcinogenesis was investigated.
The potential target genes of miR-4500 were predicted and a common
fragment domain between miR-4500 and BZW1 was identified (Fig. 5A). The dual-luciferase reporter
assay suggested that the luciferase activities of the WT group were
significantly reduced by miR-4500 mimic compared with miR-NC,
whereas miR-4500 mimic did not alter the luciferase activities of
the Mut group (Fig. 5B and C). The
protein expression levels of BZW1 were evaluated via western
blotting. miR-4500 mimic decreased BZW protein expression levels in
both CAOV3 and ES-2 cells compared with miR-NC (Fig. 5D and E), which indicated that BZW1
was directly targeted by miR-4500.
BZW1 overexpression reverses
miR-4500-mediated effects on OC cell behavior
Based on the molecular mechanism between miR-4500
and BZW1, subsequent assays were performed to identify the
regulatory function between miR-4500 and BZW1. The transfection
efficiency of the BZW1 overexpression vector is presented in
Fig. 6A. Subsequently, CAOV3 and
ES-2 cells were transfected with miR-NC + pcDNA, miR-4500 + pcDNA
or miR-4500 + BZW1 overexpression vector. The protein expression
levels of BZW1, which were significantly inhibited by miR-4500
mimic, were restored by BZW1 overexpression (Fig. 6B and C). Moreover, BZW1
overexpression inhibited the repressive effect of miR-4500 mimic on
CAOV3 and ES-2 cell proliferation (Fig. 6D and E). Similarly, colony
formation, which was significantly inhibited by miR-4500 mimic, was
rescued following BZW1 overexpression (Figs. 6F and S2A). In addition, the rate of miR-4500
mimic-induced cell apoptosis was decreased by BZW1 overexpression
(Figs. 6G and S2B). The Transwell assay indicated that
BZW1 overexpression partially reversed the inhibitory effects of
miR-4500 mimic on OC cell migration and invasion (Figs. 6H, I, S2C and D). Furthermore, miR-4500
mimic-mediated inhibition of glycolysis was recovered by BZW1
overexpression (Fig. 6J and K).
Collectively, the results indicated that miR-4500 mimic-mediated
effects on cell behavior were reversed by BZW1 overexpression.
 | Figure 6.BZW1 overexpression reverses miR-4500
mimic-mediated effects on OC cell behavior. (A) Transfection
efficiency of BZW1 overexpression. CAOV3 and ES-2 cells were
transfected with miR-NC + pcDNA, miR-4500 + pcDNA or miR-4500 +
BZW1. BZW1 protein expression levels in (B) CAOV3 and (C) ES-2
cells. The CCK-8 assay was performed to assess (D) CAOV3 and (E)
ES-2 cell proliferation at 72 h. (F) Colony formation was evaluated
by performing a colony formation assay. (G) OC cell apoptosis was
determined via flow cytometry. Transwell assays were performed to
investigate the function of miR-4500 and BZW1 in (H) CAOV3 and (I)
ES-2 cell migration and invasion. ECAR in (J) CAOV3 and (K) ES-2
cells was determined using an XF96 metabolic flux analyzer.
*P<0.05, as indicated. BZW1, basic leucine zipper and W2
domain-containing protein 1; miR, microRNA; OC, ovarian cancer; NC,
negative control; CCK-8, Cell Counting Kit-8; ECAR, extracellular
acidification rate; OM, oligomycin; 2-DG, 2-deoxyglucose. |
BZW1 expression is co-regulated by
NEAT1 and miR-4500
Based on the biological roles of NEAT1, miR-4500 and
BZW1 in OC pathogenesis, the underlying modulatory mechanism was
investigated. The present study aimed to investigate how BZW1
expression was regulated in OC. Following transfection with si-NC,
si-NEAT1, si-NEAT1 + anti-miR-NC or si-NEAT1 + anti-miR-4500, the
mRNA expression levels of BZW1 in CAOV3 and ES-2 cells were
determined via RT-qPCR. The results indicated that NEAT1 knockdown
significantly decreased the mRNA expression levels of BZW1 compared
with si-NC, which was reversed by miR-4500 inhibitor (Fig. 7A and B). Furthermore, the protein
expression levels of BZW1 were measured by performing western
blotting and displayed a similar pattern to mRNA expression levels
(Fig. 7C and D). In summary, the
results indicated that the mRNA and protein expression levels of
BZW1 were modulated by NEAT1 and miR-4500 in OC cell lines.
Discussion
lncRNAs participate in the progression of various
tumors, with a number of lncRNAs serving as sponges of miRNAs
(32). For example, it has been
reported that growth arrest-specific 5 is closely associated with
multiple types of cancer, such as breast and gastric cancer
(33–35). NEAT1, the target lncRNA
investigated in the present study, responds to proteasome
inhibition (36) and is associated
with the poor survival of patients with breast cancer (37). In the present study, NEAT1
expression levels were significantly increased in OC tissues and
cell lines (CAOV3 and ES-2) compared with control tissues and cell
lines, which was consistent with a previous study (15). Furthermore, it has been reported
that NEAT1 mediates oncogenic growth by altering the epigenetic
landscape of target genes, thereby promoting oncogenic gene
transcription in prostate cancer (38). However, the functions of NEAT1
during human OC development require further investigation. Based on
the high expression of NEAT1 in OC, the specific role of NEAT1
knockdown on OC cell behaviors, such as cell proliferation, colony
formation, apoptosis, migration, invasion and glycolysis, was
investigated in the present study. NEAT1 knockdown enhanced cell
apoptosis, but inhibited cell proliferation, colony formation,
migration, invasion and glycolysis in vitro. The results
indicated that NEAT1 may serve as an oncogene in OC pathogenesis
and NEAT1 knockdown may suppress the progression of OC. Although it
has been reported that NEAT1 is a sponge of miR-34a-5p in OC
(39), further molecular functions
of NEAT1 require investigation.
Several studies have demonstrated that lncRNAs
regulate target miRNAs (40,41).
NEAT1 may monitor tumorigenesis and malignant progression by
sponging miRNAs (42). miR-4500
has been identified as a tumor suppressor in a number of diseases,
including non-small cell lung (25) and colorectal (26) cancer. In the present study, the
interaction between miR-4500 and NEAT1 was predicted and verified.
The results suggested that NEAT1 regulated cell functions by
targeting miR-4500. In addition, miR-4500 inhibitor reversed NEAT1
knockdown-mediated effects on OC cell behaviors, including cell
proliferation, colony formation, apoptosis, migration, invasion and
glycolysis. Based on the function of miR-4500 during OC
progression, the target genes of miR-4500 need to be
identified.
A previous study demonstrated that BZW1, which
belongs to the bZIP superfamily of transcription factors, serves a
crucial role in salivary mucoepidermoid carcinoma (28). In the present study, BZW1 was
identified as a target gene of miR-4500. The effects of miR-4500
mimic on OC cell functions were reversed by BZW1 overexpression.
The aforementioned results indicated that BZW1 may serve a
promoting role during OC progression, which was consistent with a
previous study (43).
Collectively, the present study indicated that NEAT1
may serve as an oncogene during OC progression. NEAT1 knockdown
facilitated cell apoptosis, and inhibited cell proliferation,
colony formation, migration, invasion and glycolysis. The effects
of NEAT1 knockdown were reversed by miR-4500 knockdown and the
tumor suppressive effects of miR-4500 were reversed by BZW1
overexpression. miR-4500 was identified as target of NEAT1 and
directly targeted BZW1. The results indicated that NEAT1 mediated
cell behavior and tumor progression via the miR-4500/BZW1 axis in
human OC. However, the present study did not identify the
regulatory mechanism underlying NEAT1 in OC progression and
initiation; therefore, the biological role of NEAT1 in human
diseases requires further investigation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HX conceived and designed the study. XS and YH
performed the experiments. QS and ML analyzed the data and wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of Jingzhou Hospital of Traditional Chinese Medicine, The
Third Clinical Medical College of Yangtze University. Written
informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:3347–30. 2018. View Article : Google Scholar
|
|
2
|
Eisenhauer EA: Real-world evidence in the
treatment of ovarian cancer. Ann Oncol. 28 (Suppl 8):VIII61–VVIII5.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rustin GJ, Van Der Burg MEL, Griffin CL,
Guthrie D, Lamont A, Jayson GC, Kristensen G, Mediola C, Coens C,
Qian W, et al: Early versus delayed treatment of relapsed ovarian
cancer (MRC OV05/EORTC 55955): A randomised trial. Lancet.
376:1155–1163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kroeger Jr PT and Drapkin R: Pathogenesis
and heterogeneity of ovarian cancer. Curr Opin Obstet Gynecol.
29:26–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mattick JS and Rinn JL: Discovery and
annotation of long noncoding RNAs. Nat Struct Mol Biol. 22:5–7.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z and Wang J: LncRNA HOXA11-AS promotes
proliferation and invasion of gastric cancer by scaffolding the
chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res.
76:6299–6310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peter S, Borkowska E, Drayton RM, Rakhit
CP, Noon A, Chen W and Catto JW: Identification of differentially
expressed long noncoding RNAs in bladder cancer. Clin Cancer Res.
20:5311–5321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang K, Liu CY, Zhou LY, Wang JX, Wang M,
Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, et al: APF lncRNA
regulates autophagy and myocardial infarction by targeting
miR-188-3p. Nat Commun. 6:67792015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang H, Yu T, Han Y, Jiang H, Wang C, You
T, Zhao X, Shan H, Yang R, Yang L, et al: LncRNA PTAR promotes EMT
and invasion-metastasis in serous ovarian cancer by competitively
binding miR-101-3p to regulate ZEB1 expression. Mol Cancer.
17:1192018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|
|
14
|
Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW,
Jin HY, Zhang Y, Li Q and Hua KQ: Overexpression of long non-coding
RNA HOTAIR predicts poor patient prognosis and promotes tumor
metastasis in epithelial ovarian cancer. Gynecol Oncol.
134:121–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen ZJ, Zhang Z, Xie BB and Zhang HY:
Clinical significance of up-regulated lncRNA NEAT1 in prognosis of
ovarian cancer. Eur Rev Med Pharmacol Sci. 20:3373–3377.
2016.PubMed/NCBI
|
|
16
|
An J, Lv W and Zhang Y: LncRNA NEAT1
contributes to paclitaxel resistance of ovarian cancer cells by
regulating ZEB1 expression via miR-194. Onco Targets Ther.
10:5377–5390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chai Y, Liu J, Zhang Z and Liu L:
HuR-regulated lnc RNA NEAT 1 stability in tumorigenesis and
progression of ovarian cancer. Cancer Med. 5:1588–1598. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zuberi M, Khan I, Mir R, Gandhi G, Ray PC
and Saxena A: Utility of serum miR-125b as a diagnostic and
prognostic indicator and its alliance with a panel of tumor
suppressor genes in epithelial ovarian cancer. PLoS One.
11:e01539022016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cai M, Chen Q, Shen J, Lv C and Cai L:
Epigenetic silenced miR-125a-5p could be self-activated through
targeting Suv39H1 in gastric cancer. J Cell Mol Med. 22:4721–4731.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu J, Miao J, Ding Y, Zhang Y, Huang X,
Zhou X and Tang R: MiR-4458 inhibits breast cancer cell growth,
migration, and invasiveness by targeting CPSF4. Biochem Cell Biol.
97:722–730. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jima DD, Zhang J, Jacobs C, Richards KL,
Dunphy CH, Choi WW, Au WY, Srivastava G, Czader MB, Rizzieri DA, et
al: Deep sequencing of the small RNA transcriptome of normal and
malignant human B cells identifies hundreds of novel microRNAs.
Blood. 116:e118–e127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Griffiths-Jones S, Grocock RJ, Van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang L, Qian J, Qiang Y, Huang H, Wang C,
Li D and Xu B: Down-regulation of miR-4500 promoted non-small cell
lung cancer growth. Cell Physiol Biochem. 34:1166–1174. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu FY, Tu Y, Deng Y, Guo C, Ning J, Zhu Y,
Lv X and Ye H: MiR-4500 is epigenetically downregulated in
colorectal cancer and functions as a novel tumor suppressor by
regulating HMGA2. Cancer Biol Ther. 17:1149–1157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mitra P, Vaughan PS, Stein JL, Stein GS
and van Wijnen AJ: Purification and functional analysis of a novel
leucine-zipper/nucleotide-fold protein, BZAP45, stimulating cell
cycle regulated histone H4 gene transcription. Biochemistry.
40:10693–10699. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li S, Chai Z, Li Y, Liu D, Bai Z, Li Y and
Situ Z: BZW1, a novel proliferation regulator that promotes growth
of salivary muocepodermoid carcinoma. Cancer Lett. 284:86–94. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kurman RJ, Carcangiu ML, Herrington CS and
Young RH: WHO classification of tumours of female reproductive
organs, fourth edition. Lyon: IACR; 2014
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun M, Liu X, Lu K, Nie F, Xia R, Kong R,
Yang J, Xu T, Liu Y, Zou Y, et al: EZH2-mediated epigenetic
suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell
proliferation and metastasis by affecting the
epithelial-mesenchymal transition. Cell Death Dis. 5:e12982014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mourtada-Maarabouni M, Pickard M, Hedge V,
Farzaneh F and Williams G: GAS5, a non-protein-coding RNA, controls
apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun M, Jin FY, Xia R, Kong R, Li JH, Xu
TP, Liu YW, Zhang EB, Liu XH and De W: Decreased expression of long
noncoding RNA GAS5 indicates a poor prognosis and promotes cell
proliferation in gastric cancer. BMC Cancer. 14:3192014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pickard MR and Williams GT: Regulation of
apoptosis by long non-coding RNA GAS5 in breast cancer cells:
Implications for chemotherapy. Breast Cancer Res Treat.
145:359–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hirose T, Virnicchi G, Tanigawa A,
Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Bénard M, Fox AH
and Pierron G: NEAT1 long noncoding RNA regulates transcription via
protein sequestration within subnuclear bodies. Mol Biol Cell.
25:169–183. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Choudhry H, Albukhari A, Morotti M, Haider
S, Moralli D, Smythies J, Schödel J, Green CM, Camps C, Buffa F, et
al: Tumor hypoxia induces nuclear paraspeckle formation through
HIF-2α dependent transcriptional activation of NEAT1 leading to
cancer cell survival. Oncogene. 34:4482–4490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chakravarty D, Sboner A, Nair SS,
Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K,
Kossai M, et al: The oestrogen receptor alpha-regulated lncRNA
NEAT1 is a critical modulator of prostate cancer. Nat Commun.
5:53832014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ding N, Wu H, Tao T and Peng E: NEAT1
regulates cell proliferation and apoptosis of ovarian cancer by
miR-34a-5p/BCL2. Onco Targets Ther. 10:4905–4915. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing miRNA-lncRNA interactions. Methods Mol Biol.
1402:271–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jalali S, Bhartiya D, Lalwani MK,
Sivasubbu S and Scaria V: Systematic transcriptome wide analysis of
lncRNA-miRNA interactions. PLoS One. 8:e538232013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li JH, Zhang SQ, Qiu XG, Zhang SJ, Zheng
SH and Zhang DH: Long non-coding RNA NEAT1 promotes malignant
progression of thyroid carcinoma by regulating miRNA-214. Int J
Oncol. 50:708–716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu F, Zhao H, Gong L, Yao L, Li Y and
Zhang W: MicroRNA-129-3p functions as a tumor suppressor in serous
ovarian cancer by targeting BZW1. Int J Clin Exp Pathol.
11:5901–5908. 2018.PubMed/NCBI
|