Introduction
Sebaceous gland carcinoma (SGC) of the eyelid is a
highly malignant tumor that most frequently arises from the
meibomian gland and Zeis gland in the periocular region (1). According to clinical reports, it is
uncommon in Caucasians populations, accounting for less than a few
percent of cases of malignant eyelid tumors, but relatively common
in Asian populations, accounting for ~30% of malignant eyelid
tumors (2,3). Early diagnosis of SGC is difficult
because the disease can mimic benign inflammatory conditions such
as chalazion, unilateral conjunctivitis, blepharitis, tarsitis and
blepharoconjunctivitis (1,4). SGC tends to be histopathologically
misdiagnosed as squamous cell carcinoma or basal cell carcinoma
(5,6), but immunohistological stains for
adipophilin (7,8) and androgen receptor (9,10) and
Oil Red O staining can be helpful to confirm a diagnosis.
First-line treatments are surgery and cryotherapy followed by
chemotherapy and radiotherapy (11), but some patients have poor
prognosis. Delayed diagnosis may result in metastasis to lymph
nodes and other organs, leading to metastasis-related mortality in
~6–9% of cases (12–14). Therefore, early and accurate
diagnostic markers unique to SGC are needed to improve the
prognosis.
The molecular mechanisms underlying the pathogenesis
and progression of SGC remain to be fully elucidated. As is the
case with other cancers, most SGC have point mutations in the p53
tumor suppressor gene (15) and
overexpressed anti-apoptotic proteins including X-linked inhibitor
of apoptosis (XIAP) (16) and BAG
cochaperone 3 (BAG3) (17). High
expression of growth factor receptors such as vascular endothelial
growth factor receptor-2, epidermal growth factor receptor, and
platelet-derived growth factor receptor are also known as
clinicopathologic features of SGC (18). Moreover, expression levels of
prognosis factors such as zinc finger E-box binding homeobox 2
(ZEB2) (19), human epidermal
growth factor receptor 2 (20) and
aldehyde dehydrogenase 1 (21) were
known to be used for the prognosis prediction. In our previous
study, we revealed the mRNA expression profiles of SGC and
identified the gene network consisting of cell cycle related genes
including cyclin dependent kinase inhibitor 2A (CDKN2A),
cyclin dependent kinase 1 (CDK1) and cyclin E1
(CCNE1) (22). To date,
although a number of studies have been conducted to explore novel
therapeutic targets, no specific protein expression patterns have
been identified for either primary or metastatic lesions of SGC
(18).
MicroRNAs (miRNAs) are small non-coding RNAs that
bind to complementary sequences of multiple target mRNAs, resulting
in post-transcriptional inhibition of gene expression. In humans,
more than 2,000 miRNAs controlling complex cellular processes such
as proliferation, apoptosis, development and differentiation have
been identified (23). In many
cancers, miRNAs play roles as upstream regulators of tumorigenesis
by contributing to alterations in the gene expression of oncogenes
and tumor suppressor genes (24,25)
and exploring miRNA-mRNA interactions will thus be critically
important to improve our understanding of pathogenesis. Expression
patterns of miRNAs vary critically depending on the types of
cancers, and a number of clinical trials are currently underway to
examine the use of circulating miRNAs as molecular biomarkers for
cancer diagnosis (26,27). A few previous studies have shown
expression changes in only a limited number of miRNA in SGC samples
(28–30), and whole picture of the miRNA-mRNA
network of SGC are not fully understood.
In the present study, a small RNA-sequencing
analysis were performed to reveal the miRNA expression profiles of
SGC and to identify differentially expressed miRNAs common to the
tumor samples from three patients with SGC compared to a sebaceous
adenoma control sample. In addition, we conducted integrated
bioinformatics analyses to identify biological functions, canonical
pathways and miRNA-mRNA networks of SGC using the data of mRNA
expression profiles obtained from the same tumor sample sets in our
previous study (22).
Materials and methods
Patient and tissue samples
This study was performed with the approval of the
internal review board of the University of Toyama (no. 27-51), and
the procedures conformed to the tenets of the World Medical
Association's Declaration of Helsinki. Written informed consent was
obtained from all patients prior to enrollment in the present
study. Tissues were obtained by surgical excision of tumors from
four patients: a 74-year-old woman (case 1), an 83-year-old woman
(case 2) and a 58-year-old woman (case 3) with SGC of the eyelid;
and a 92-year-old man with sebaceous adenoma of the eyelid
(control) for comparison. Tissue samples were immediately frozen
and stored at −80°C after sampling for RNA extraction.
RNA extraction and quality
control
Total RNA including miRNA was extracted from tissue
samples using a NucleoSpin miRNA kit (Macherey-Nagel GmbH &
Co.) following the manufacturer's instructions. The quality and
quantity of the miRNA were analyzed using a Bioanalyzer 2100 with
an RNA 6000 Nano kit (Agilent Technologies) (31).
Library preparation and small
RNA-sequencing
Small RNA libraries were prepared using a NEBNext
Multiplex Small RNA Library Prep Set for Illumina Set 1 (New
England BioLabs). In brief, 1 µg of total RNA per sample was
ligated with 3′ and 5′ adaptors and reverse transcribed into
first-strand cDNA. Each library was labeled with indexed primers by
15 cycles of PCR amplification and cleaned up using a QIAquick PCR
purification kit (Qiagen). Appropriate fractions of 140–150 bp were
size-selected by polyacrylamide electrophoresis on the Novex TBE
PAGE gel 6% (Invitrogen; Thermo Fisher Scientific, Inc.) and then
the purity and concentration were checked using a Bioanalyzer 2100
with a High Sensitivity DNA kit (Agilent Technologies). The pooled
libraries were sequenced (2×150 bp) on the HiSeq X Ten platform
(Illumina) by Genewiz, Inc. All low sequence data analyzed in this
study were deposited in the DNA Data Bank of Japan database under
the accession number DRA009187.
Row read data processing
The first 50 bp sequences were extracted from the
raw 150 bp sequence reads using Seqkit. Adaptor sequences were
trimmed from 50 bp reads using cutadapt. Low-quality (less than
Q20) and short-length (less than 10 bp) sequences were removed from
processed reads using the FASTX-tool kit. The filtered reads were
mapped with hg19, and miRNA annotation was performed using Strand
NGS Ver. 3.3.
Microarray data and miRNA-mRNA
interaction analyses
In our previous study, mRNA expression profile data
of the same sample set were obtained using the GeneChip system with
Clariom S human arrays (Affymetrix) (22). Briefly, the raw intensity data (Gene
Expression Omnibus; accession no. GSE125582) were normalized using
GeneSpring GX 14.9 software (Agilent Technologies). To examine the
molecular functions and interaction networks of differentially
expressed miRNA and mRNA, the combined data from our present and
previous studies were analyzed using Ingenuity Pathways Analysis
(IPA) software (Ingenuity Systems).
Results
Identification of differentially
expressed miRNAs and mRNAs
To reveal the miRNA expression profiles of SGC
samples, a total of 280,241,626 raw reads were obtained in this
study, including at least 20,000,000 reads for each sample. Raw
sequencing reads were quality checked, and the low-quality
sequences and adaptors were removed; the reads were then aligned
against the human miRBase using Strand NGS. We obtained read counts
of over 2,600 miRNAs, and then identified miRNAs that were at least
2.0-fold differentially expressed compared with the control sample.
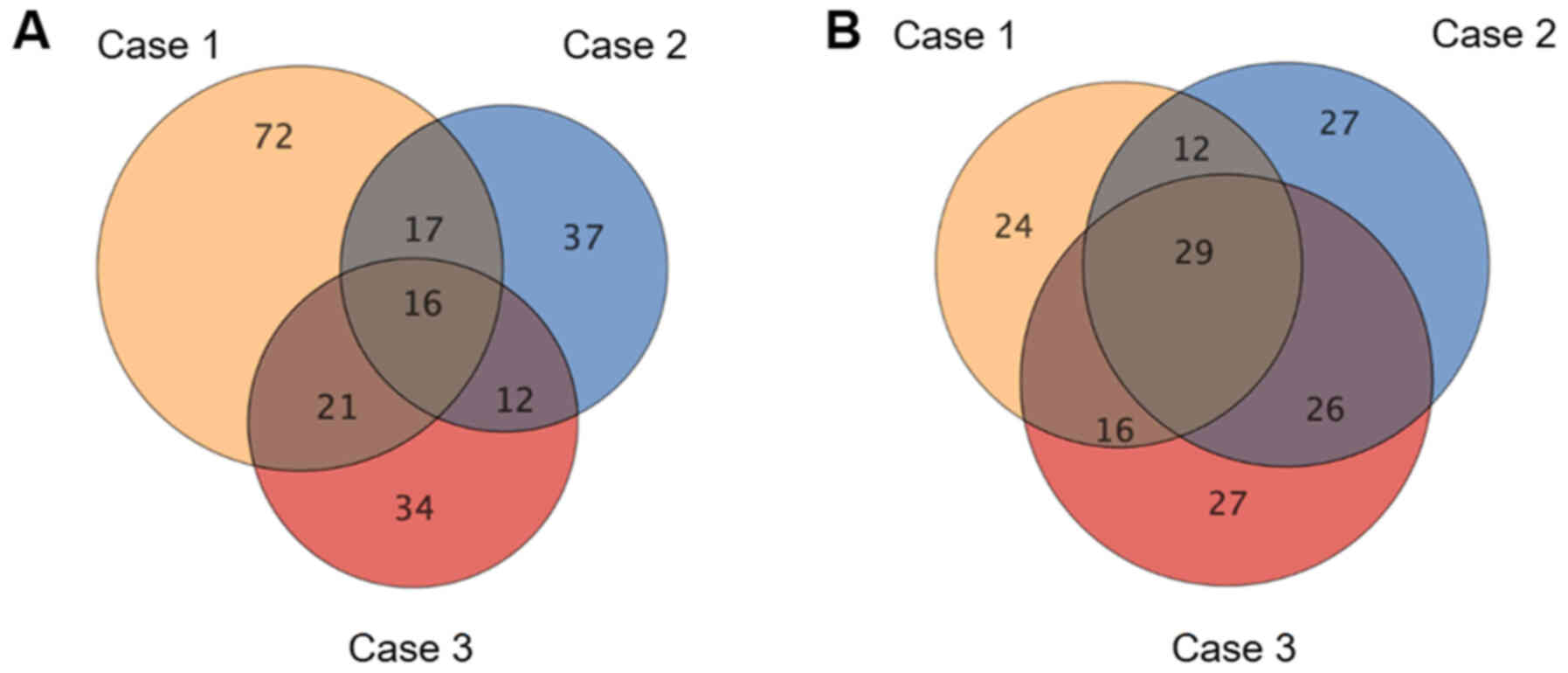
As shown in the Venn-diagrams in Fig.
1, 16 upregulated and 29 downregulated miRNAs were common to
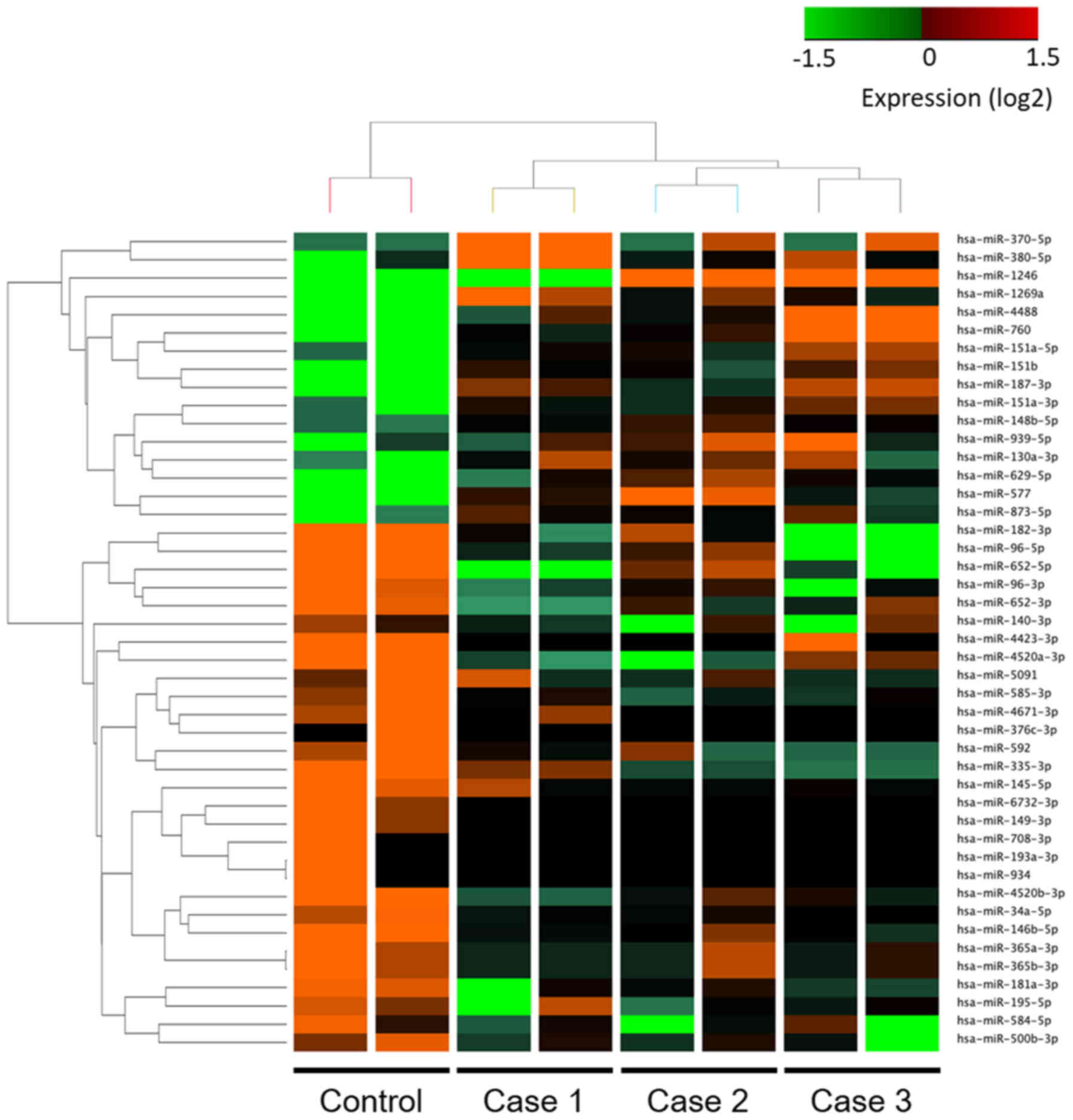
all three tumor samples; these 45 miRNAs are listed in Tables SI and SII. Hierarchical clustering showed that
there were distinct expression profiles of miRNAs in SGC and the
control sebaceous adenoma sample (Fig.
2). Similarly, 194 upregulated and 516 downregulated mRNAs with
at least 2.0-fold change were also identified from our previous
study (22).
Functional analyses of differentially
expressed miRNAs and mRNAs
To explore the biological functions and canonical
pathways involving the miRNAs and mRNAs that were differentially
expressed between the SGC and control samples, two integrated
miRNA-mRNA data sets including 16 upregulated miRNAs with 516
downregulated mRNAs and 29 downregulated miRNAs with 194
upregulated mRNAs were analyzed by IPA software. The top 5
biological functions with positive z-scores in the differentially
expressed miRNAs and mRNAs are summarized in Tables I and II. Most biological functions that were
significantly enriched in the 16 upregulated miRNAs with 516
downregulated mRNAs were related to the decreasing of the lipid
metabolism (i.e., ‘Synthesis of lipid’ and ‘Fatty acid
metabolism’). In contrast, 29 downregulated miRNAs with 194
upregulated mRNAs were associated with the increasing of the cell
survival and proliferation (i.e., ‘Cell viability of tumor cell
lines’, ‘Cell viability’ and ‘Cell proliferation of tumor cell
lines’). As shown in Figs. S1 and
S2, the top 10 canonical pathways
that were differentially activated or suppressed in two integrated
miRNA-mRNA data sets are presented, respectively. Multiple
annotations associated with cholesterol biosynthesis (i.e.,
‘Superpathway of Cholesterol Biosynthesis’ and ‘Cholesterol
Biosynthesis I’ to ‘Cholesterol Biosynthesis III’) were redundantly
listed in 16 upregulated miRNAs with 516 downregulated mRNAs.
Additionally, 29 downregulated miRNAs with 194 upregulated mRNAs
were mainly involved in DNA damage-induced cell cycle regulation
pathways (i.e., ‘DNA damage-induced 14-3-3δ Signaling’, ‘GADD45
Signaling’ and ‘Cell Cycle: G2/M DNA Damage Checkpoint Regulation’)
with no activity pattern available.
Construction of molecular interaction
networks of differentially expressed miRNAs and mRNAs
To further understand the regulatory interaction of
differentially expressed miRNAs and mRNAs in SGC samples, target
prediction analyses were conducted based on the miRNA target filter
tool on the IPA software. We identified two miRNA-mRNA networks
including 7 upregulated miRNAs that downregulated 8 mRNAs related
to decreasing of the synthesis of lipid (Fig. 3) and 5 downregulated miRNAs that
upregulated 9 mRNAs related to increasing of the cell proliferation
of tumor cell lines (Fig. 4). Based
on the results shown in Fig. 3,
miR-130a-3p and miR-939-5p were identified as the key hub nodes
connected with 3 target mRNAs in the network. In addition, the
results shown in Fig. 4
demonstrated that downregulation of miR-146a-5p, miR-149-3p,
miR-193a-3p, miR-195-5p and miR-4671-3p played regulatory roles in
the promotion of cell proliferation.
Discussion
SGC of the eyelid is a rare aggressive tumor with a
relatively high rate of metastasis and mortality. One of the
treatments for SGC is a complete surgical resection, but the
disease occasionally recurs with poor prognosis (6,32). The
pathogenesis of SGC remains unclear; therefore, a detailed
understanding of the molecular mechanisms will be crucial for
improvement of the disease diagnosis, treatment and prognosis. In
the present study, we determined the relevant miRNA expression
profiles and identified 16 miRNAs that were upregulated and 29
miRNAs that were downregulated in SGC samples compared with the
control sebaceous adenoma sample. We then explored the biological
functions and canonical pathways and miRNA-mRNA networks related to
the clinicopathological characteristics of SGC using integrated
miRNA-mRNA data sets. To the best of our knowledge, there are only
three previous studies about miRNA expression of SGC. In the first,
Bhardwaj et al (28) showed
that underexpression of miR-200c and miR-141 were correlated with
clinicopathological parameters in SGC, but in our present study we
did not observe the downregulation of either of these miRNAs. In
the second, Bladen et al (29) demonstrated miRNA expression profiles
of SGC using miRNA arrays with 800 probe sets and identified
overexpression of miR-16-5p and miR-34a-5p, which were
downregulated in the present study. These inconsistencies might be
attributable to the control samples used in each experiment: the
previous studies used normal tissues (adjacent normal epidermis and
tarsal plate) as the control samples while we used sebaceous
adenoma. In the third study, Tetzlaff et al (30) examined the expression of 387 miRNAs
in SGC by real-time polymerase chain reaction techniques and found
that miR-486-5p and miR-184 were overexpressed, while miR-211 and
miR-195 were downregulated. They used formalin-fixed
paraffin-embedded tissue of sebaceous adenoma as the control
samples, and their finding that miR-195 was downregulated was in
agreement with our present results. Unlike these previous studies,
our present analysis revealed comprehensive expression profiles of
over 2,600 miRNAs in SGC using next-generation sequence techniques,
and thus our results could provide novel findings of molecular
mechanisms of SGC.
One of the interesting findings of this study is
that the 16 upregulated miRNAs and 516 downregulated mRNAs in SGC
samples were highly associated with the downregulation of lipid
metabolism functions and enriched in cholesterol biosynthesis
pathways. The main origins of SGC tumors, the meibomian gland and
Zeis gland, produce oily substances to protect the periocular
regions. As mentioned above, lipid accumulation in the cytoplasm,
which is detectable by immunohistochemical staining for adipophilin
and Oil Red O staining, is a practical pathological marker of SGC
(7,8), which supports the idea that a
malfunction in lipid metabolism of the sebaceous glands is involved
in the pathogenesis of SGC. It is noted that the gene expressions
of both thyroid hormone responsive spot14 (THRSP) and MID1
interacting protein 1 (MID1IP1), a ligand/receptor pair that
regulates fatty acid synthesis in non-hepatic cells (33) and lipogenic cancer cells (34,35),
were downregulated in this study. In addition, downregulated genes
including low-density lipoprotein receptor (LDLR) and
glycerol-3-phosphate dehydrogenase 1 (GPD1) were regulatory factors
in the synthesis of cholesterol and triglycerides (36,37).
Our network demonstrated that miR-130a-3p and miR-939-5p were
upstream regulators controlling the expression of these genes
related to lipid metabolism. In particular, miR-130 suppressed
adipogenesis in human adipocytes in association with a decrease in
the gene expression of peroxisome proliferator-activated receptor γ
(PPARγ) (38), implying that
upregulation of miR-130a-3p could be an index marker related to
loss of lipid metabolism functions.
On the other hand, our results also indicated that
functions related to cell survival and proliferation were activated
in the 29 downregulated miRNAs and 194 upregulated mRNAs in SGC
samples. Considering that DNA damage-induced cell cycle regulation
pathways were also significantly enriched in the data set, there
should be abnormalities in the G2/M cell cycle checkpoint resulting
in cell-cycle progression in SGC. Several studies previously showed
that high expressions of cell cycle regulatory proteins including
p21, p27, cyclin E and p16 (39,40),
but hypermethylation of promoter region of CDKN2A were found
in half of the cases of SGC tumor (41). Our network showed that upregulation
of cell cycle-related genes, including CCNE1, CCNE2 and
cyclin dependent kinase inhibitor 3 (CDKN3), was caused by
downregulation of miR146a-5p, miR-195-5p and miR-4671-3p. Most
importantly, miR-195 is also known to inhibit cell proliferation in
association with a decrease in protein expression of cyclin D1 in
human cervical cancer cells (42,43),
human glioma cells (44) and
squamous cell lung cancer (45). In
addition, miR-149 acts as a tumor suppressor miRNA controlling cell
proliferation and invasion in medullary thyroid carcinoma (46) and renal cell carcinoma (47), and it also plays important roles in
regulating the expression of multiple genes in SGC. Overexpression
and point mutation of the p53 gene were detected in two-thirds of
SGC samples (15,39), suggesting that dysregulation of the
cell cycle with downregulation of these miRNA was one of the
critical mechanisms in the tumorigenesis and development of SGC
tumors.
In conclusion, the present study provides the first
comprehensive description of the differentially expressed miRNAs
and miRNA-mRNA interaction networks in SGC. We also identified
several changes in the expression of miRNAs that control important
functional alterations in SGC, including loss of lipid metabolism
and promotion of the cell proliferation. These results could
improve our understanding of the pathophysiological mechanisms of
SGC and provide novel clues for earlier and more accurate
diagnosis. Further studies will be needed to confirm the functional
roles of these miRNA-mRNA networks in the pathogenesis of SGC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by JSPS KAKENHI (grant nos.
JP16K20309, JP17K01353, JP18K09442 and JP19K19406).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TH, TY, YF and YT designed the present study,
performed the experimental analysis and wrote the manuscript. TY
and AH performed the surgical procedures. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was performed with the approval of the
Internal Review Board of the University of Toyama (no. 27-51), and
the procedures conformed to the tenets of the World Medical
Association's Declaration of Helsinki. Written informed consent was
obtained from all patients prior to enrollment in the present
study.
Patient consent for publication
Written informed consent was obtained from the
patients after they were provided with sufficient information about
the procedures and the publication of results.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCNE1
|
cyclin E1
|
|
CDK1
|
cyclin dependent kinase 1
|
|
CDKN2A
|
cyclin dependent kinase inhibitor
2A
|
|
IPA
|
Ingenuity Pathways Analysis
|
|
miRNA
|
microRNA
|
|
SGC
|
sebaceous gland carcinoma
|
References
|
1
|
Shields JA, Demirci H, Marr BP, Eagle RC
Jr and Shields CL: Sebaceous carcinoma of the ocular region: A
review. Surv Ophthalmol. 50:103–122. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cook BE Jr and Bartley GB: Epidemiologic
characteristics and clinical course of patients with malignant
eyelid tumors in an incidence cohort in Olmsted County, Minnesota.
Ophthalmology. 106:746–750. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takamura H and Yamashita H:
Clinicopathological analysis of malignant eyelid tumor cases at
Yamagata university hospital: Statistical comparison of tumor
incidence in Japan and in other countries. Jpn J Ophthalmol.
49:349–354. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Buitrago W and Joseph AK: Sebaceous
carcinoma: the great masquerader: emgerging concepts in diagnosis
and treatment. Dermatol Ther. 21:459–466. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai TF, Huilgol SC, Selva D and James CL:
Eyelid sebaceous carcinoma masquerading as in situ squamous cell
carcinoma. Dermatol Surg. 30:222–225. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kan LW, Leu YS, Tzen CY and Wu CH:
Recurrent sebaceous gland carcinoma of eyelid previously diagnosed
as basal cell carcinoma: Case report. Am J Otolaryngol. 32:620–623.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jakobiec FA and Mendoza PR: Eyelid
sebaceous carcinoma: Clinicopathologic and multiparametric
immunohistochemical analysis that includes adipophilin. Am J
Ophthalmol. 157:186–208.e2. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Milman T, Schear MJ and Eagle RC Jr:
Diagnostic utility of adipophilin immunostain in periocular
carcinomas. Ophthalmology. 121:964–971. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mulay K, White VA, Shah SJ and Honavar SG:
Sebaceous carcinoma: Clinicopathologic features and diagnostic role
of immunohistochemistry (including androgen receptor). Can J
Ophthalmol. 49:326–332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yunoki T, Miyakoshi A, Otsuka M and
Hayashi A: Clinicopathological features of considerable reduction
in androgen receptor expression in sebaceous gland carcinoma of the
eyelid. Int Ophthalmol. 39:1703–1708. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shields JA, Demirci H, Marr BP, Eagle RC
Jr and Shields CL: Sebaceous carcinoma of the eyelids: Personal
experience with 60 cases. Ophthalmology. 111:2151–2157. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zürcher M, Hintschich CR, Garner A, Bunce
C and Collin JR: Sebaceous carcinoma of the eyelid: A
clinicopathological study. Br J Ophthalmol. 82:1049–1055. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Muqit MM, Roberts F, Lee WR and Kemp E:
Improved survival rates in sebaceous carcinoma of the eyelid. Eye
(Lond). 18:49–53. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song A, Carter KD, Syed NA, Song J and
Nerad JA: Sebaceous cell carcinoma of the ocular adnexa: Clinical
presentations, histopathology, and outcomes. Ophthal Plast Reconstr
Surg. 24:194–200. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kiyosaki K, Nakada C, Hijiya N, Tsukamoto
Y, Matsuura K, Nakatsuka K, Daa T, Yokoyama S, Imaizumi M and
Moriyama M: Analysis of p53 mutations and the expression of p53 and
p21WAF1/CIP1 protein in 15 cases of sebaceous carcinoma of the
eyelid. Invest Ophthalmol Vis Sci. 51:7–11. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jayaraj P, Sen S, Dhanaraj PS, Jhajhria R,
Singh S and Singh VK: Immunohistochemical expression of X-linked
inhibitor of apoptosis in eyelid sebaceous gland carcinoma predicts
a worse prognosis. Indian J Ophthalmol. 65:1109–1113. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yunoki T, Tabuchi Y and Hayashi A:
Expression of anti-apoptotic protein BAG3 in human sebaceous gland
carcinoma of the eyelid. Anticancer Res. 37:1931–1934. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Erovic BM, Al Habeeb A, Harris L,
Goldstein DP, Kim D, Ghazarian D and Irish JC: Identification of
novel target proteins in sebaceous gland carcinoma. Head Neck.
35:642–648. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bhardwaj M, Sen S, Sharma A, Kashyap S,
Chosdol K, Pushker N, Bajaj MS and Bakhshi S: ZEB2/SIP1 as novel
prognostic indicator in eyelid sebaceous gland carcinoma. Hum
Pathol. 46:1437–1442. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee MJ, Kim N, Choung HK, Choe JY, Khwarg
SI and Kim JE: Increased gene copy number of HER2 and concordant
protein overexpression found in a subset of eyelid sebaceous gland
carcinoma indicate HER2 as a potential therapeutic target. J Cancer
Res Clin Oncol. 142:125–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim N, Choung HK, Lee MJ, Khwarg SI and
Kim JE: Cancer stem cell markers in eyelid sebaceous gland
carcinoma: High expression of ALDH1, CD133, and ABCG2 correlates
with poor prognosis. Invest Ophthalmol Vis Sci. 56:1813–1819. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yunoki T, Hirano T, Tabuchi Y, Furusawa Y,
Torigoe M, Nakajima T, Imura J and Hayashi A: CDKN2A, CDK1, and
CCNE1 overexpression in sebaceous gland carcinoma of eyelid. Int
Ophthalmol. 40:343–350. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Olive V, Minella AC and He L: Outside the
coding genome, mammalian microRNAs confer structural and functional
complexity. Sci Signal. 8:re22015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bhardwaj M, Sen S, Chosdol K, Sharma A,
Pushker N, Kashyap S, Bakhshi S and Bajaj MS: miRNA-200c and
miRNA-141 as potential prognostic biomarkers and regulators of
epithelial-mesenchymal transition in eyelid sebaceous gland
carcinoma. Br J Ophthalmol. 101:536–542. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bladen JC, Wang J, Sangaralingam A,
Moosajee M, Fitchett C, Chelala C, Beaconsfield M, O'Toole EA,
Philpott MP and Ezra DG: MicroRNA and transcriptome analysis in
periocular Sebaceous Gland Carcinoma. Sci Rep. 8:75312018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tetzlaff MT, Curry JL, Yin V,
Pattanaprichakul P, Manonukul J, Uiprasertkul M, Manyam GC, Wani
KM, Aldape K, Zhang L, et al: Distinct pathways in the pathogenesis
of sebaceous carcinomas implicated by differentially expressed
microRNAs. JAMA Ophthalmol. 133:1109–1116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Furusawa Y, Yunoki T, Hirano T, Minagawa
S, Izumi H, Mori H, Hayashi A and Tabuchi Y: Identification of
genes and genetic networks associated with BAG3 dependent cell
proliferation and cell survival in human cervical cancer HeLa
cells. Mol Med Rep. 18:4138–4146. 2018.PubMed/NCBI
|
|
32
|
Kaliki S, Ayyar A, Dave TV, Ali MJ, Mishra
DK and Naik MN: Sebaceous gland carcinoma of the eyelid:
Clinicopathological features and outcome in Asian Indians. Eye
(Lond). 29:958–963. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Q, Yang J, Lin X, Huang Z, Xie C and
Fan H: Spot14/Spot14R expression may be involved in MSC adipogenic
differentiation in patients with adolescent idiopathic scoliosis.
Mol Med Rep. 13:4636–4642. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wells WA, Schwartz GN, Morganelli PM, Cole
BF, Gibson JJ and Kinlaw WB: Expression of ‘Spot 14’ (THRSP)
predicts disease free survival in invasive breast cancer:
Immunohistochemical analysis of a new molecular marker. Breast
Cancer Res Treat. 98:231–240. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Donnelly C, Olsen AM, Lewis LD, Eisenberg
BL, Eastman A and Kinlaw WB: Conjugated linoleic acid (CLA)
inhibits expression of the Spot 14 (THRSP) and fatty acid synthase
genes and impairs the growth of human breast cancer and liposarcoma
cells. Nutr Cancer. 61:114–122. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Goldstein JL and Brown MS: The LDL
receptor. Arterioscler Thromb Vasc Biol. 29:431–438. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Basel-Vanagaite L, Zevit N, Har Zahav A,
Guo L, Parathath S, Pasmanik-Chor M, McIntyre AD, Wang J,
Albin-Kaplanski A, Hartman C, et al: Transient infantile
hypertriglyceridemia, fatty liver, and hepatic fibrosis caused by
mutated GPD1, encoding glycerol-3-phosphate dehydrogenase 1. Am J
Hum Genet. 90:49–60. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim
MM, Srikantan S, Martindale JL, Hutchison ER, Kim HH, Marasa BS, et
al: miR-130 suppresses adipogenesis by inhibiting peroxisome
proliferator-activated receptor gamma expression. Mol Cell Biol.
31:626–638. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim N, Kim JE, Choung HK, Lee MJ and
Khwarg SI: Expression of cell cycle regulatory proteins in eyelid
sebaceous gland carcinoma: Low p27 expression predicts poor
prognosis. Exp Eye Res. 118:46–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bell WR, Singh K, Rajan Kd A and Eberhart
CG: Expression of p16 and p53 in intraepithelial periocular
sebaceous carcinoma. Ocul Oncol Pathol. 2:71–75. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liau JY, Liao SL, Hsiao CH, Lin MC, Chang
HC and Kuo KT: Hypermethylation of the CDKN2A gene promoter is a
frequent epigenetic change in periocular sebaceous carcinoma and is
associated with younger patient age. Hum Pathol. 45:533–539. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Z, Wang H, Wang Z and Cai H: MiR-195
inhibits the proliferation of human cervical cancer cells by
directly targeting cyclin D1. Tumour Biol. 37:6457–6463. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhong J, Yuan H, Xu X and Kong S: MicroRNA
195 inhibits cell proliferation, migration and invasion by
targeting defective in cullin neddylation 1 domain containing 1 in
cervical cancer. Int J Mol Med. 42:779–788. 2018.PubMed/NCBI
|
|
44
|
Hui W, Yuntao L, Lun L, WenSheng L,
ChaoFeng L, HaiYong H and Yueyang B: MicroRNA-195 inhibits the
proliferation of human glioma cells by directly targeting cyclin D1
and cyclin E1. PLoS One. 8:e549322013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu H, Chen Y, Li Y, Li C, Qin T, Bai M,
Zhang Z, Jia R, Su Y and Wang C: miR-195 suppresses metastasis and
angiogenesis of squamous cell lung cancer by inhibiting the
expression of VEGF. Mol Med Rep. 20:2625–2632. 2019.PubMed/NCBI
|
|
46
|
Ye X and Chen X: miR-149-5p inhibits cell
proliferation and invasion through targeting GIT1 in medullary
thyroid carcinoma. Oncol Lett. 17:372–378. 2019.PubMed/NCBI
|
|
47
|
Jin L, Li Y, Liu J, Yang S, Gui Y, Mao X,
Nie G and Lai Y: Tumor suppressor miR-149-5p is associated with
cellular migration, proliferation and apoptosis in renal cell
carcinoma. Mol Med Rep. 13:5386–5392. 2016. View Article : Google Scholar : PubMed/NCBI
|