Introduction
Human defensins are widely distributed in epithelial
tissues; the defensins family consists of small (2–6 kDa) cationic
antimicrobial peptides between 20–50 amino acids with six
evolutionary conserved cysteine residues (1). They form disulphide bridges in three
pairs, giving rise to three anti-parallel beta sheets structure
that assume evolutionary conserved structural fold (2,3).
Apart from the six cysteine residues, members of the defensin
family have low sequence homology. This observation was believed to
result in the difference of characters between all the family
members. In humans, 6 α-defensins and 11 human β-defensins (HBDs)
have been isolated (4). HBDs are
produced by epithelial cells lining the body surface and acts as
natural antibiotics and immune regulators thus, providing the first
line of defence against infection, inflammation and wound healing
(4). HBDs have wide-spectrum of
antimicrobial and biological activities with little risk of
developing resistances. They can also inhibit many steps in viral
infection as well as growth of microbes (5). The expression of HBDs is either
constitutive or inducible in response to infection or tissue injury
(6,7). When induced they normally result in
their most effective site-specific response.
HBDs demonstrate proinflammatory activity by binding
to certain receptors. For example, HBD2 and HBD1 bind to chemokine
receptor 6 (CCR6) leading to increment in chemo-attraction of both
CD4+ memory T-helper cells and immature dendritic cells (8). HBDs can also play a role in
carcinogenesis of epithelial tumours. Changes in expression of HBDs
were observed in epithelial-derived cancers such as prostatic
cancer, basal cell carcinoma, oral squamous cell carcinoma (OSCC)
and renal cell carcinoma (9). The
variation in expression pattern of β-defensins makes it a suitable
tool to investigate its effects in pterygium.
A pterygium is a wedge-shaped fibro-vascular
proliferative conjunctival tissue that typically starts on the
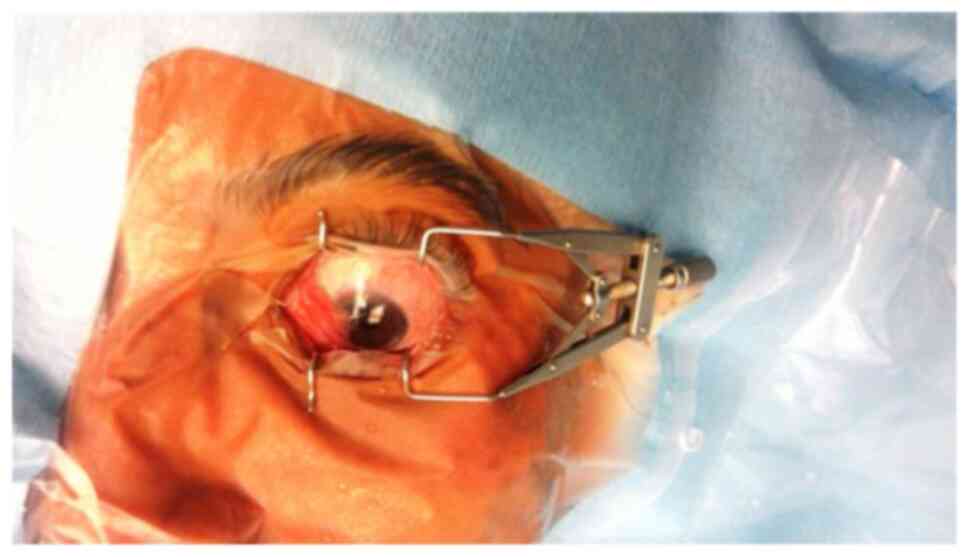
nasal conjunctiva and extends laterally onto the cornea (10). Typical cases with pterygium are
shown in Fig. 1. Pterygium refers
to the shape of the tissue, which looks similar to an insect
wing.
The prevalence rate of pterygium as reported in
different studies varies widely with age, gender and geographical
location. They are mostly observed in people from tropical
climates, but pterygium can be found in over 200 million people
worldwide (11). The exact
aetiology of pterygium is not fully understood. Previous studies
suggested that pterygium was highly associated with ultraviolet
radiation (UVR) exposure. Molecular alterations associated with
pterygium include loss of heterozygosity (LOH), point mutations of
proto-oncogenes (K-ras) and alterations in the expression of tumour
suppressor genes (p53 or p63) and nuclear factor (cyclic AMP
response element-binding protein CREB) (12). Other findings in pterygium include
the frequent detection of HPV DNA, over expression of various
ocular surface proteins, including defensins and phospholipases D,
as well as the up-regulation of growth factors, such as bFGF or
VEGF (13).
Pterygium can cause irregular corneal astigmatism,
corneal scarring and restriction of ocular motility. In some severe
cases, pterygium may result in visual impairment if it approaches
visual axis or chronic ocular surface inflammation (14,15).
Pterygium management usually depends on the size and extent of the
pterygium. A small pterygium can be treated with mild steroid eye
drops (16) while a large size
would require surgery (17–19)
which is normally enhanced by the use of antimetabolites. Current
progress in the biochemical and molecular pathogenesis of pterygium
has helped in the use of minimally invasive methods of treatment
like minimally invasive pterygium excision MIPE (17). HBDs may influence the pathogenesis
of pterygium however; its expression has not been established and
has become the focus of this study. In the present study, we
examined the expression of the HBD defensin β1 (DEFB1), DEFB4A and
DEFB109 genes in pterygium and normal conjunctiva epithelial cells
to investigate their role in pterygium development.
Materials and methods
The present study was approved by the Ethics
Committee of the Faculty of Medicine and Health Sciences (FMHS),
Universiti Putra Malaysia (Serdang, Malaysia). Written informed
consent to participate in this study was also obtained from each
patient.
The study population consisted of 18 patients all
whom underwent pterygium surgery in Hospital Serdang. Based on the
inclusion criteria; individuals who had no previous history of
pterygium surgery with more than 40 years of age and agreed to
participate in the study, were included. Demographic data
including, age, gender and the eye affected are shown in Table I. Nine of the patients were male
and nine were female. The average mean age of the patients was 57.5
years ranged (40–78 years). Out of 30 patients with pterygium, ten
were on the right eye and eight on the left eye.
 | Table I.Demographics of the patients. |
Table I.
Demographics of the patients.
| Sample number | Sex | Age (years) | Eye |
|---|
| 1 | Female | 68 | R |
| 2 | Male | 68 | L |
| 3 | Female | 67 | L |
| 4 | Male | 56 | R |
| 5 | Male | 42 | R |
| 6 | Female | 59 | R |
| 7 | Male | 49 | R |
| 8 | Male | 74 | R |
| 9 | Female | 55 | L |
| 10 | Male | 53 | L |
| 11 | Female | 59 | R |
| 12 | Male | 52 | R |
| 13 | Male | 63 | L |
| 14 | Female | 40 | R |
| 15 | Male | 60 | R |
| 16 | Female | 78 | R |
| 17 | Female | 62 | R |
| 18 | Female | 52 | L |
Collection of samples from
patients
Pterygium tissues were obtained from 18 patients
undergoing pterygium surgery in Serdang Hospital. The tissues were
placed in chilled phosphate-buffered saline (PBS).
Conjunctival impression cytology (CIC) for normal
samples (from the same patients) were obtained as described by
Tseng's modified method (20,21).
Lignocaine hydrochloride 2% (w/v) (Ain Medicare Sdn Bhd, Kota
Bharu, Malaysia) was first injected in the eye, then a D-shaped
halve autoclaved cellulosenitrate filter membrane (Sartorius
StedimBiotech, Goettingen, Germany) was placed on the normal
conjunctiva for 10 sec. The membrane was gently removed and placed
into 0.5 ml chilled PBS in 1.5 ml Eppendorf tube. In order to avoid
sample contamination, the normal conjunctiva was obtained at the
opposite site of the pterygium lesion before excision of the
pterygium. Both samples were transported on ice to the laboratory
within 3 h after excision, and then stored at −80°C until used. The
tissues were disrupted using syringe and needle then homogenized
with QIAshredder (Qiagen, Hilden, Germany).
RNA extraction
The RNA extraction was achieved using RNeasy mini
kit (cat. nos. 74104 and 74106) for each sample, according to the
manufacturer's manual (Qiagen). The concentration and quality were
measured by using Eppendorf Bio spectrometer. A260/A280 was
measured for each sample and quality.
cDNA synthesis
The RNA was reverse transcribed into complementary
DNA (cDNA) using QuantiTech Rev. Transcription kit (Qiagen)
according to the manufacturer's instruction. Briefly, genomic DNA
wipe-out buffer was mixed with RNA on ice and incubated at 42°C for
2 min, and then placed on ice immediately, Quantscript RT buffer
5X, Quantiscript RT (enzyme) and RT primer mix was also prepared
and mixed with the RNA mixture. The final volume of 20 µl was
incubated at 42°C for 20 min, followed by 95°C for 3 min using
(Bio-Rad thermal cycler; Bio-Rad Laboratories, Inc., Hercules, CA,
USA) and stored at −20°C till used.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The RT-qPCR reaction of DEFB1, DEFB4A and DEFB109
was conducted using SYBR-Green master mix (Qiagen) using C1000
Thermal Cycler and CFX96 real time cycler (Bio-Rad Laboratories,
Inc.). The total volume of PCR (20 µl) contained 3 µl of cDNA, 1 µl
of 20 µM/µl of each primers, 10 µl of 2X SYBR-Green and 5 µl of
RNase-free water. Negative control (NTC) was also run in each
experiment. The list of primers used is shown in Table II. The optimum thermal cycling
condition was initially activated at 95°C for 3 min, followed by 40
cycles of denaturation at 95°C for 20 sec, annealing/extension at
60°C for 30 sec, melting curve analysis at 70 to 90°C for 10 min.
GAPDH and β-ACTIN were used as reference genes. The relative
quantification of mRNA was done using 2-∆∆Cq method as
described by Livak and Schmittgen (22) using Bio-Rad CFX manager software
version 3.1 (Bio-Rad Laboratories, Inc.).
 | Table II.List of human β-defensin primers. |
Table II.
List of human β-defensin primers.
| Primer name | Sequence
(5′-3′) | Product size
(bp) |
|---|
| Human β-defensin
genes |
|
|
|
DEFB1 | F:
AGCGTCTCCCCAGTTCCTGAAATCCT | 273 |
|
| R:
TCTTCTGGTCACTCCCAGCTCACTTG |
|
|
DEFB4A | F:
CATCAGCCATGAGGGTCTTG | 199 |
|
| R:
GGCTTTTTGCAGCATTTTGT |
|
|
DEFB109 | F:
TGCAGTAAGAGGTGATTTGG | 174 |
|
| R:
TGACATGATAAGTGGTGTTGG |
|
| Reference
genes |
|
|
|
β-actin | F:
CTCCTTAATGTCACGCAGGATTTC | 520 |
|
| R:
GTGGGGCGCCCCAGGCACCA |
|
|
GAPDH | F:
CCCATCACCATCTTCCAGAGC | 473 |
|
| R:
CCAGTGAGCTTCCCGTTCAGC |
|
Statistical analysis
Data was expressed as the mean ± standard error of
the mean, and were statistically analysed using SPSS (version 22.0;
IBM Corp., Armonk, NY, USA) with significance set at P<0.05
using Student's t-test. Normalised expressions of DEFB1, DEFB4A and
DEFB109 in pterygium were compared with normalised expressions of
DEFB1, DEFB4A and DEFB109 in normal conjunctiva, respectively.
Results
Table III
summarizes the expression of DEFB1, DEFB4A and DEFB109 results of
18 patients using qPCR. Gene expression analysis was carried out on
raw Cq values using ΔΔCq method, and the normalization was
performed using multiple reference genes, GAPDH and β-actin. The
overall results of mRNA expression of DEFB1, DEFB4A and DEFB109 are
shown in Figs. 2–4, respectively. The results are presented
as normalized fold changes which were further converted to log2
values to facilitate data presentation.
 | Table III.DEFB1, DEFB4A and DEFB109 expression
in the studied subjects. |
Table III.
DEFB1, DEFB4A and DEFB109 expression
in the studied subjects.
| Patient | Genes | Normal
conjunctiva | Pterygium |
|---|
| T1 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | NE | Downregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T2 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Downregulated | Upregulated |
| T3 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T4 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | NE | Upregulated |
|
| DEFB109 | Downregulated | Upregulated |
| T5 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T6 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T7 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T8 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T9 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T10 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T11 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T12 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T13 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Downregulated | Upregulated |
| T14 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T15 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T16 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | Downregulated | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T17 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | NE | Downregulated |
|
| DEFB109 | Upregulated | Downregulated |
| T18 | DEFB1 | Downregulated | Upregulated |
|
| DEFB4A | NE | Upregulated |
|
| DEFB109 | Upregulated | Downregulated |
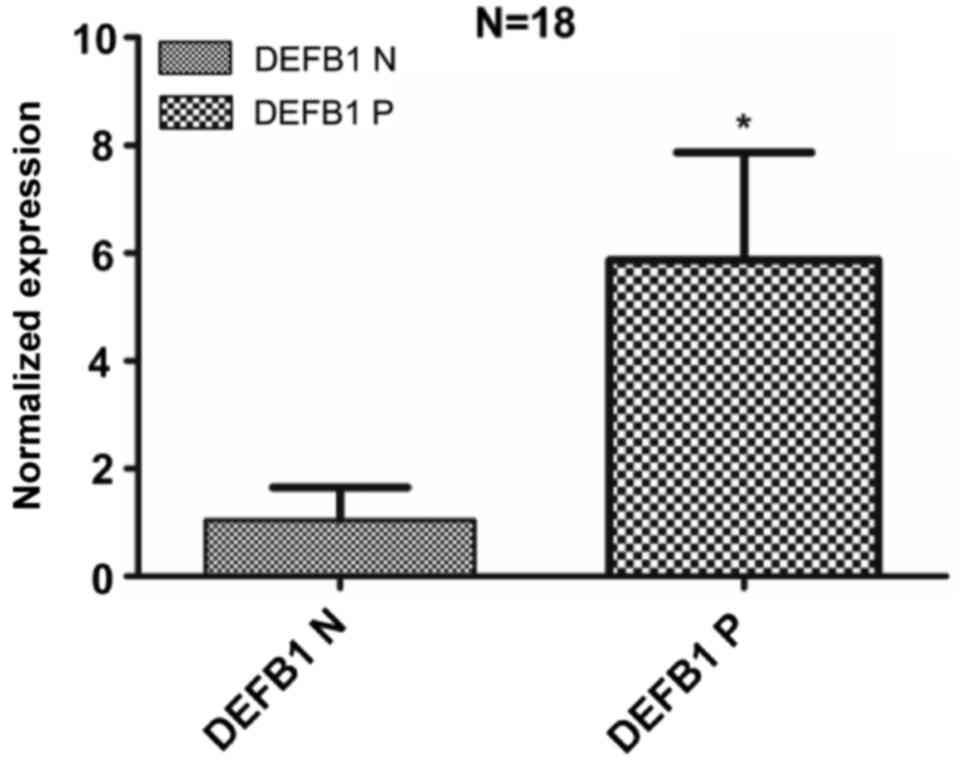
DEFB1 showed low levels of expression in most normal
conjunctiva samples. Its expression was up-regulated in
corresponding pterygium samples from the same patient. The
expression was significantly increased compared with normal
conjunctiva samples (P=0.015); there was an overall 5-fold increase
in the expression of DEFB1 in pterygium samples compared to normal
conjunctiva samples.
DEFB4A showed low level of expression in most normal
conjunctiva samples. Its expression was upregulated in
corresponding pterygium samples from the same patient. The
expression of DEFB4A was elevated in pterygium compared to normal
conjunctiva samples, although statistically not significant
(P=0.064), however DEFB4A was not detected in 4 samples of normal
conjunctiva within the 40 cycles. There was an approximately 6-fold
increase in the expression of DEFB4A in pterygium samples relative
to normal conjunctiva samples.
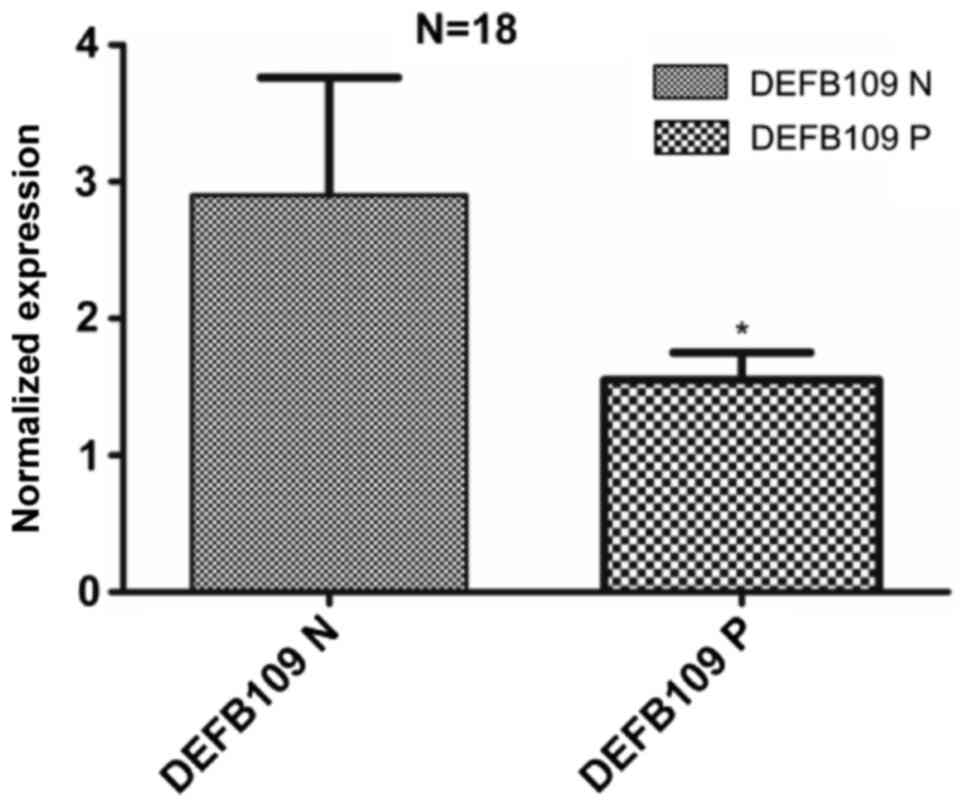
DEFB109 showed high level of expression in most
normal conjunctiva samples. Its expression was down regulated in
corresponding pterygium samples of the same patient. The expression
of DEFB109 was significantly decreased in pterygium compared with
normal conjunctiva samples (P=0.037), There was an overall −1.3
fold change in the DEFB109 expression in pterygium compared to
normal conjunctiva samples.
Discussion
β-Defensins play important roles in both innate and
adaptive immune response (2).
β-defensins are mainly expressed in different types of epithelial
cells such as intestinal epithelial, respiratory epithelia,
genitourinary tissues, nasolacrimal duct, mammary gland and
sometimes in immune cells such as dendritic cells (2,23–27).
On the ocular surface, β-defensins are endogenously produced by
epithelial cells. It has been reported that DEFB1, DEFB4A and
DEFB109 mRNA expression were detected in scraped corneal epithelial
cells and whole conjunctiva tissues (28,29).
Messenger RNA expression of three HBD genes (DEFB1,
DEFB4A and DEFB109) in pterygium and normal conjunctiva of the same
patient was investigated using qPCR. Though, the present study is
the first to report the effect of these β-defensins expression in
pterygium. The expression of DEFB1 mRNA was detected in all normal
conjunctival and pterygium samples in the present study, which was
in agreement with previous reports that DEFB1 was constitutively
expressed in epithelial cells (24,28,30–32).
However, the expression of DEFB4A was positive in 14 normal
conjunctiva samples, and positive in all 18 pterygium samples. This
finding was in agreement with a number of studies that reported
DEFB4A expression by epithelial cells is variable as its expression
by normal epithelial occurs only occasionally (inducible) (33,34).
DEFB109 was positive in all examined normal conjunctival and
pterygium samples (constitutively expressed) which also agrees with
previous studies (29,35).
This study also showed that human normal conjunctiva
epithelial cells expressed low level of DEFB1 and DEFB4A and high
level of DEFB109 mRNA. However, DEFB1 and DEFB4A mRNA expression
were up regulated in pterygium while DEFB109 mRNA was down
regulated relative to normal conjunctiva samples. Though, the exact
mechanisms that regulates the expression of these defensins at the
ocular surface are yet to be determined, it has been shown that the
expression of β-defensins on the ocular surface is part of the
innate immune response in preventing infection or invasion by
microorganism (bacteria, viruses and fungi) (28,36).
DEFB1 and DEFB4A are antimicrobial peptides
identified at the ocular surface (31,37),
which produces potent antimicrobial effects against common ocular
pathogens in vitro. The DEFB1 exists as a single copy gene
with several SNPs that have been implicated in the pathogenesis of
some chronic inflammatory diseases, including asthma and chronic
obstructive pulmonary disease (38–40).
Genomic variations in DEFB1 (1)
may also contribute to the pathogenesis of pterygium in the
presence of specific haplotypes associated with either increased
susceptibility to tumors associated inflammation, or protection
from severe infection (viruses). It has been demonstrated that
DEFB4A expression is up regulated in response to infection by Gram
negative bacteria e.g., P. aeruginosa and their products
such as lipopolysaccharide, peptidoglycan and lipoproteins,
including inflammatory cytokines which consist of IL-α and tumor
necrosis factor (23,34). Another study found that human
rhinovirus infection induces airway epithelial cell production of
DEFB4A both in vitro and in vivo (23,41).
The expression of this β-defensin was also up regulated in the
cornea in response to tissue injury and in conjunctiva epithelium
of patients with dry eye (42).
Moreover, DEFB4A has been found to stimulate human corneal
epithelial cell migration and proliferation (36). Also, DEFB4A was recognized as a
potential natural antibiotic. The up regulation of DEFB4A in the
present study was considered to indicate an inflammatory response
caused by UV radiation and viral infection in pterygium tissue.
Also, absence of detectable DEFB4A in some normal conjunctiva
samples (4) indicates that DEFB4A
may have a specific influence on innate immune response within the
pterygium.
In a recent study by Abedin et al (43), expression of DEFB109, was detected
in the ocular surface epithelia, and interestingly its expression
was down regulated, thus resulting to decrease in inflammation and
infection. Abedin et al (43) demonstrated the down-regulation and
constitutive expression of DEFB109, in some of the diseases
affecting the ocular surface such as bacterial
keratoconjunctivitis, viral keratitis, acanthamoeba keratitis, and
dry eye disease. Similarly, the down regulation of DEFB109 was
reported after in vitro stimulation of gingival
keratinocytes with Candida albicans (29,44).
The increase in mRNA expression levels of DEFB1, DEFB4A with
decrease in mRNA expression DEFB109 in pterygium epithelial
suggests a potential role for the three defensins in the
development of the resistant susceptible phenotype. Previous
studies reported that β-defensins provide an initial block to a
variety of pathogens on the epithelial surface (2,45,46).
It is now well recognized that many antimicrobial peptides possess
dual roles as they are capable of killing bacteria or viruses and
are able to modulate mammalian cell functions such as migration,
proliferation and cytokine production (25). Previous findings indicate that
innate and adaptive immune response in the ocular surface is an
intricate process likely involving so many processes such as
anatomical, biochemical or cellular and humoral factors (34,47).
The underlying cause of increased DEFB1 and DEFB4A
expressions, decreased DEFB109 expression in pterygium is not
apparent. It is also unclear whether these genes could be
functioning as an antimicrobial, pro inflammatory, immune/cellular
modulator or both, and whether functionality of these genes affects
clinical symptoms of pterygia. Oncogenic viruses, including HPV,
CMV, HSV or EBV (48) are being
investigated in pterygium but, HPV DNA has been shown to localize
quite specifically to pterygium in several studies. Taking into
account that HPV infection may be associated with pterygium
development. A mechanism that might explain the results was
proposed based on some recent findings, and the published studies
of others. The increase in expressions of DEFB1 and DEFB4A with
decrease of DEF109 in pterygium may suggest an immune response to
microbial derived stimuli impinging from the systemic explosion of
the ocular surface to HPV infection.
Even though, identification of pathogenic
determinants in pterygium, including HPV viruses has been an
inconsistent finding, and no single pathogenic agent has been
categorically identified as sole contributor to pterygium
development apart from UV radiation. Nonetheless, the functional
role that DEFB1, DEFB4A and DEFB109 play in the development of
pterygium could be influenced by viral stimuli within the
pterygium.
Finally, qPCR technique provided evidence that both
DEFB1 and DEFB4A were constitutively expressed in pterygia and
disparately up-regulated, DEFB109 was also constitutively expressed
and down regulated in pterygium.
HBDs are involved in various cellular processes.
Multitude of functions makes HBDs a promising tool for certain
clinical applications. Results, revealed that the expression of
HBD1 and HBD2 was significantly higher and up-regulated in some
pterygium samples when compared with normal conjunctiva samples
from the same patient (P<0.05) while in HBD9 no significant
changed was observed. Thus to our knowledge, this is the first
study to determine the expression human β-defensins in pterygium
specifically. HBDs can be a potential target in understanding
pterygium development. Furthermore, more research is needed to
determine the exact mechanism of defensins in pterygia.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Fundamental
Research Grant Scheme, Ministry of Education Malaysia (grant no.
5524402).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MMI, SAA and NO conceived and designed the study.
SAA and TSW performed the experiments and collected the data. SAA
drafted the manuscript. All authors revised and approved the final
version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Faculty of Medicine and Health Sciences (FMHS),
Universiti Putra Malaysia (Serdang, Malaysia). Written informed
consent to participate in the present study was also obtained from
each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu L, Zhao C, Heng HH and Ganz T: The
human beta-defensin-1 and alpha-defensins are encoded by adjacent
genes: Two peptide families with differing disulfide topology share
a common ancestry. Genomics. 43:316–320. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jia HP, Schutte BC, Schudy A, Linzmeier R,
Guthmiller JM, Johnson GK, Tack BF, Mitros JP, Rosenthal A, Ganz T
and McCray PB Jr: Discovery of new human beta-defensins using a
genomics-based approach. Gene. 263:211–218. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ganz T: Defensins: Antimicrobial peptides
of innate immunity. Nat Rev Immunol. 3:710–720. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou YS, Webb S, Lettice L, Tardif S,
Kilanowski F, Tyrrell C, MacPherson H, Semple F, Tennant P, Baker
T, et al: Partial deletion of chromosome 8 β-defensin cluster
confers sperm dysfunction and infertility in male mice. PLoS Genet.
9:e10038262013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wilson SS, Wiens ME and Smith JG:
Antiviral mechanisms of human defensins. J Mol Biol. 425:4965–4980.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dhople V, Krukemeyer A and Ramamoorthy A:
The human beta-defensin-3, an antibacterial peptide with multiple
biological functions. Biochim Biophys Acta. 1758:1499–1512. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wehkamp J, Wang G, Kübler I, Nuding S,
Gregorieff A, Schnabel A, Kays RJ, Fellermann K, Burk O, Schwab M,
et al: The Paneth cell alpha-defensin deficiency of ileal Crohn's
disease is linked to Wnt/Tcf-4. J Immunol. 179:3109–3118. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang D, Chertov O, Bykovskaia SN, Chen Q,
Buffo MJ, Shogan J, Anderson M, Schröder JM, Wang JM, Howard OM and
Oppenheim JJ: Beta-defensins: Linking innate and adaptive immunity
through dendritic and T cell CCR6. Science. 286:525–528. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Rayahi IA and Sanyi RH: The overlapping
roles of antimicrobial peptides and complement in recruitment and
activation of tumor-associated inflammatory cells. Front Immunol.
6:22015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Masters JS and Harris DJ Jr: Low
recurrence rate of pterygium after excision with conjunctival
limbal Autograft: A retrospective study with long-term follow-up.
Cornea. 34:1569–1572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lucas RM, McMichael AJ, Armstrong BK and
Smith WT: Estimating the global disease burden due to ultraviolet
radiation exposure. Int J Epidemiol. 37:654–667. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nubile M, Curcio C, Lanzini M, Calienno R,
Iezzi M, Mastropasqua A, Di Nicola M and Mastropasqua L: Expression
of CREB in primary pterygium and correlation with cyclin D1, ki-67,
MMP7, p53, p63, Survivin and Vimentin. Ophthalmic Res. 50:99–107.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Detorakis ET and Spandidos DA:
Pathogenetic mechanisms and treatment options for ophthalmic
pterygium: Trends and perspectives (Review). Int J Mol Med.
23:439–447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu L, Wu J, Geng J, Yuan Z and Huang D:
Geographical prevalence and risk factors for pterygium: A
systematic review and meta-analysis. BMJ Open. 3:e0037872013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Julio G, Lluch S, Pujol P and Merindano D:
Ocular discomfort in pterygium patients. Optom Vis Sci. 90:269–274.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rachmiel R, Leiba H and Levartovsky S:
Results of treatment with topical mitomycin C 0.02% following
excision of primary pterygium. Br J Ophthalmol. 79:233–236. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bozkir N, Yilmaz S and Maden A: Minimally
invasive pterygium surgery: A new approach for prevention of
recurrence. Eur J Ophthalmol. 18:27–31. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ozkurt YB, Kocams O, Comez AT, Uslu B and
Dogan OK: Treatment of primary pterygium. Optom Vis Sci.
86:1178–1181. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Varssano D, Shalev H, Lazar M and Fischer
N: Pterygium excision with conjunctival autograft: True survival
rate statistics. Cornea. 32:1243–1250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nelson JD: Impression cytology. Cornea.
7:71–81. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Singh R, Joseph A, Umapathy T, Tint NL and
Dua HS: Impression cytology of the ocular surface. Br J Ophthalmol.
89:1655–1659. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vora P, Youdim A, Thomas LS, Fukata M,
Tesfay SY, Lukasek K, Michelsen KS, Wada A, Hirayama T, Arditi M
and Abreu MT: Beta-defensin-2 expression is regulated by TLR
signaling in intestinal epithelial cells. J Immunol. 173:5398–5405.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Alp S, Skrygan M, Schlottmann R, Kreuter
A, Otte JM, Schmidt WE, Brockmeyer NH and Bastian A: Expression of
beta-defensin 1 and 2 in nasal epithelial cells and alveolar
macrophages from HIV-infected patients. Eur J Med Res. 10:1–6.
2005.PubMed/NCBI
|
|
25
|
Musumeci G, Carnazza ML, Leonardi R and
Loreto C: Expression of β-defensin-4 in ‘an in vivo and ex vivo
model’ of human osteoarthritic knee meniscus. Knee Surg Sports
Traumatol Arthrosc. 20:216–222. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang XF, Cao RM, Li J, Wu J, Wu SM and
Chen TX: Identification of sociodemographic and clinical factors
associated with the levels of human β-defensin-1 and human
β-defensin-2 in the human milk of Han Chinese. Br J Nutr.
111:867–874. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suarez-Carmona M, Hubert P, Delvenne P and
Herfs M: Defensins: ‘Simple’ antimicrobial peptides or
broad-spectrum molecules? Cytokine Growth Factor Rev. 26:361–370.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ikeda A, Sakimoto T, Shoji J and Sawa M:
Expression of alpha-and beta-defensins in human ocular surface
tissue. Jpn J Ophthalmol. 49:73–78. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Otri AM, Mohammed I, Al-Aqaba MA, Fares U,
Peng C, Hopkinson A and Dua HS: Variable expression of human Beta
defensins 3 and 9 at the human ocular surface in infectious
keratitis. Invest Ophthalmol Vis Sci. 53:757–761. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Haynes RJ, McElveen JE, Dua HS, Tighe PJ
and Liversidge J: Expression of human beta-defensins in intraocular
tissues. Invest Ophthalmol Vis Sci. 41:3026–3031. 2000.PubMed/NCBI
|
|
31
|
Huang LC, Jean D, Proske RJ, Reins RY and
McDermott AM: Ocular surface expression and in vitro activity of
antimicrobial peptides. Curr Eye Res. 32:595–609. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jarczak J, Kościuczuk EM, Lisowski P,
Strzałkowska N, Jóźwik A, Horbańczuk J, Krzyżewski J, Zwierzchowski
L and Bagnicka E: Defensins: Natural component of human innate
immunity. Human Immunol. 74:1069–1079. 2013. View Article : Google Scholar
|
|
33
|
McNAMARA NA, Van R, Tuchin OS and Fleiszig
SM: Ocular surface epithelia express mRNA for human beta
defensin-2. Exp Eye Res. 69:483–490. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
McDermott AM: The role of antimicrobial
peptides at the ocular surface. Ophthalmic Res. 41:60–75. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mohammed I, Suleman H, Otri AM, Kulkarni
BB, Chen P, Hopkinson A and Dua HS: Localization and gene
expression of human beta-defensin 9 at the human ocular surface
epithelium. Invest Ophthalmol Vis Sci. 51:4677–4682. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Garreis F, Schlorf T, Worlitzsch D, Steven
P, Bräuer L, Jäger K and Paulsen FP: Roles of human beta-defensins
in innate immune defense at the ocular surface: Arming and alarming
corneal and conjunctival epithelial cells. Histochem Cell Biol.
134:59–73. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pazgier M, Hoover DM, Yang D, Lu W and
Lubkowski J: Human beta-defensins. Cell Mol Life Sci. 63:1294–1313.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Matsushita I, Hasegawa K, Nakata K, Yasuda
K, Tokunaga K and Keicho N: Genetic variants of human
beta-defensin-1 and chronic obstructive pulmonary disease. Biochem
Biophys Res Commun. 291:17–22. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Levy H, Raby BA, Lake S, Tantisira KG,
Kwiatkowski D, Lazarus R, Silverman EK, Richter B, Klimecki WT,
Vercelli D, et al: Association of defensin beta-1 gene
polymorphisms with asthma. J Allergy Clin Immunol. 115:252–258.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Andresen E, Günther G, Bullwinkel J, Lange
C and Heine H: Increased expression of beta-defensin 1 (DEFB1) in
chronic obstructive pulmonary disease. PLoS One. 6:e218982011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Proud D, Sanders SP and Wiehler S: Human
rhinovirus infection induces airway epithelial cell production of
human beta-defensin 2 both in vitro and in vivo. J Immunol.
172:4637–4645. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Meloni M, De Servi B, Marasco D and Del
Prete S: Molecular mechanism of ocular surface damage: Application
to an in vitro dry eye model on human corneal epithelium. Mol Vis.
17:113–126. 2011.PubMed/NCBI
|
|
43
|
Abedin A, Mohammed I, Hopkinson A and Dua
HS: A novel antimicrobial peptide on the ocular surface shows
decreased expression in inflammation and infection. Invest
Ophthalmol Vis Sci. 49:28–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Premratanachai P, Joly S, Johnson GK,
McCray PB Jr, Jia HP and Guthmiller JM: Expression and regulation
of novel human beta-defensins in gingival keratinocytes. Oral
Microbiol Immunol. 19:111–117. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
McDermott AM: Defensins and other
antimicrobial peptides at the ocular surface. Ocul Surf. 2:229–247.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Machado LR and Ottolini B: An evolutionary
history of defensins: A role for copy number variation in
maximizing host innate and adaptive immune responses. Front
Immunol. 6:1152015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bolaños-Jiménez R, Navas A,
López-Lizárraga EP, de Ribot FM, Peña A, Graue-Hernández EO and
Garfias Y: Ocular surface as barrier of innate immunity. Open
Ophthalmol J. 9:49–55. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chalkia AK, Spandidos DA and Detorakis ET:
Viral involvement in the pathogenesis and clinical features of
ophthalmic pterygium (Review). Int J Mol Med. 32:539–543. 2013.
View Article : Google Scholar : PubMed/NCBI
|