Introduction
Ovarian cancer (OC) is a major malignant tumor of
the female reproductive system and has a high mortality rate. A
study demonstrated that 47% of women diagnosed with ovarian cancer
survive five years after diagnosis worldwide (1). As the early symptoms are not obvious,
the disease onset is hidden and metastasis occurs earlier, which
poses a great threat to women's health (2). The invasion and metastasis of OC is a
multifactor and multistep process that is regulated by multiple
factors such as lncRNA PTAR (3),
and miR-320 (4) and oncoprotein
Rab23 (5). However, currently, the
molecular mechanisms are not completely elucidated.
Long non-coding (lnc.) RNAs are non-coding RNAs with
specific sequences >200 nucleotides in length, which are
conserved during mammalian evolution (6,7).
Previous studies have reported that lncRNAs participate in several
biological processes, such as proliferation (8), metabolism (9) and metastasis (10). Moreover, lncRNAs, such as HOTTIP
(11), DNM3OS (12) and PTAR (3), are suggested to have a vital role in
the development and progression of OC. LncRNA nuclear enriched
abundant transcript 1 (NEAT1) has been revealed to be dysregulated
in various human cancer types, such as cervical cancer (13), breast cancer (14) and melanoma (15). Inhibition of NEAT1 is known to
attenuate the proliferation, migration and invasion of melanoma
cells (16); for example, a study
has previously shown that NEAT1 is upregulated in papillary thyroid
carcinoma tissues and cell lines, and can promote proliferation,
invasion and migration (17).
However, the function of NEAT1 in OC is currently unknown.
Epithelial-mesenchymal transition (EMT) is a unique
biological process, which refers to the process of transforming
polar epithelial cells into mesenchymal cells with mobility
(18). From the perspective of
molecular biology, EMT mainly involves the downregulated expression
of epithelial cell markers, E-cadherin and β-catenin, and the
upregulated expression of the interstitial cell markers,
fibronectin and Vimentin (19). In
recent years, the relationship between lncRNA and tumor EMT has
been widely reported, and numerous lncRNAs have been shown to
promote the EMT process via different molecular mechanisms in
several tumors. For instance, lncRNA SNHG5 induces zinc finger
E-box binding homeobox 1 expression by competitively inhibiting
microRNA (miRNA/miR)-205-5p, thus promoting the EMT process in
clear cell renal cell carcinoma (20). Furthermore, lncRNA AFAP1-AS1
promotes the EMT process of liver cancer by acting on the oncogenic
protein CRKL-mediated Ras/mitogen-activated protein kinase
kinase/c-Jun and the cadherin/vimentin signaling pathway (21), while lncRNA ROR1-AS1 promotes the
EMT process of cervical cancer via the Wnt/β-catenin pathway
(22).
The role of miRNA in the progression of
carcinogenesis has been well characterized (23,24).
Previous studies have reported that lncRNAs can regulate the
expression of miRNAs as competitive endogenous RNAs (25). For example, HOXD-AS1 was found to
inhibit the expression of miR-133a-3p and subsequently enhance the
EMT process of OC (26), while
lncRNA H19 was determined to be an endogenous competitive RNA that
inhibits miR-370-3p expression and further promotes the EMT process
(27). However, to the best of our
knowledge, there are currently no reports as to whether NEAT1
affects the biological features of OC cells via miRNA.
A study suggested that more than 90% of malignant
ovarian tumors are of epithelial origin (28). Tight junction protein 3 (TJP3),
also named ZO-3, is a scaffolding protein (29) that has a significant role in
intercellular communication (30)
and epithelial differentiation (31). Thus, we hypothesized that TJP3
plays a crucial role in OC. In fact, the role of TJP3 in tumor
progression has been clarified. TJP3 is required for
4-(furan-2-yl)-2-(pyridin-2-yl)-5, 6-dihydro-1, 10-phenanthroline
(FPDHP)-regulated cancer therapy (32). In prostate cancer, hepatocyte
growth factor (HGF) modulates metastasis through TJ proteins
including TJP3 (33). TJP3 is
proposed as a diagnostic tool for lung cancer (34) and is associated with poor prognosis
in patients with breast cancer (35). Moreover, in endometrial cancer,
TJP3 functions as a molecular marker (36). However, the potential role of TJP3
in the progression of OC remains unknown.
Previous studies demonstrated lncRNA NEAT1 has an
important significance on OC (37,38).
Bioinformatics analysis indicated miR-1321 is a target of NEAT. It
has been reported that Cordyceps militaris could reduce the
severity of acute lung injury in mice via miR-1321- and
miR-3188-mediated C-X-C motif chemokine receptor 2 inhibition
(39). miR-1321 was also revealed
to be abnormally expressed in pediatric glioma using miRNA
microarray analysis (40). Based
on the aforementioned findings, it was hypothesized that miR-1321
has an important role in the process of cancer development. In this
study, the role of NEAT1 in OC cell invasion and metastasis were
well characterized. This study also demonstrated the underlying
mechanism by which NEAT1 modulates OC cell invasion and
metastasis.
Materials and methods
Specimens collection
Surgical samples (tumor tissues and matched adjacent
healthy tissues) were collected from 36 female patients with
ovarian cancer from 05/2018-02/2019 in Department of Gynecology,
Yunnan Tumor Hospital & The Third Affiliated Hospital of
Kunming Medical University. The age range age of patients was 25–65
years old with an average age of 42.56±12.12 years old. All
specimen diagnoses were confirmed by a qualified pathologist after
surgery. The study was approved by the Research Ethics Committee of
Kunming Medical University and written informed consent was
obtained from each patient.
Cell culture
The epithelial normal cell line (IOSE80) and four OC
cell lines (OVCAR-3, SKOV3, ES-2 and A2780) were purchased from
American Type Culture Collection. 293T cells was purchased from
Thermo Fisher Scientific, Inc. All the cell lines were cultured in
DMEM (cat. no. 12100; Beijing Solarbio Science & Technology
Co., Ltd.) supplemented with 10% FBS (cat. no. 11041-8611; Beijing
Solarbio Science & Technology Co., Ltd.), 100 µg/ml
streptomycin and 100 IU/ml penicillin at 37°C in a 5%
CO2 chamber.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was isolated from the OC cells using
TRIzol® reagent (cat. no. 15596026; Invitrogen; Thermo
Fisher Scientific, Inc.). 0.5 µg total RNA was reverse transcribed
to cDNA via PrimeScript™ RT Master Mix (cat. no. RR036A;
Takara Bio, Inc.). The cDNA was diluted 1:10. The reaction
conditions were as follows: 6 cycles of 15 min at 37°C, 85°C for 5
sec. The detection of NEAT1 expression was achieved using
SYBR® Premix Ex Taq™ II kit (cat. no. RR820A;
Takara Bio, Inc.) with the LightCycler® 96 instrument
(Roche Diagnostics). GAPDH was used as an internal control to
standardize the results. The reaction conditions were as follows:
95°C for 30 sec; 40 cycles of 5 sec at 95°C, 30 sec at 60°C and 15
sec at 65°C, and 10 min at 72°C. The sequences of the specific
primers are as follows: lncRNA NEAT1 forward,
5′-TCGGGTATGCTGTTGTGAAA-3′ and reverse,
5′-TGACGTAACAGAATTAGTTCTTACCA-3′; and GAPDH forward,
5′-GGTCTCCTCTGACTTCAACA-3′ and reverse, 5′-GTGAGGGTCTCTCTCTTCCT-3′.
miR-1321 forward, 5′-GCGGCGGCAGGGAGGTGAATGTG-3′ and reverse,
5′-ATCCAGTGCAGGGTCCGAGG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-GCAGGGTCCGAGGTATTC−3′. The PureLink™ miRNA
Isolation kit (cat. no. K157001; Invitrogen; Thermo Fisher
Scientific, Inc.) was used for miR-1321 acquisition, and the
quantification of miRNA expression was performed with a TaqMan
MicroRNA Assay kit (cat. no. 4366593; Applied Biosystems; Thermo
Fisher Scientific, Inc.). The expression was measured using the
2−ΔΔCq method (41).
Western blotting
OC cell lysates were harvested using RIPA lysis
buffer (cat. no. R0010; Beijing Solarbio Science & Technology
Co., Ltd.). The protein concentrations were determined using a
bicinchoninic acid kit (cat. no. P0012, Beyotime Institute of
Biotechnology) and 20 µg proteins were loaded in to each well.
Proteins were separated using 10% SDS-PAGE and transferred to PVDF
membranes (cat. no. IPVH00010; Merck KGaA). The membranes were
blocked with 10% milk at room temperature for 1 h. Cells were
blotted with specific primary antibodies against E-cadherin
(1:3,000; cat. no. ab15148; Abcam), N-cadherin (1:3,000; cat. no.
ab18203; Abcam), vimentin (1:3,000; cat. no. ab11256; Abcam), TJP3
(1:2,000; cat. no. 710857; Thermo Fisher Scientific, Inc.) or GAPDH
(1:3,000; cat. no. ab9485; Abcam) at 4°C overnight and then
incubated with a horseradish peroxide-conjugated goat anti-rabbit
IgG H&L secondary antibody (1:10,000; cat. no. ab205718; Abcam)
for 1 h at room temperature. The signal of proteins was visualized
using ECL detection reagent (cat. no. 36208ES60, Shanghai Yeasen
Biotechnology Co., Ltd.) with a Tanon-5200 imaging system (Tanon
Science & Technology Co., Ltd.). ImageJ 1.8.0 software
(National Institutes of Health) was used to quantify the western
blots.
Cell transfection
The small interfering (si)RNA, specifically
targeting the lncRNA NEAT1 (si-NEAT1; sense:
5′-AGUUUAGCGCCAAACCUAGAG−3′; antisense:
5′-CUAGGUUUGGCGCUAAACUCU−3′), and TJP3 (si-TJP3; sense:
5′-AAUUAGCUGGGCAUAGUGGUG−3′; antisense:
5′-CCACUAUGCCCAGCUAAUUUU−3′) were obtained from RiboBio. miR-1321
mimics and the pcDNA-TJP3 overexpression plasmid (OE-TJP3) were
constructed by Cytiva. Cells were plated on 6-well plates
(2×105 cells/well). Plasmids (2 µg) and siRNAs
(1.5 µg) were diluted in 50 µl medium with 1 µl
Lipofectamine® 2000 (cat. no. 11668-027; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The mixture was added to the wells after 15 min. Cells
were co-cultured with the plasmid and/or siRNA and
Lipofectamine® 2000 mixture for 24 h in a humidified 5%
CO2 incubator at 37°C and then replaced with complete
medium containing 10% FBS.
Cell proliferation assays
Transfected cells and their corresponding controls
were cultured in 96-well plates (2×103 cells/well).
Proliferation was assessed using the Cell Counting Kit-8 assay
(CCK-8; Beyotime Institute of Biotechnology) at 0, 24, 48, 72 and
96 h according to the manufacturer's protocol. The absorbance at
450 nm was detected with a microplate reader (Bio-Rad Laboratories
Inc.).
Transwell assays
Transwell assays were performed using Millipore
Transwell chambers (8-µm pore size; cat. no. MCEP06H48; EMD
Millipore). Transfected ES-2 and A2780 cells (2×104
cells/well) were seeded in the upper chambers of a 12-well plate
(Corning, Inc.) in 500 µl serum-free medium with 500 µl complete
medium added to the bottom chamber. The chamber was incubated for
24 h in a humidified 5% CO2 incubator at 37°C. At the
end of incubation, cotton-tipped swabs were used to remove the
non-migratory and non-invading cells. For invasion assays, the
upper chamber was precoated with Matrigel (EMD Millipore) for 30
min at 37°C. Then, the remaining cells were fixed with formaldehyde
and stained using 0.1% crystal violet at 37°C for 1 h. The images
were captured using an Olympus CKX31 inverted microscope (Olympus
Corporation) at ×100 magnification and quantified by ImageJ 1.8.0
software (National Institutes of Health).
Wound-healing assays
Transfected ES-2 and A2780 cells and their
corresponding controls (cell density of 3×105/ml) were
cultured in 6-well plates (Sigma-Aldrich; Merck KGaA) in DMEM
supplemented with 10% FBS, overnight to form a confluent monolayer.
The 500 µm scratch was performed on this monolayer with a 1-ml
pipette tip under sterile conditions and then the cells were washed
twice with serum-free culture media. The cells were allowed to
migrate freely across the scratch. Images were captured with a
digital camera (Canon, Inc.) after 24 h. Marks were made at the
bottom of the dish to ensure that all images were captured at the
same site. The area within the scratch occupied by cells was taken
at 0 and 24 h under an inverted microscope (Olympus) at ×100
magnification and calculated with ImageJ 1.8.0 software (National
Institutes of Health). The percentage of gap closure was used as an
indicator of cell migration.
Dual-luciferase reporter assays
The bioinformatics prediction website starBase
(http://starbase.sysu.edu.cn) (42,43)
was used to predict the complementary binding sites between NEAT1
or TJP3 and miR-1321. A fragment of the wild-type (WT) and mutated
(MUT) 3′-untranslated region (UTR) NEAT1 or TJP3 containing the
predicted miR-1321 targeting regions was amplified (Guangzhou
FulenGen Co. Ltd.) and cloned into the pGL3-reporter
luciferase reporter vector (Sigma-Aldrich; Merck KGaA) at
Kpn I and Hind III restriction sites or Hind
III and Xho I restriction sites by PCR. The PCR
thermocycling protocol was as follows: 98°C for 3 min; 30 cycles of
30 sec at 98°C, 30 sec at 57°C and 3 min at 72°C; 10 min at 72°C.
The PCR sequences were as follows: NEAT1 forward,
5′-TCGACACCCCTTCTTCCTCCCTT-3′ and reverse,
3′-AGCTAAGGGAGGAAGAAGGGGTG-5′, TJP3 forward,
5′-TCGACCCACAACCTTACCTCCCTC−3′ and reverse,
3′-AGCTGAGGGAGGTAAGGTTGTGGG−5′.
293T cells (Thermo Fisher Scientific, Inc.) were
co-transfected with either 20 nM TJP3-3′UTR-wt or TJP3-3′UTR-mut
with miR-1321 mimic or negative control (NC) mimics using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and incubated at 37°C in a 5% CO2
chamber for 48 h. the luciferase activity of the harvested cells
was measured by Dual-Luciferase Assay reagent (Promega Corporation)
according to the manufacturer's protocol. Renilla luciferase
was normalized to firefly luciferase activity.
Statistical analysis
Data are presented as the mean ± standard deviation
of ≥3 independent repeats. Unpaired Student's t-test was used to
analyze the differences between the two groups. One-way ANOVA with
Tukey's post hoc test was used to analyze the differences between
multiple groups. A Pearson correlation coefficient test was used to
detect the relationship between lncRNA NEAT1, miR-1321 and TJP3.
P<0.05 was considered to indicate a statistically significant
difference.
Results
lncRNA NEAT1 expression and function
in OC
NEAT1 expression in normal epithelial cells (IOSE80)
and four OC cell lines (SKOV3, OVCAR-3, ES-2 and A2780) was
measured by RT-qPCR. NEAT1 was highly expressed in OC cells,
especially in ES-2 and A2780 cells (P<0.05; Fig. 1A). Therefore, ES-2 and A2780 were
selected for the subsequent experiments.
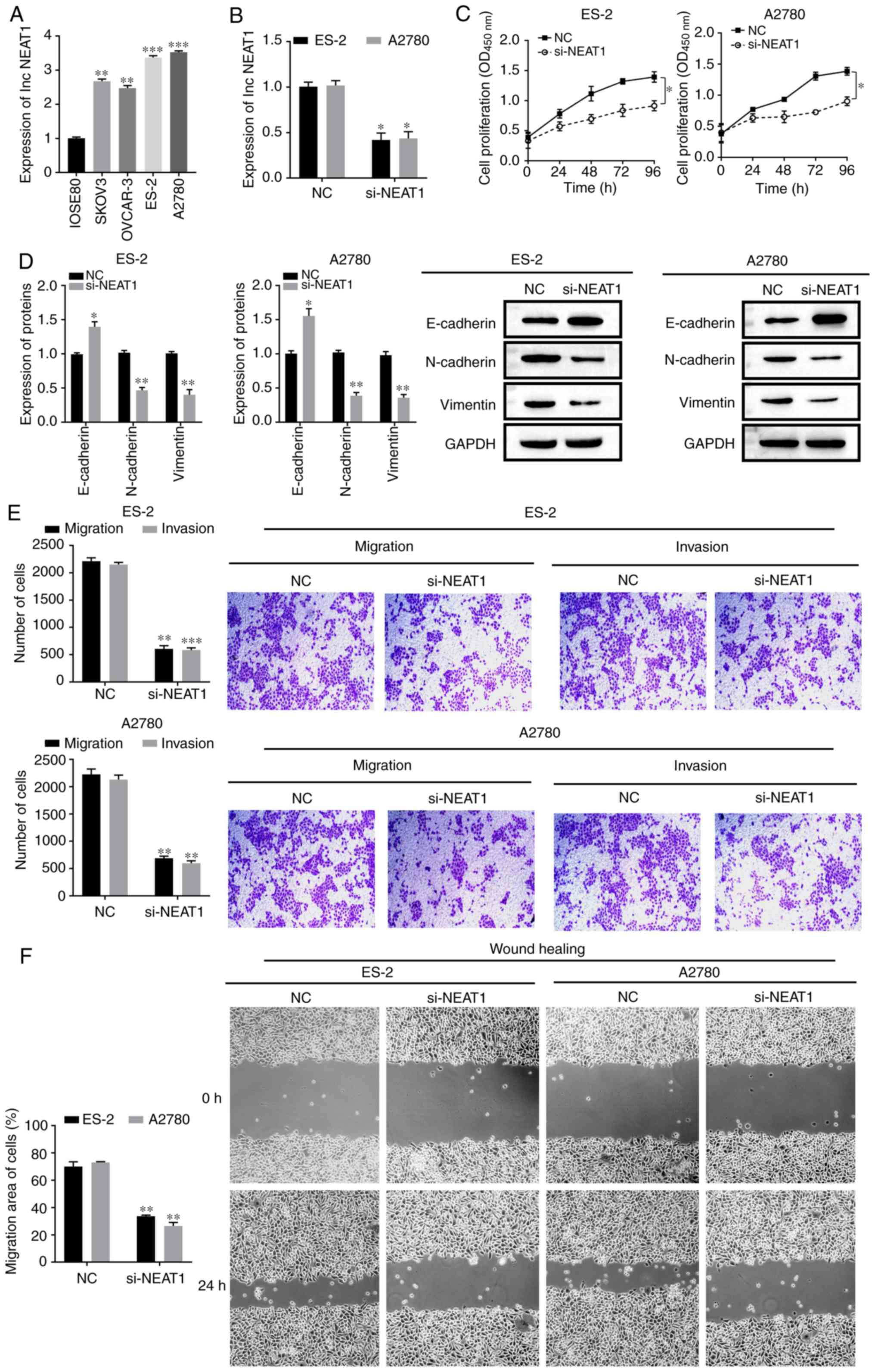 | Figure 1.NEAT1 expression and function in OC.
(A) NEAT1 expression in normal epithelial cells (IOSE80) and OC
cells (SKOV3, OVCAR-3, ES-2 and A2780) and (B) transfection
efficiency of si-NEAT1 in ES-2 and A2780 cells were detected using
reverse transcription-quantitative PCR. (C) Proliferative abilities
of ES-2 and A2780 cells were measured using Cell Counting Kit-8
assays. (D) E-cadherin, N-cadherin and Vimentin protein expression
level in ES-2 and A2780 cells were assessed using western blotting.
(E) Migration and invasive abilities of ES-2 and A2780 cells were
measured using Transwell assays. Scale bar, 100 µm. (F) Migration
abilities of ES-2 and A2780 cells were measured using wound-healing
assays. Scale bar, 100 µm. All data are representative of ≥3
independent experiments. Data are presented as the mean ± SD.
*P<0.05 vs. control group; **P<0.01 vs. control group;
***P<0.001 vs. control group, as determined by Student's t-tests
or ANOVA followed by a Tukey's post hos test. OC, ovarian cancer;
NEAT1, nuclear enriched abundant transcript1; si, small interfering
RNA; NC, negative control; OD, optical density. |
The function of NEAT1 in OC cells was subsequently
analyzed via NEAT1 knockdown in ES-2 and A2780 cells. NEAT1
expression was successfully depleted in ES-2 and A2780 cells
transfected with si-NEAT1 compared with the NC group (P<0.05;
Fig. 1B). CCK-8 results
demonstrated that cell proliferation was decreased following
downregulation of NEAT1 (P<0.05; Fig. 1C). Moreover, it was found that
NEAT1 knockdown led to the upregulation of the E-cadherin protein
and downregulation of N-cadherin and vimentin expression levels
(P<0.05; Fig. 1D), which in
turn, suggested that NEAT1 inhibition attenuated cell metastasis.
Transwell assays and wound-healing assays also indicated that NEAT1
knockdown significantly reduced the migratory and invasive
abilities of OC cells (P<0.05; Fig.
1E and F). These results demonstrated that NEAT1 may serve a
regulatory role in cell EMT, as well as the migratory and invasive
potential of OC.
Interrelation of lncRNA NEAT1,
miR-1321 and TJP3
The present study found that NEAT1 and miR-1321 had
complementary base sequences using the bioinformatics prediction
website starBase (Fig. 2A).
miR-1321 has been shown to be related to cancer genesis (40), but the function of miR-1321 in
cancer remains unknown. To assess the relationship between NEAT1
and miR-1321, a dual-luciferase reporter assay was performed after
the transfection efficiency of the miR-1321 mimics was verified
(P<0.05; Fig. 2B). It was
demonstrated that luciferase activity was decreased in 293T cells
co-transfected with NEAT1 WT and miR-1321 mimics (P<0.05;
Fig. 2C), while there was no
change in NEAT1 MUT. The target genes of miR-1321 were also
investigated using starBase (Fig.
2D), and TJP3 was found to be a target gene of miR-1321.
Furthermore, the results indicated that transfection with miR-1321
mimics decreased the luciferase activity of TJP3 WT (P<0.05;
Fig. 2E) but did not influence the
luciferase activity of TJP3 MUT.
 | Figure 2.Interrelation of NEAT1, miR-1321 and
TJP3. (A) StarBase website predicted the complementary binding
sites between NEAT1 and miR-1321. (B) Transfection efficiency of
miR-1321 mimics was detected using RT-qPCR. (C) Targeting
relationship of NEAT1 and miR-1321 was identified using
dual-luciferase reporter gene assays. (D) StarBase website
predicted the complementary binding site between miR-1321 and TJP3.
(E) Targeting relationship of miR-1321 and TJP3 was identified
using dual-luciferase reporter gene assays. (F) Expression of
miR-1321 in ES-2 and A2780 cells was measured using RT-qPCR. (G)
Effect of NEAT1 on miR-1321 expression was determined using
RT-qPCR. (H) Effect of miR-1321 on expression of TJP3 protein was
measured using western blotting. Correlations of (I) lncRNA NEAT1
and miR-1321, (J) miR-1321 and TJP3, and (K) lncRNA NEAT1 and TJP3
on ovarian cancer specimens were investigated using a Pearson
Correlation Coefficient. Data are representative of ≥3 independent
experiments. Data are presented as the mean ± SD. *P<0.05 vs.
control group; **P<0.01 vs. control group as determined by
Student's t-tests or ANOVA followed by a Tukey's post hos test.
NEAT1, nuclear enriched abundant transcript1; TJP3, tight junction
protein 3; miR, microRNA; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; lncRNA, long non-coding RNA; NC,
negative control; WT, wild-type; MUT, mutant; si, small interfering
RNA. |
RT-qPCR results demonstrated that miR-1321
expression was downregulated in ES-2 and A2780 cells (P<0.05;
Fig. 2F). Then, the relationship
between lncRNA NEAT1, miR-1321 and TJP3 was analyzed. The results
identified that miR-1321 expression was upregulated in ES-2 and
A2780 cells following knockdown of NEAT1 (P<0.01; Fig. 2G). Furthermore, transfection with
an miR-1321 mimic led to the downregulation of TJP3 protein
expression in ES-2 and A2780 cells (P<0.01; Fig. 2H).
Pearson's correlation analysis was conducted to
assess the relationship between NEAT1, miR-1321 and TJP3 in OC
specimens. The results demonstrated a weak negative correlation
between miR-1321 and NEAT1 expression levels, as well as between
miR-1321 and TJP3 (Fig. 2I and J),
and a weak positive correlation between NEAT1 and TJP3 (Fig. 2K). The aforementioned findings
indicated that NEAT1 regulated TJP3 expression by sponging
miR-1321.
TJP3 expression and function in
OC
To determine the role of TJP3 in OC, the expression
of TJP3 was first detected in normal epithelial cells (IOSE80) and
OC cells (ES-2 and A2780). The protein expression of TJP3 was
enhanced in ES-2 and A2780 cells compared with IOSE80 cells
(P<0.05; Fig. 3A). Then, ES-2
and A2780 cells were transfected with si-TJP3. The results
demonstrated that, compared with the NC group, the protein
expression of TJP3 was decreased following si-TJP3 transfection in
ES-2 and A2780 cells (P<0.05; Fig.
3B). In addition, TJP3 knockdown led to a reduction in the cell
proliferation compared with the NC (P<0.05; Fig. 3C). The protein expression of the
EMT marker E-cadherin was increased after knockdown of TJP3, but
the protein expression levels of N-cadherin and vimentin were
decreased in both ES-2 and A2780 cells (P<0.05; Fig. 3D). In addition, Transwell and
wound-healing assays indicated that the migratory and invasive
abilities of ES-2 and A2780 cells were reduced following the
knockdown of TJP3 (P<0.05; Fig. 3E
and F). These findings suggested that TJP3 regulated the EMT,
migratory and invasive potential of OC cells.
lncRNA NEAT1 promotes cell EMT,
migration and invasion of OC by regulating TJP3
To further investigate whether NEAT1-induced EMT in
OC cells was mediated by TJP3, ES-2 and A2780 cells were
transfected with si-NEAT1 and/or OE-TJP3. NEAT1 was downregulated
in ES-2 and A2780 cells using si-NEAT1 transfection, and TJP3 was
found to be overexpressed following treatment with OE-TJP3
(P<0.05; Figs. 4A and S1).
Western blotting was used to measure the effect of
NEAT1 on TJP3 expression. The results identified that protein
expression of TJP3 was significantly decreased by knocking down
NEAT1 in ES-2 and A2780 cells; however, the inhibitory effect was
alleviated by overexpression of TJP3 (P<0.05; Fig. 4B). Cell proliferative abilities
were also decreased by NEAT1 knockdown, but TJP3 overexpression
partly rescued the proliferative abilities of ES-2 and A2780 cells
(P<0.05; Fig. 4C). The protein
expression of E-cadherin was enhanced by si-NEAT1 transfection,
while the protein expression levels of N-cadherin and vimentin were
decreased (P<0.05; Fig. 4D).
However, these EMT-related protein expression levels in the
si-NEAT1 + OE-TJP3 group were the same as those in the NC group
(P>0.05; Fig. 4D), which
indicated that NEAT1 was acting independently from TJP3 in the
regulation of these EMT-related proteins. Compared with the NC
group, si-NEAT1 transfection resulted in reduced cell migratory and
invasive abilities according to the Transwell and wound-healing
assay results. However, cell migratory and invasive abilities were
significantly rescued by si-NEAT1 + OE-TJP3 (P<0.05; Fig. 4E and F). Therefore, the results
suggested that NEAT1 promoted EMT, migration and invasion of OC
cells by regulating TJP3, but also potentially via regulating
proteins that are involved in cell migration and invasion, which
were not included in the present study.
Discussion
The present study demonstrated that NEAT1 was highly
expressed in OC cells and knockdown of NEAT1 attenuated their
migratory and invasive abilities. Moreover, the present study
provided evidence that NEAT1 sponges miR-1321 to enhance TJP3
protein expression, thus promoting tumor invasion and migration.
The significance of the NEAT1/miR-1321/TJP3 axis in the
pathogenesis of OC was also identified, which suggested the
potential role of NEAT1 in OC diagnosis and treatment.
Previous studies have reported the involvement of
lncRNAs in the occurrence and development of a variety of cancer
types, including OC (10–12). A recent study found that knockdown
of NEAT1 suppressed cell proliferation, migration and invasion of
melanoma cells (15). In
colorectal cancer, NEAT1 has been shown to activate Wnt/β-catenin
and promote cancer progression (44). Additionally, NEAT1 has been
revealed to be responsible for tumor chemoresistance (45,46).
The present results identified that NEAT1 was also significantly
upregulated in OC. In addition, it was found that knockdown of
NEAT1 led to the inhibition of migration and invasion in OC
cells.
EMT is a unique biological process that promotes the
metastasis of breast cancer (47),
bladder cancer (48) and OC
(49). It was reported that forced
upregulation of NEAT1 could induce metastasis in hepatocellular
cancer types via EMT (50), while
another study has shown that NEAT1 affected the migratory and
invasive abilities of gastric cancer cells via EMT (51). Consistent with previous studies,
the present results demonstrated that knockdown of NEAT1 had an
impact on EMT-associated proteins, including E-cadherin, vimentin
and N-cadherin, which indicated that NEAT1 regulates OC cell
invasion and migration via EMT.
lncRNAs have been reported to sponge miRNAs via a
competitive endogenous mechanism, thus antagonizing the function of
miRNAs (52). NEAT1 upregulates
Homeobox A13 by targeting miR-34a-5p and regulates the development
of osteosarcoma (53). NEAT1 also
promotes cervical cancer cell proliferation by sponging miR-9-5p
(13). Thus, it was hypothesized
that NEAT1 may act as a competitive endogenous RNA and interact
with miRNAs in OC. Bioinformatic analysis and luciferase analysis
were performed to screen miRNAs that directly bind to NEAT1
transcripts. NEAT1 was found to directly bind to miR-1321 and there
was a mutual inhibitory effect between NEAT1 and miR-1321 in OC
cells. Collectively, these data indicated that NEAT1 affects the
biological characteristics of OC cells by regulating miR-1321
function.
Normal epithelial cells act as sentries in most
living systems, providing a protective barrier to various organs
and the surrounding environment, helping to maintain homeostasis
(54). The intercellular
connections of this protective barrier are divided into TJs,
adhesion junctions and desmosome junctions. Among them, TJs are
mainly composed of occludin, claudins and junction adhesion
molecules (55). The constituent
proteins of TJs participate in the modulation of intercellular
communication and paracellular transport (56). Among them, TJPs, such as TJP1, 2
and 3, are framework-forming proteins that connect transmembrane
proteins and the actin cytoskeleton (57). Downregulation of TJPs have been
reported to be related to tumor development, such as a decrease in
TJP3 expression leading to an increased capacity for migration of
pancreatic cancer cells (58). A
study indicated that TJP3 mRNA is upregulated in endometrial
carcinoma (36). To the best of
our knowledge, the present study was the first to identify that
TJP3 was a direct target of miR-1321. Considering the interaction
of NEAT1 and miR-1321, it was hypothesized that NEAT1 could
regulate TJP3 expression in OC. As expected, the present results
demonstrated that TJP3 expression was positively correlated with
NEAT1 in OC cells. It was also found that TJP3 attenuated the
influence of NEAT1 knockdown on OC cells. These results suggested
that NEAT1 regulated TJP3 expression by interacting with miR-1321.
However, in the current research, NEAT1 overexpression experiments
were not performed to confirm the regulatory relationship between
NEAT1 and TJP3. Therefore, further studies will be conducted to
further confirm the regulation between NEAT1 and TJP3.
In conclusion, the present data suggested that NEAT1
was upregulated in OC. Thus, the current study has provided
evidence that NEAT1 promotes OC cell invasion and metastasis by
downregulating miR-1321 and activating TJP3 function. Therefore,
the NEAT1/miR-1321/TJP3 axis may represent a new prognostic
biomarker and therapeutic target in OC.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Malin Li
(Yunnan University of Chinese Medicine) for her advice on this
project.
Funding
The present study was funded by the National Natural
Science Foundation (grant no. 81860463), the Science and Technology
Department of Yunnan Province and Kunming Medical University
Project Foundation [grant no. 2018FE001(−003)] and the Science and
Technology Department of Yunnan Province and Kunming Medical
University Project Foundation [grant no. 2017FE468(−067)].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CQ designed the research. ML performed the research,
analyzed the data and wrote the manuscript. LZ collected clinical
specimens and acquired the clinical data. HY participated in
analyzing the clinical specimens data. KL perfomed the cell
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Research Ethics
Committee of Kunming Medical University and written informed
consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:3429–34. 2019. View Article : Google Scholar
|
|
2
|
Zhao L, Wang W, Huang S, Yang Z, Xu L,
Yang Q, Zhou X, Wang J, Shen Q, Wang C, et al: The RNA binding
protein SORBS2 suppresses metastatic colonization of ovarian cancer
by stabilizing tumor-suppressive immunomodulatory transcripts.
Genome Biol. 19:352018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liang H, Yu T, Han Y, Jiang H, Wang C, You
T, Zhao X, Shan H, Yang R, Yang L, et al: lncRNA PTAR promotes EMT
and invasion-metastasis in serous ovarian cancer by competitively
binding miR-101-3p to regulate ZEB1 expression. Mol Cancer.
17:1192018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang W, Yang J, Xiang YY, Pi J and Bian J:
Overexpression of hsa-miR-320 is associated with invasion and
metastasis of ovarian cancer. J Cell Biochem. 118:3654–3661. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao L, Zheng M, Guo Q, Nie X, Li X, Hao Y,
Liu J, Zhu L and Lin B: Downregulation of Rab23 inhibits
proliferation, invasion, and metastasis of human ovarian cancer.
Int J Biochem Cell Biol. 116:1056172019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhan L, Li J and Wei B: Long non-coding
RNAs in ovarian cancer. J Exp Clin Cancer Res. 37:1202018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bu D, Luo H, Jiao F, Fang S, Tan C, Liu Z
and Zhao Y: Evolutionary annotation of conserved long non-coding
RNAs in major mammalian species. Sci China Life Sci. 58:787–798.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen QN, Chen X, Chen ZY, Nie FQ, Wei CC,
Ma HW, Wan L, Yan S, Ren SN and Wang ZX: Long intergenic non-coding
RNA 00152 promotes lung adenocarcinoma proliferation via
interacting with EZH2 and repressing IL24 expression. Mol Cancer.
16:172017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nolte W, Weikard R, Brunner RM, Albrecht
E, Hammon HM, Reverter A and Kühn C: Biological network approach
for the identification of regulatory long non-coding RNAs
associated with metabolic efficiency in cattle. Front Genet.
10:11302019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yao Z, Xiong Z, Li R, Liang H, Jia C and
Deng M: Long non-coding RNA NRON is downregulated in HCC and
suppresses tumour cell proliferation and metastasis. Biomed
Pharmacother. 104:102–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zou T, Wang PL, Gao Y and Liang WT: Long
noncoding RNA HOTTIP is a significant indicator of ovarian cancer
prognosis and enhances cell proliferation and invasion. Cancer
Biomark. 25:133–139. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mitra R, Chen X, Greenawalt EJ, Maulik U,
Jiang W, Zhao Z and Eischen CM: Decoding critical long non-coding
RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat
Commun. 8:16042017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie Q, Lin S, Zheng M, Cai Q and Tu Y:
Long noncoding RNA NEAT1 promotes the growth of cervical cancer
cells via sponging miR-9-5p. Biochem Cell Biol. 97:100–108. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang X, Zhou Y, Sun AJ and Xue JL: NEAT1
contributes to breast cancer progression through modulating miR-448
and ZEB1. J Cell Physiol. 233:8558–8566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xia Y, Zhou Y, Han H, Li P, Wei W and Lin
N: lncRNA NEAT1 facilitates melanoma cell proliferation, migration,
and invasion via regulating miR-495-3p and E2F3. J Cell Physiol.
234:19592–19601. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ding F, Lai J, Gao Y, Wang G, Shang J,
Zhang D and Zheng S: NEAT1/miR-23a-3p/KLF3: A novel regulatory axis
in melanoma cancer progression. Cancer Cell Int. 19:2172019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Cai Y, Zheng L, Zhang Z, Lin X
and Jiang N: Long noncoding RNA NEAT1 regulate papillary thyroid
cancer progression by modulating miR-129-5p/KLK7 expression. J Cell
Physiol. 233:6638–6648. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aiello NM and Kang Y: Context-dependent
EMT programs in cancer metastasis. J Exp Med. 216:1016–1026. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiang W, Lv L, Zhou G, Wu W, Yuan J, Zhang
C and Jiang G: The lncRNA SNHG5-mediated miR-205-5p downregulation
contributes to the progression of clear cell renal cell carcinoma
by targeting ZEB1. Cancer Med. 9:4251–4264. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abdul S, Majid A, Wang J, Liu Q, Sun MZ
and Liu S: Bidirectional interaction of lncRNA AFAP1-AS1 and CRKL
accelerates the proliferative and metastatic abilities of
hepatocarcinoma cells. J Adv Res. 24:121–130. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang L, Yao HR, Liu SK and Song LL: Long
noncoding RNA ROR1-AS1 overexpression predicts poor prognosis and
promotes metastasis by activating Wnt/β-catenin/EMT signaling
cascade in cervical cancer. Eur Rev Med Pharmacol Sci.
24:2928–2937. 2020.PubMed/NCBI
|
|
23
|
Lai X, Eberhardt M, Schmitz U and Vera J:
Systems biology-based investigation of cooperating microRNAs as
monotherapy or adjuvant therapy in cancer. Nucleic Acids Res.
47:7753–7766. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wilk G and Braun R: Integrative analysis
reveals disrupted pathways regulated by microRNAs in cancer.
Nucleic Acids Res. 46:1089–1101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wei L, Wang X, Lv L, Liu J, Xing H, Song
Y, Xie M, Lei T, Zhang N and Yang M: The emerging role of microRNAs
and long noncoding RNAs in drug resistance of hepatocellular
carcinoma. Mol Cancer. 18:1472019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y, Dun Y, Zhou S and Huang XH:
lncRNA HOXD-AS1 promotes epithelial ovarian cancer cells
proliferation and invasion by targeting miR-133a-3p and activating
Wnt/β-catenin signaling pathway. Biomed Pharmacother. 96:1216–1221.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Huang Y, Deng X, Luo M, Wang X, Hu
H, Liu C and Zhong M: Long noncoding RNA H19 promotes transforming
growth factor-β-induced epithelial-mesenchymal transition by acting
as a competing endogenous RNA of miR-370-3p in ovarian cancer
cells. Onco Targets Ther. 11:427–440. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Colombo N, Sessa C, du Bois A, Ledermann
J, McCluggage WG, McNeish I, Morice P, Pignata S, Ray-Coquard I,
Vergote I, et al: ESMO-ESGO consensus conference recommendations on
ovarian cancer: Pathology and molecular biology, early and advanced
stages, borderline tumours and recurrent disease†. Ann Oncol.
30:672–705. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kiener TK, Selptsova-Friedrich I and
Hunziker W: Tjp3/zo-3 is critical for epidermal barrier function in
zebrafish embryos. Dev Biol. 316:36–49. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kausalya PJ, Reichert M and Hunziker W:
Connexin 45 directly binds to ZO-1 and localizes to the tight
junction region in epithelial MDCK cells. FEBS Lett. 505:92–96.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Haskins J, Gu L, Wittchen ES, Hibbard J
and Stevenson BR: ZO-3, a novel member of the MAGUK protein family
found at the tight junction, interacts with ZO-1 and occludin. J
Cell Biol. 141:199–208. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim SG, Yooun JH, Kim DE, Lee ES, Kwon TK,
Kim S and Park JW: A novel anti-cancer agent, FPDHP, induces
anoikis in various human cancer cells through activation of
calpain, and downregulation of anoikis-related molecules. J Cell
Biochem. 119:5620–5631. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martin TA, Mason MD and Jiang WG: HGF and
the regulation of tight junctions in human prostate cancer cells.
Oncol Rep. 32:213–224. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Paschoud S, Bongiovanni M, Pache JC and
Citi S: Claudin-1 and claudin-5 expression patterns differentiate
lung squamous cell carcinomas from adenocarcinomas. Mod Pathol.
20:947–954. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martin TA, Watkins G, Mansel RE and Jiang
WG: Loss of tight junction plaque molecules in breast cancer
tissues is associated with a poor prognosis in patients with breast
cancer. Eur J Cancer. 40:2717–2725. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Colas E, Perez C, Cabrera S, Pedrola N,
Monge M, Castellvi J, Eyzaguirre F, Gregorio J, Ruiz A, Llaurado M,
et al: Molecular markers of endometrial carcinoma detected in
uterine aspirates. Int J Cancer. 129:2435–2444. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu Y, Wang Y, Fu X and Lu Z: Long
non-coding RNA NEAT1 promoted ovarian cancer cells' metastasis
through regulation of miR-382-3p/ROCK1 axial. Cancer Sci.
109:2188–2198. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen ZJ, Zhang Z, Xie BB and Zhang HY:
Clinical significance of up-regulated lncRNA NEAT1 in prognosis of
ovarian cancer. Eur Rev Med Pharmacol Sci. 20:3373–3377.
2016.PubMed/NCBI
|
|
39
|
Liu S, Tang J, Huang L, Xu Q, Ling X and
Liu J: Cordyceps militaris alleviates severity of murine acute lung
injury through miRNAs-mediated CXCR2 inhibition. Cell Physiol
Biochem. 36:2003–2011. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu F, Xiong Y, Zhao Y, Tao L, Zhang Z,
Zhang H, Liu Y, Feng G, Li B, He L, et al: Identification of
aberrant microRNA expression pattern in pediatric gliomas by
microarray. Diagn Pathol. 8:1582013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou KR, Liu S, Cai L and Bin L: ENCORI:
The Encyclopedia of RNA Interactomes. http://starbase.sysu.edu.cn/index.phpApril
15–2020
|
|
43
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((Database Issue)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan
C, Xu M, Sun H, Liu C, Wei P and Du X: The lncRNA NEAT1 activates
Wnt/β-catenin signaling and promotes colorectal cancer progression
via interacting with DDX5. J Hematol Oncol. 11:1132018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dong P, Xiong Y, Yue J, Xu D, Ihira K,
Konno Y, Kobayashi N, Todo Y and Watari H: Long noncoding RNA NEAT1
drives aggressive endometrial cancer progression via
miR-361-regulated networks involving STAT3 and tumor
microenvironment-related genes. J Exp Clin Cancer Res. 38:2952019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shin VY, Chen J, Cheuk IW, Siu MT, Ho CW,
Wang X, Jin H and Kwong A: Long non-coding RNA NEAT1 confers
oncogenic role in triple-negative breast cancer through modulating
chemoresistance and cancer stemness. Cell Death Dis. 10:2702019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Di L, Liu LJ, Yan YM, Fu R, Li Y, Xu Y,
Cheng YX and Wu ZQ: Discovery of a natural small-molecule compound
that suppresses tumor EMT, stemness and metastasis by inhibiting
TGFβ/BMP signaling in triple-negative breast cancer. J Exp Clin
Cancer Res. 38:1342019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Q, Mao Z and Sun J: NF-κB
inhibitor, BAY11-7082, suppresses M2 tumor-associated macrophage
induced EMT potential via miR-30a/NF-κB/Snail signaling in bladder
cancer cells. Gene. 710:91–97. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mitra T, Prasad P, Mukherjee P, Chaudhuri
SR, Chatterji U and Roy SS: Stemness and chemoresistance are
imparted to the OC cells through TGFβ1 driven EMT. J Cell Biochem.
119:5775–5787. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zheng X, Zhang Y, Liu Y, Fang L, Li L, Sun
J, Pan Z, Xin W and Huang P: HIF-2α activated lncRNA NEAT1 promotes
hepatocellular carcinoma cell invasion and metastasis by affecting
the epithelial-mesenchymal transition. J Cell Biochem.
119:3247–3256. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fu JW, Kong Y and Sun X: Long noncoding
RNA NEAT1 is an unfavorable prognostic factor and regulates
migration and invasion in gastric cancer. J Cancer Res Clin Oncol.
142:1571–1579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Du Z, Sun T, Hacisuleyman E, Fei T, Wang
X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW and Liu XS:
Integrative analyses reveal a long noncoding RNA-mediated sponge
regulatory network in prostate cancer. Nat Commun. 7:109822016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ji S, Wang S, Zhao X and Lv L: Long
noncoding RNA NEAT1 regulates the development of osteosarcoma
through sponging miR-34a-5p to mediate HOXA13 expression as a
competitive endogenous RNA. Mol Genet Genomic Med. 7:e6732019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gibson MC and Perrimon N: Apicobasal
polarization: Epithelial form and function. Curr Opin Cell Biol.
15:747–752. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jurkiewicz D, Michalec K, Skowronek K and
Nałęcz KA: Tight junction protein ZO-1 controls organic
cation/carnitine transporter OCTN2 (SLC22A5) in a protein kinase
C-dependent way. Biochim Biophys Acta Mol Cell Res. 1864:797–805.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Abraham V, Chou ML, George P, Pooler P,
Zaman A, Savani RC and Koval M: Heterocellular gap junctional
communication between alveolar epithelial cells. Am J Physiol Lung
Cell Mol Physiol. 280:L1085–L1093. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Guillemot L, Paschoud S, Pulimeno P,
Foglia A and Citi S: The cytoplasmic plaque of tight junctions: A
scaffolding and signalling center. Biochim Biophys Acta.
1778:601–613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Doi Y, Yashiro M, Yamada N, Amano R, Noda
S and Hirakawa K: VEGF-A/VEGFR-2 signaling plays an important role
for the motility of pancreas cancer cells. Ann Surg Oncol.
19:2733–2743. 2012. View Article : Google Scholar : PubMed/NCBI
|


















