Introduction
Ulcerative colitis (UC) is a type of inflammatory
bowel disease that is characterized by chronic, non-infectious
inflammation of the colon (1). To
date, the etiology of UC has remained to be fully elucidated.
Prolonged duration of active disease is associated with a high risk
of UC-associated neoplasia (UCAN) (2). Pathologically, UCAN exhibits a broad
range of severity, including low-grade dysplasia (LGD), high-grade
dysplasia (HGD) and invasive carcinoma (3). It was reported that ~18% of patients
with UC may develop colorectal cancer (CRC) after 30 years,
particularly patients with extensive colitis (4). As the detailed mechanisms that
participate in the pathogenesis of UCAN remain elusive, associated
research is warranted.
MicroRNAs (miRNAs/miRs) are a class of non-coding
RNAs that are single-stranded, consist of 19–24 nucleotides and
regulate approximately one-third of human genes at the
post-transcriptional level via binding to their 3′ untranslated
region (5). Previous studies
demonstrated that miRNAs have a key role in the inflammatory
process associated with UC (6,7).
Although several studies have proven that miRNAs are involved in
CRC (8,9), the pathogenesis of UCAN is not
consistent with that of CRC. Furthermore, there is little research
on how miRNAs regulate tumorigenesis in UC.
The aim of the present study was to identify
differentially expressed miRNAs (DEMs) and downstream genes in
patients with UCAN using bioinformatics analysis and verify the
regulatory effect of candidate miRNAs on their target genes, for
the purpose of uncovering an epigenetic regulation involving miRNAs
and associated target genes, thereby further contributing to the
elucidation of the pathogenesis of UCAN.
Materials and methods
Bioinformatics analysis
Datasets containing miRNA and mRNA expression
profiles in the colonic mucosa of patients with UC and UCAN
(GSE68306 and GSE37283) were downloaded from the Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). Linear Models for
Microarray Data (10) was applied
to identify differentially expressed genes (DEGs). Fold change
>2.0 and P<0.05 were used as the cutoff values. Heatmap,
volcano and scatter plots were generated to describe the GEO data
visually using ggplot2 and heatmap packages in R. Gene Ontology
(GO) (11) and Kyoto Encyclopedia
of Gene and Genomes (KEGG) pathway (12) enrichment analyses were performed
using the Database for Annotation, Visualization and Integrated
Discovery (DAVID) online tool (13,14).
The miRNA target genes were predicted with the miRWalk tool
(mirwalk.umm.uni-heidelberg.de), which
integrates the miRNA resources from TargetScan, miRDB and
miRTarBase.
Patients for validation
A total of 50 patients with UC were enrolled in the
present study as the validation cohort. All of the patients were
admitted to Tianjin University General Hospital (Tianjin, China)
between January 2000 and December 2018. The inclusion criteria were
as follows: Patients aged ≥18 and <60 years, with UC of any
extent, including proctitis, left-sided colitis and extensive
colitis. The exclusion criteria were as follows: Patients with UC
who were diagnosed with malignant tumors or other autoimmune
diseases, and patients without biopsy as the pathological sample.
The Ethics Committee at Tianjin Medical University General Hospital
(Tianjin, China) approved the study protocol and informed consent
was obtained from all the enrolled patients. The patients were
divided into three groups as follows: UC patients without neoplasia
(UC group; n=20), UC patients with low-grade dysplasia (UC-LGD
group; n=20) and UC patients with high-grade dysplasia (UC-HGD
group; n=10). Colonic mucosa samples were retrieved from archived
formalin-fixed paraffin-embedded (FFPE) tissues obtained from these
patients by endoscopic biopsy.
RNA extraction and quality check
Total RNA was extracted from the 10-mm sections of
the FFPE tissue after deparaffinization as described previously
(15) and from the cultured cells.
RNA was extracted using the Ambion RecoverAll kit (Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. The RNA
quality check was performed by Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Complementary DNA (cDNA) was reverse-transcribed
from the extracted RNA using the M-MuLV First Strand cDNA Synthesis
System (Thermo Fisher Scientific, Inc.) and the miRNA First Strand
cDNA Synthesis System (Poly A Tailing; Thermo Fisher Scientific,
Inc.). The expression level of candidate miRNAs and mRNAs was
determined using the miRNA Quantitation PCR kit (Thermo Fisher
Scientific, Inc.) and the SGExcel FastSYBR Mixture (Thermo Fisher
Scientific, Inc.). GAPDH and U6 were used as internal controls. The
sequences of the primers used are listed in Table SI. The relative fold changes in
miRNA and target gene expression were calculated using the
2−ΔΔCq method (16).
Cell culture
The human colon cancer cell line SW480 was obtained
from the American Type Culture Collection. SW480 cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in a humidified atmosphere containing 5%
CO2.
Cell transfection
miR-31 mimics and miR negative controls were
obtained from Gene Pharma Co. The SW480 cells were seeded in a
6-well plate and at 70–80% confluence, they were transfected with
miR-31 mimics and negative controls using the
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol for 48 h
at 37°C. The cells were then seeded in 96-well plates at a density
of 2×104 per well for further experiments.
Cell proliferation assay
Cells were seeded in 96-well plates at a density of
2×104 per well and cultured at 37°C for 24, 48, 72 or 96
h. A total of 10 µl Cell Counting kit-8 (CCK-8) solution
(Sigma-Aldrich; Merck KGaA) was then added to each well. After
incubation for 1 h at 37°C, the optical density at 450 nm was
detected to evaluate the cell proliferation with Fisherbrand™
accuSkan™ GO ultraviolet/Vis Microplate Spectrophotometer (Thermo
Fisher Scientific, Inc.).
Statistical analysis
The experimental results are presented as the mean ±
standard error of the mean. The differences in among multiple
groups were analyzed using ANOVA (parametric model) followed by
Tukey's post-hoc test using GraphPad Prism 7 (GraphPad Software,
Inc.). Pearson's correlation analysis was performed to assess the
correlation between miRNA and target gene expression. P<0.05 was
considered to indicate statistical significance. Heatmaps, volcano
and scatter plots, as well as bubble charts, were generated in
R.
Results
DEGs
According to the expression profiles of patients
with UC and UCAN (GSE37283 dataset), a total of 307 DEGs
(P<0.05; fold change >2) were identified. Among these 307
DEGs, 165 were upregulated and 142 were downregulated. The volcano
plots were created according to the expression profile (Fig. 1A). A scatter plot (Fig. 1B) and heatmap (Fig. 1C) of mRNA expression levels
compared between UC and UCAN groups were also generated, which
exhibited an even distribution of gene expression data upon visual
examination.
Gene function and pathway enrichment
analysis
The functions and pathway enrichment of DEGs were
analyzed using DAVID and a false discovery rate <0.05 was used
as the cut-off. The top 10 GO terms in which the up- and
downregulated DEGs were enriched are listed in Table I. The detailed data of the GO
analysis are illustrated in Fig.
2. The result of the KEGG pathway enrichment analysis is
presented in Fig. 3. The
upregulated DEGs were mainly enriched in complement and coagulation
cascades, proteoglycans in cancer, phagosome, natural killer
cell-mediated cytotoxicity and the peroxisome
proliferator-activated receptor signaling pathway, whereas the
downregulated genes were mainly enriched in retinol metabolism,
ascorbate and aldarate metabolism, steroid hormone biosynthesis,
pentose and glucuronate interconversions, and porphyrin and
chlorophyll metabolism.
 | Table I.GO terms enriched by the
differentially expressed genes in different categories. |
Table I.
GO terms enriched by the
differentially expressed genes in different categories.
| A, Cellular
component |
|---|
|
|---|
| Direction of
differential expression | GO terms |
|---|
| Upregulated
genes | Extracellular
region; extracellular exosome; plasma membrane; proteinaceous
extracellular matrix |
| Downregulated
genes | Endoplasmic
reticulum membrane; basolateral plasma membrane |
|
| B, Molecular
function |
|
| Direction of
differential expression | GO
terms |
|
| Upregulated
genes | Collagen binding;
protein binding |
| Downregulated
genes | Protein
heterodimerization activity; Transferase activity, transferring
hexosyl groups; actin filament binding |
|
| C, Biological
process |
|
| Direction of
differential expression | GO
terms |
|
| Upregulated
genes | Leukocyte
migration; signal transduction; inflammatory response; neutrophil
chemotaxis |
| Downregulated
genes | Negative regulation
of glucuronosyltransferase activity; negative regulation of fatty
acid metabolic process; flavonoid glucuronidation; flavonoid
biosynthetic process; retinoic acid metabolic process |
DEMs and corresponding target
genes
Among all miRNAs in the expression profile (GSE68306
dataset), 38 were identified as DEMs (P<0.05; fold change >2;
Fig. 4A). A total of 15 DEMs were
downregulated, including miR-10b, miR-584, miR-655, miR-453,
miR-495, miR-634, miR-10a, miR-615, miR-548, miR-23a, miR-523,
miR-455, miR-28, miR-193a and let-7f, while 23 DEMs were
upregulated, including miR-106b, miR-34a, miR-31, miR-135b,
miR-1974, miR-151-3p, miR-186, miR-423, miR-143, miR-127, miR-1290,
miR-381, miR-152, miR-214, miR-374a, miR-140, miR-331, miR-1178,
miR-493, miR-1246, miR-141, miR-374b and miR-708. Among these
dysregulated miRNAs, miR-31 (17,18),
miR-34a (15,19,20)
and miR-106b (21) were reported
to be differentially expressed in CRC by to previous studies.
Furthermore, miR-31 was also reported to be dysregulated in UC
(8,22). These three miRNAs miR-31, miR-34a
and miR-106b, were selected as the candidate miRNAs for further
validation. As each miRNA had numerous target genes, there was an
overlap among the target genes of these three candidate DEMs and
the associations are presented in Fig.
4B.
 | Figure 4.Differentially expressed miRNAs and
corresponding target genes. (A) Volcano plots of miRNA expression
between patients with UC-associated neoplasia and UC. (B) The
overlapping association between miR-31, miR-34a, miR-106b and their
target genes, including ZBTB20, SPRED1, SAMD12, PTPN4, NR2C2, GAB1,
ERC1, CHMP7, CERS6, AK4 AND AFF4. miR, microRNA; UC, ulcerative
colitis; ZBTB20, zinc finger and BTB domain containing 20; SPRED1,
sprouty related EVH1 domain containing 1; SAMD12, sterile α motif
domain containing 12; PTPN4, protein tyrosine phosphatase
non-receptor type 4; NR2C2, nuclear receptor subfamily 2 group C
member 2; GAB1, GRB2 associated binding protein 1; ERC1,
ELKS/RAB6-interacting/CAST family member 1; CHMP7, charged
multivesicular body protein 7; CERS6, ceramide synthase 6; AK4,
adenylate kinase 4; AFF4, AF4/FMR2 family member 4. |
Patient characteristics
The characteristics of the patients enrolled in the
present study, including demographic and clinicopathological data
and laboratory parameters, are summarized in Table II. There were no statistical
differences identified in the average age and sex between the UC,
UC-LGD and UC-HGD groups.
 | Table II.Characteristics of the patients. |
Table II.
Characteristics of the patients.
| Characteristic | UC (n=20) | UC-LGD (n=20) | UC-HGD (n=10) |
|---|
| Age (years) |
43.7±11.3 |
46.5±13.2 |
47.2±15.3 |
| Sex
(male/female) | 11/9 | 12/8 | 4/6 |
| Extensive
colitis | 8 (40) | 13 (65) | 8 (80) |
| Disease duration
(years) | 12.1±6.0 | 12.3±3.1 | 13.7±4.9 |
| CRP (mg/dl) |
8.1±2.6 |
7.5±3.3 |
7.8±2.9 |
| ESR (mm/h) | 31.3±1.9 | 28.1±2.7 | 30.8±3.4 |
Validation of DEMs and corresponding
target genes in patient samples
Validation of the three candidate miRNAs (miR-31,
miR-34a and miR-106b) was performed by RT-qPCR analysis in all
samples from patients with UC (n=20), UC-LGD (n=20) and UC-HGD
(n=10). The expression of miR-31 among the three groups was
significantly different (P<0.01, Fig. 5A). In detail, the expression of
miR-31 in patients with UC-HGD was significantly higher compared
with that in patients with UC (P<0.01). By contrast, there were
no significant differences in the expression of miR-34a (Fig. 5B) and miR-106b (Fig. 5C) among the three groups.
Considering the target gene predicting results, the
expression profiling and whether the candidate genes were
previously reported to be involved in CRC or UC, three target genes
of miR-31 [special AT-rich DNA-binding protein 2 (SATB2), zinc
finger CCCH type-containing 12C (ZC3H12C) and Ras p21 protein
activator 1 (RASA1)] were selected for validation. The RT-qPCR
results revealed that the expression of SATB2 was significantly
different among the UC, UC-LGD and UC-HGD samples (P<0.0001;
Fig. 6A). The expression of SATB2
in patients with UC-LGD was significantly decreased compared with
that in patients with UC (P<0.05). A more significant difference
was observed between the UC-HGD and UC groups (P<0.0001).
Contrary to the expression profile in the dataset GSE37283, there
was no significant difference in the expression of ZC3H12C
(Fig. 6B) and RASA1 (Fig. 6C) among the UC, UC-LGD and UC-HGD
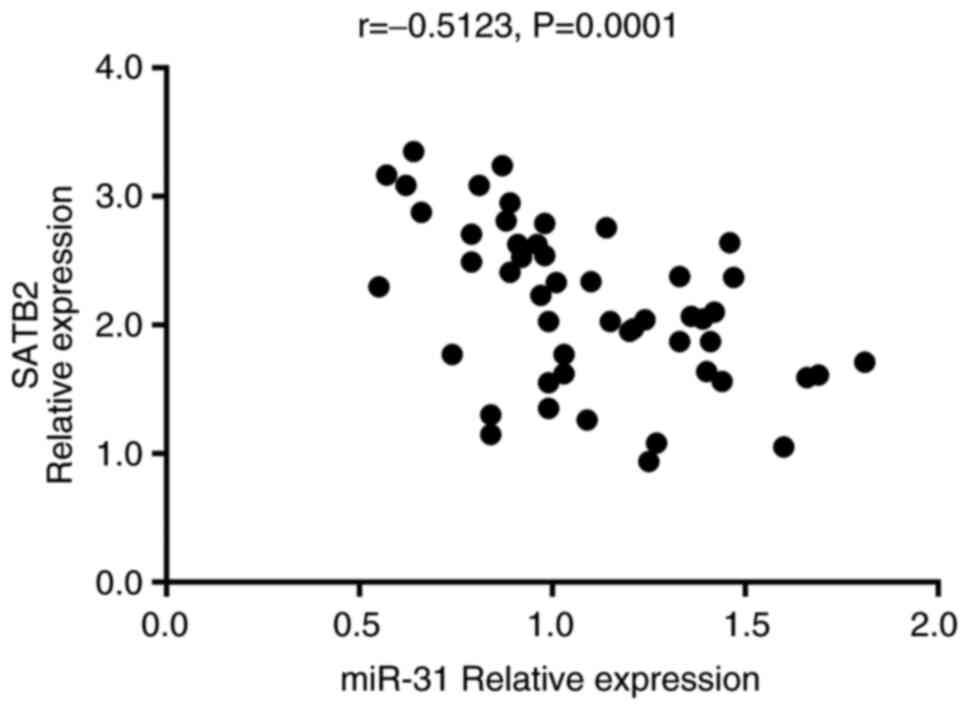
groups. In addition, the expression of SATB2 was indicated to be
negatively correlated with that of miR-31 in all patients with UC
(r=−0.5123, P=0.0001; Fig. 7).
miR-31 overexpression promotes CRC
cell proliferation via repressing SATB2 expression in vitro
To confirm whether miR-31 promotes carcinogenesis
via downregulating SATB2, CRC cells were transfected with miR-31
mimics and negative controls. Subsequently, RT-qPCR and cell
proliferation assays were performed. miR-31 expression was
confirmed to be increased in SW480 cells following transfection
(P<0.0001; Fig. 8A). In
parallel, the expression of SATB2 was downregulated in SW480 cells
transfected with miR-31 mimics (P<0.01; Fig. 8B). Furthermore, overexpression of
miR-31 promoted SW480 cell proliferation, as indicated by the CCK-8
assay (P<0.001; Fig. 8C).
Discussion
In the present study, DEMs and DEGs were screened
from expression profiles from the GEO database. The result of the
bioinformatics analysis demonstrated that the mRNA and the miRNA
expression data were evenly distributed. A visual presentation of
the results of the enrichment analysis was also performed. Although
the final candidate functional genes were not included in these
signaling pathways, valuable information was obtained for future
studies.
With a longer duration of UC, malignant
transformation is the most serious complication (2). Histologically, the UCAN lesions may
include LGD, HGD and invasive carcinoma (3). However, the pathogenesis of UCAN
differs from that of sporadic CRC. Inflammation-associated
microsatellite alterations were detected in the affected tissues of
patients with UC, which indicated the presence of genomic mutations
in the neoplastic tissue (23).
The frequency of inflammation-associated microsatellite alterations
was increased during the progression of UCAN (23). Glycosyltransferase ST6
N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (ST6GALNAC1)
was induced by M2-like macrophages, and ST6GALNAC1 alters the
glycosylation status of the oncoprotein mucin 1, thereby promoting
cancer development and progression in UC (24). However, the exact mechanisms
underlying the pathogenesis of UCAN remain elusive.
miRNAs participate in a variety of biological
processes in a large number of diseases via regulating their target
genes. Indeed, numerous studies have proven the regulatory function
of miRNAs in UC. Cytokines serve critical roles in the inflammatory
process of UC and miR-124 was indicated to promote the inflammatory
process by upregulating the expression of STAT3 (6). miR-31 and miR-155 may inhibit the
expression of IL-13 receptor α-1 (IL13RA1), thus regulating the
IL-13 signaling pathway (7).
Furthermore, miR-155 exerted an important effect on the NF-κB
signaling pathway via targeting forkhead box O3a (25). However, the current knowledge
regarding the functions of miRNAs in UCAN remains limited.
miR-31 has been implicated in a variety of diseases.
As mentioned above, miR-31 was suggested to be overexpressed in the
inflamed colonic mucosa in UC and it regulated IL-13 signaling by
targeting IL13RA1 (7). miR-31
regulated not only cytokine receptor expression, but also the Hippo
and Wnt signaling pathways, which promoted the generation of
epithelium in mice with colitis (22). In addition to UC, miR-31 was also
upregulated in tissues from patients with Crohn's disease (26). However, the regulatory role of
miR-31 is not limited to the progression of inflammation in UC. A
number of studies have attempted to elucidate the regulatory
function of miR-31 in the pathogenesis of colon cancer. miR-31 was
considered as a therapeutic target in colon cancer. Wang et
al (27) demonstrated that
inhibiting miR-31 enhanced the sensitivity of colon cancer cells to
5-fluorouracil (5-FU). In addition, the expression of miR-31 was
elevated in 5-FU-resistant colon cancer cell lines. Of note, the
miR-31 levels were indicated to be positively associated with the
clinical stage of CRC (28). In
addition, the relative expression of miR-31 was positively
correlated with the survival time of the patients (29). Therefore, miR-31 may also serve as
a prognosis-predicting index in colon cancer. The present study
demonstrated that miR-31 was overexpressed in UCAN tissues,
particularly in HGD samples, based on both the GEO dataset and the
validation results in the current patient cohort. These results
suggest that miR-31 is involved in the carcinogenesis of UC.
There was an overlap between the target genes of
miR-31 and the DEGs according to the expression profile. Among
those, SATB2, ZC3H12C and RASA1 were selected as the candidates for
further validation. Monocyte chemotactic protein-induced protein 3
(MCPIP3) is also known as ZC3H12C. A study by Suk et al
(30) indicated that MCPIP3 may
act as a potential metastasis suppressor gene in human CRC. RAS p21
activator protein 1 was indicated to be significantly downregulated
in human colon cancer RKO cells exhibiting an aggressive malignant
behavior (31). However, in the
present study, these two target genes of miR-31 were not
differentially expressed in patients with UCAN compared with
patients with UC.
SATB2 is a member of the SATB family of proteins.
SATB2, a nuclear matrix-associated protein, serves as a key
regulator of high-order chromatin organization (32). SATB2 and hepatocyte paraffin 1
expression may be helpful for distinguishing between an inflamed
and architecturally altered ileal pouch and rectal cuff mucosa
(33). Based on the expression of
SATB2 in different tissues, a large number of studies have reported
the involvement of SATB2 in numerous cancer types, including CRC
(34), bone (35), and head and neck cancers (36), with the majority of the studies
focusing on the role of SATB2 in CRC. SATB2 was identified as a
sensitive biomarker for CRC diagnosis with considerable accuracy
(37,38). Particularly when combined with
cytokeratin 20, SATB2 identified >95% of all CRCs (39). In addition, SATB2 expression was
also indicated to be associated with the prognosis of CRC. A number
of epidemiological studies suggested that patients with colon
cancer exhibiting upregulated SATB2 expression had a better
prognosis. According to various studies, including that by Wang
et al (34) downregulated
SATB2 expression was associated with CRC invasion and metastasis
(39,40). Mechanistically, SATB2 expression
was also indicated to be negatively associated with microsatellite
instability that may impair 5-FU sensitivity in CRC (41). Furthermore, low SATB2 expression
was determined to be common and to serve as a biomarker of
UC-associated colorectal dysplasia (42). These results indicate that SATB2
may be involved not only in the progression of CRC, but also in the
carcinogenesis of UCAN. The expression of SATB2 is regulated by
multiple factors (43,44). It has been proven that miR-31
overexpression may repress SATB2 expression, resulting in CRC
progression (43). Therefore, in
the present study, SATB2 was selected as the target gene of miR-31
to be validated. It was observed that the expression of SATB2
decreased in a stepwise manner from inflamed epithelium to
high-grade dysplasia and was negatively correlated with the
expression of miR-31. This indicated that miR-31 promotes
carcinogenesis in UCAN via downregulating SATB2. However, the
downstream mechanism of SATB2 in the carcinogenesis of UCAN remains
elusive. SATB1 is another member of the SATB protein family, which
shares a high degree of sequence homology with SATB2. It has been
reported that the dynamic balance between these two proteins
regulates tumor invasion and metastasis (32). SATB1 acts as a positive regulator
during these processes, whereas SATB2 serves the opposite role.
SATB1 reportedly promotes tumor progression and metastasis through
Wnt/β-catenin (45),
retinoblastoma protein-E2 transcription factor signaling (46) and epithelial-to-mesenchymal
transition-associated transcription factors (47). These regulating functions of SATB1
mentioned above may provide a starting point to investigate the
downstream regulatory mechanism of SATB2 in the future, as further
research on the downstream mechanism of SATB2 in UC is
required.
In conclusion, in the present study, the DEMs and
target genes were screened with the profiles from the GEO database.
The regulatory function of miR-31 on SATB2 was investigated and it
was demonstrated that miR-31 promotes tumorigenesis through
downregulating SATB2. This result may provide a theoretical basis
to further elucidate the pathogenesis of UCAN. However, there were
certain limitations to the present study. The microarray data were
from the GEO database rather than our institution. The presence of
selection bias cannot be excluded. In addition, the difference in
the expression of miR-31 was not particularly notable, which may be
attributed to inter-individual differences, although it was
determined to be statistically significant. Further studies are
required to confirm the present results and to further elucidate
the pathogenesis of UCAN.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KJ and BMW conceived the study and interpreted the
data. WTL and RL designed the study. YS performed the experiments,
analyzed the data and wrote the manuscript. RL also generated the
figures and tables. BMW approved the manuscript to be published.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from all of the
patients. This study was approved by the Ethics Committee of
Tianjin Medical University General Hospital (Tianjin, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Huang R, Wang K, Gao L and Gao W: TIMP1 is
a potential key gene associated with the pathogenesis and prognosis
of ulcerative colitis-associated colorectal cancer. OncoTargets
Ther. 12:8895–8904. 2019. View Article : Google Scholar
|
|
2
|
Kawachi H: Histopathological diagnosis of
ulcerative colitis-associated neoplasia. Dig Endosc. 1 (Suppl
31):31–35. 2019. View Article : Google Scholar
|
|
3
|
Riddell RH, Goldman H, Ransohoff DF,
Appelman HD, Fenoglio CM, Haggitt RC, Ahren C, Correa P, Hamilton
SR, Morson BC, et al: Dysplasia in inflammatory bowel disease:
Standardized classification with provisional clinical applications.
Hum Pathol. 14:931–968. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ungaro R, Mehandru S, Allen PB,
Peyrin-Biroulet L and Colombel JF: Ulcerative colitis. Lancet.
389:1756–1770. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koukos G, Polytarchou C, Kaplan JL,
Morley-Fletcher A, Gras-Miralles B, Kokkotou E, Baril-Dore M,
Pothoulakis C, Winter HS and Iliopoulos D: MicroRNA-124 regulates
STAT3 expression and is down-regulated in colon tissues of
pediatric patients with ulcerative colitis. Gastroenterology.
145:842–852.e842. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gwiggner M, Martinez-Nunez RT, Whiteoak
SR, Bondanese VP, Claridge A, Collins JE, Cummings JRF and
Sanchez-Elsner T: MicroRNA-31 and MicroRNA-155 are overexpressed in
ulcerative colitis and regulate IL-13 signaling by targeting
interleukin 13 receptor α-1. Genes (Basel). 9:852018. View Article : Google Scholar
|
|
8
|
Zhang XF, Tu R, Li K, Ye P and Cui X:
Tumor suppressor PTPRJ is a target of miR-155 in colorectal cancer.
J Cell Biochem. 118:3391–3400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Haidari A, Algaber A, Madhi R, Syk I
and Thorlacius H: miR-155-5p controls colon cancer cell migration
via post-transcriptional regulation of human antigen R (HuR).
Cancer Lett. 421:145–151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
The Gene Ontology Consortium: Expansion of
the gene ontology knowledgebase and resources. Nucleic Acids Res.
45:D331–D338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pekow J, Meckel K, Dougherty U, Huang Y,
Chen X, Almoghrabi A, Mustafi R, Ayaloglu-Butun F, Deng Z, Haider
HI, et al: miR-193a-3p is a key tumor suppressor in ulcerative
colitis-associated colon cancer and promotes carcinogenesis through
upregulation of IL17RD. Clin Cancer Res. 23:5281–5291. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gutierrez-Uribe JA, Salinas-Santander M,
Serna-Guerrero D, Serna-Saldivar SRO, Rivas-Estilla AM and
Rios-Ibarra CP: Inhibition of miR31 and miR92a as oncological
biomarkers in RKO colon cancer cells treated with
kaempferol-3-O-glycoside isolated from black bean. J Med Food.
23:50–55. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li T, Luo W, Liu K, Lv X and Xi T: miR-31
promotes proliferation of colon cancer cells by targeting E2F2.
Biotechnol Lett. 37:523–532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xie ZY, Wang FF, Xiao ZH, Liu SF, Tang SL
and Lai YL: Overexpressing microRNA-34a overcomes ABCG2-mediated
drug resistance to 5-FU in side population cells from colon cancer
via suppressing DLL1. J Biochem. 167:557–564. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li X, Li C, Li D, Yang L, Jin J and Zhang
B: lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in
colon cancer by targeting the miR-34a/ATG4B pathway. OncoTargets
Ther. 12:2649–2660. 2019. View Article : Google Scholar
|
|
21
|
Zhuang M, Zhao S, Jiang Z, Wang S, Sun P,
Quan J, Yan D and Wang X: MALAT1 sponges miR-106b-5p to promote the
invasion and metastasis of colorectal cancer via SLAIN2 enhanced
microtubules mobility. EBioMedicine. 41:286–298. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tian Y, Xu J, Li Y, Zhao R, Du S, Lv C, Wu
W, Liu R, Sheng X, Song Y, et al: MicroRNA-31 reduces inflammatory
signaling and promotes regeneration in colon epithelium, and
delivery of mimics in microspheres reduces colitis in mice.
Gastroenterology. 156:2281–2296.e2286. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Munakata K, Koi M, Kitajima T,
Tseng-Rogenski S, Uemura M, Matsuno H, Kawai K, Sekido Y, Mizushima
T, Toiyama Y, et al: Inflammation-associated microsatellite
alterations caused by MSH3 dysfunction are prevalent in ulcerative
colitis and increase with neoplastic advancement. Clin Transl
Gastroenterol. 10:e001052019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kvorjak M, Ahmed Y, Miller ML, Sriram R,
Coronnello C, Hashash JG, Hartman DJ, Telmer CA, Miskov-Zivanov N,
Finn OJ and Cascio S: Cross-talk between colon cells and
macrophages increases ST6GALNAC1 and MUC1-sTn expression in
ulcerative colitis and colitis-associated colon cancer. Cancer
Immunol Res. 8:167–178. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Min M, Peng L, Yang Y, Guo M, Wang W and
Sun G: MicroRNA-155 is involved in the pathogenesis of ulcerative
colitis by targeting FOXO3a. Inflamm Bowel Dis. 20:652–659. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin J, Welker NC, Zhao Z, Li Y, Zhang J,
Reuss SA, Zhang X, Lee H, Liu Y and Bronner MP: Novel specific
microRNA biomarkers in idiopathic inflammatory bowel disease
unrelated to disease activity. Mod Pathol. 27:602–608. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang CJ, Stratmann J, Zhou ZG and Sun XF:
Suppression of microRNA-31 increases sensitivity to 5-FU at an
early stage, and affects cell migration and invasion in HCT-116
colon cancer cells. BMC Cancer. 10:6162010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nakagawa Y, Kuranaga Y, Tahara T,
Yamashita H, Shibata T, Nagasaka M, Funasaka K, Ohmiya N and Akao
Y: Induced miR-31 by 5-fluorouracil exposure contributes to the
resistance in colorectal tumors. Cancer Sci. 110:2540–2548. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cui Q: Significance of miR-27a and miR-31
in early diagnosis and prognosis of colorectal cancer. Oncol Lett.
18:3092–3096. 2019.PubMed/NCBI
|
|
30
|
Suk FM, Chang CC, Lin RJ, Lin SY, Chen YT
and Liang YC: MCPIP3 as a potential metastasis suppressor gene in
human colorectal cancer. Int J Mol Sci. 19:13502018. View Article : Google Scholar
|
|
31
|
Gong B, Liu WW, Nie WJ, Li DF, Xie ZJ, Liu
C, Liu YH, Mei P and Li ZJ: MiR-21/RASA1 axis affects malignancy of
colon cancer cells via RAS pathways. World J Gastroenterol.
21:1488–1497. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Naik R and Galande S: SATB family
chromatin organizers as master regulators of tumor progression.
Oncogene. 38:1989–2004. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Groisman G, Cai Z, Sabo E and Harpaz N:
SATB2 and Hep Par 1 immunohistochemistry is helpful in
distinguishing between inflamed and architecturally altered Ileal
pouch and rectal cuff mucosa. Int J Surg Pathol. 27:159–165. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang S, Zhou J, Wang XY, Hao JM, Chen JZ,
Zhang XM, Jin H, Liu L, Zhang YF, Liu J, et al: Down-regulated
expression of SATB2 is associated with metastasis and poor
prognosis in colorectal cancer. J Pathol. 219:114–122. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seong BKA, Lau J, Adderley T, Kee L,
Chaukos D, Pienkowska M, Malkin D, Thorner P and Irwin MS: SATB2
enhances migration and invasion in osteosarcoma by regulating genes
involved in cytoskeletal organization. Oncogene. 34:3582–3592.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu TR, Xu LH, Yang AK, Zhong Q, Song M,
Li MZ, Hu LJ, Chen FJ, Hu ZD, Han P and Zeng MS: Decreased
expression of SATB2: A novel independent prognostic marker of worse
outcome in laryngeal carcinoma patients. PLoS One. 7:e407042012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ma C, Olevian D, Miller C, Herbst C,
Jayachandran P, Kozak MM, Chang DT and Pai RK: SATB2 and CDX2 are
prognostic biomarkers in DNA mismatch repair protein deficient
colon cancer. Mod Pathol. 32:1217–1231. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin F, Shi J, Zhu S, Chen Z, Li A, Chen T,
Wang HL and Liu H: Cadherin-17 and SATB2 are sensitive and specific
immunomarkers for medullary carcinoma of the large intestine. Arch
Pathol Lab Med. 138:1015–1026. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Magnusson K, de Wit M, Brennan DJ, Johnson
LB, McGee SF, Lundberg E, Naicker K, Klinger R, Kampf C, Asplund A,
et al: SATB2 in combination with cytokeratin 20 identifies over 95%
of all colorectal carcinomas. Am J Surg Pathol. 35:937–948. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang MH, Yu J, Jiang DM, Li WL, Wang S and
Ding YQ: microRNA-182 targets special AT-rich sequence-binding
protein 2 to promote colorectal cancer proliferation and
metastasis. J Transl Med. 12:1092014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Eberhard J, Gaber A, Wangefjord S, Nodin
B, Uhlén M, Ericson Lindquist K and Jirström K: A cohort study of
the prognostic and treatment predictive value of SATB2 expression
in colorectal cancer. Br J Cancer. 106:931–938. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ma C, Henn P, Miller C, Herbst C, Hartman
DJ and Pai RK: Loss of SATB2 expression is a biomarker of
inflammatory bowel disease-associated colorectal dysplasia and
adenocarcinoma. Am J Surg Pathol. 43:1314–1322. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang MH, Yu J, Chen N, Wang XY, Liu XY,
Wang S and Ding YQ: Elevated microRNA-31 expression regulates
colorectal cancer progression by repressing its target gene SATB2.
PLoS One. 8:e853532013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang YQ, Jiang DM, Hu SS, Zhao L, Wang L,
Yang MH, Ai ML, Jiang HJ, Han Y, Ding YQ and Wang S: SATB2-AS1
suppresses colorectal carcinoma aggressiveness by inhibiting
SATB2-dependent snail transcription and epithelial-mesenchymal
transition. Cancer Res. 79:3542–3556. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mir R, Pradhan SJ, Patil P, Mulherkar R
and Galande S: Wnt/β-catenin signaling regulated SATB1 promotes
colorectal cancer tumorigenesis and progression. Oncogene.
35:1679–1691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Agrelo R, Kishimoto H, Novatchkova M,
Peraza V, Paolino M, Souabni A and Wutz A: SATB1 collaborates with
loss of p16 in cellular transformation. Oncogene. 32:5492–5500.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wan F, Cheng C, Wang Z, Xiao X, Zeng H,
Xing S, Chen X, Wang J, Li S, Zhang Y, et al: SATB1 overexpression
regulates the development and progression in bladder cancer through
EMT. PLoS One. 10:e01175182015. View Article : Google Scholar : PubMed/NCBI
|