Introduction
With social and economic development and the aging
population, the occurrence of cardiovascular events is increasing
each year (1). Among cardiovascular
diseases, acute myocardial infarction (AMI), which is associated
with high lethality and high disability, has become the leading
cause of human mortality globally (2). AMI refers to myocardial necrosis
caused by acute or continuous ischemia and hypoxia in the coronary
arteries (3). Myocardial ischemia
is a pathological state in which blood perfusion of the heart is
decreased, resulting in decreased oxygen supply to the heart and
abnormal energy metabolism of the myocardial cells, which cannot
support the normal function of the heart (4). Persistent and acute ischemia of the
heart can develop into AMI (5).
At present, the treatment strategies for AMI
primarily include thrombolysis, percutaneous coronary intervention
and surgical bypass surgery. The primary purpose of the therapeutic
strategies is to restore the blood supply to the heart, rescue
myocardial cells in the infarction area and prevent further damage
caused by ischemia (6). However,
the current treatment strategies have limited therapeutic effects
on damaged cardiomyocytes, as cardiomyocytes in adults cannot be
naturally regenerated once they are lost (7). Therefore, rescuing myocardial cells at
the near-death state in the marginal area of MI by investigating
the function of certain specific long non-coding RNAs (lncRNAs) or
proteins is a novel research topic for treating AMI.
lncRNAs are transcripts >200 nucleotides in
length, lacking a specific complete open reading frame and protein
coding functions (8). lncRNAs
function as crucial regulators via regulating gene expression at
both the transcriptional and post-transcriptional levels (9). lncRNAs can regulate microRNAs
(miRNAs/miRs) by serving as miRNA sponges, adsorbing the
corresponding miRNA and exerting transcriptional regulation
(10). A number of studies have
demonstrated that lncRNAs serve important roles in the occurrence
and development of human diseases, including cardiovascular
diseases (11,12).
lncRNA discrimination antagonizing non-protein
coding RNA (Dancr) encodes human chromosome 4q12, and was first
identified as an epidermal cell differentiation suppressor
(13). Further investigation
revealed that Dancr functions as an oncogene in various types of
cancer, including hepatocellular carcinoma (14), non-small cell lung carcinoma
(15,16) and osteosarcoma (17). Recently, Dancr has been reported to
alleviate hypoxia-induced H9c2 cardiomyocyte damage by upregulating
hypoxia inducible factor-1α (18).
In addition, Dancr decreased hypoxia- and hypoglycemia-induced
damage of cerebral microvascular endothelial cells via regulating
miR-33a-5p/spliced X-box-binding protein 1 (Xbp1s) (19). However, to the best of our
knowledge, the effect of Dancr in myocardial ischemia and
myocardial infarction has not been previously reported.
In the case of MI/reperfusion (R), endoplasmic
reticulum (ER) homeostasis is destroyed, resulting in the
accumulation of a large number of unfolded or misfolded proteins in
the ER, triggering the unfolded protein response (UPR) and ER
stress (ERS) (20). Early ERS
exerts a compensatory protective effect, but excessive ERS is
involved in the pathophysiological process of various
cardiovascular diseases (21).
Tunicamycin (Tm) is a commonly used ERS inducer (22). Previous studies have demonstrated
that low doses of Tm can produce moderate ERS, which displays a
certain protective effect on MI/R (23,24).
Moderate ERS can also induce autophagy to help the degradation of
unfolded or misfolded proteins, thus alleviating ERS (25). However, excessive ERS may initiate
the apoptosis reaction and inflammatory pathways, ultimately
participating in the deterioration of cardiovascular disease
(26). ERS in response to various
adverse stimuli has been detected and has been reported to be
associated with the pathogenesis of MI/R injury, myocardial
hypertrophy, ischemic cardiomyopathy, diabetic cardiomyopathy and
cardiac fibrosis (27). For
example, miR-711 mimic could induce cardiomyocyte apoptosis after
ERS-induced MI via upregulating Xbp1 (28). Xu et al (29) demonstrated that inhibition of ERS
and the cell apoptosis signaling pathway could protect
cardiomyocytes against MI-induced injury.
Autophagy is an important metabolic process that
degrades senescent or damaged proteins and organelles into amino
groups (30). Autophagy is
activated in response to nutritional deficiencies or metabolic
stress to maintain tissue function and homeostasis (31). Basic autophagy has been reported to
be essential for maintaining normal heart function. Meanwhile,
under ischemic stress, autophagy is activated to protect
cardiomyocytes from ischemia or I/R injury (32). The beneficial role of autophagy in
AMI in the alleviation of MI under ischemic and ischemia/R injuries
has been extensively reported (33,34).
Therefore, taking advantage of autophagy provides a potential
strategy for the development of novel drugs or therapies for AMI
(35).
The aforementioned studies indicated that the
moderate enhancement of ERS and autophagy, together with the
repression of excessive ERS and ERS-mediated apoptosis might serve
as a valuable therapeutic strategy for relieving MI-induced injury.
The present study aimed to investigate the roles and molecular
mechanism underlying Dancr in ERS-induced cardiomyocytes to provide
a novel target for the diagnosis and therapy of AMI.
Materials and methods
Cell culture and treatment
The H9c2 rat embryonic cardiomyocyte cell line
(American Type Culture Collection) was cultured in DMEM (Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) and 100 units/ml penicillin at 37°C in a
humidified atmosphere of 5% CO2. The medium was replaced
every other day. At 70–80% confluence, cells were digested with
trypsin and EDTA.
Tm (MedChemExpress) was utilized to induce ERS. H9C2
cells were treated Tm (0.1, 0.5, 2.5 or 12.5 µM) for 6 h at 37°C
and control cells were cultured in normal medium (36).
Cell transfection
Prior to transfection, the medium was replaced with
serum- and antibiotic-free DMEM. To overexpress lncRNA Dancr, the
recombinant full-length rat Dancr cDNA [overexpression (Oe)-Dancr]
was cloned into the pcDNA3.1 vector (Thermo Fisher Scientific,
Inc.). The pcDNA3.1 empty vector was used as a negative control
(NC; Oe-NC). H9C2 cells (60-70% confluence) were transfected with
oe-Dancr, oe-NC, miR-6324 mimic (5′-AGUAGGCCAGACAGCAAGC-3′;
Sigma-Aldrich; Merck KGaA) or mimic-NC
(5′-GGUUCGUACGUACACUGUUCA-3′; Sigma-Aldrich; Merck KGaA) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Briefly, Lipofectamine 2000 was mixed with 50 nM
plasmids or mimics, added to the cells and incubated for 6 h at
37°C. Subsequently, cells were cultured in DMEM for 24 h and then
used for subsequent experiments. Transfection efficiency was
evaluated via reverse transcription-quantitative PCR (RT-qPCR).
Cell Counting Kit-8 (CCK-8)
The CCK-8 assay was performed to assess cell
viability. H9c2 cells (1×104 cells/well) were cultured
in 96-well plates and subjected to corresponding treatments.
Subsequently, 10 µl CCK-8 reagent (Beyotime Institute of
Biotechnology) was added to each well and incubated at 37°C for 2 h
in the dark. The optical density of each well was measured at a
wavelength of 450 nm using a microplate reader.
TUNEL staining
Apoptotic H9c2 cardiomyocytes were visualized by
performing TUNEL staining (Nanjing KeyGen Biotech Co., Ltd.)
according to the manufacturers protocol. Briefly, H9c2 cells were
cultured on cover slips. After the corresponding treatment, cells
were fixed with 4% neutral buffered formalin solution for 30 min at
room temperature. Subsequently, 50 µl TUNEL reaction mixture was
added and incubated for 1 h at 37°C. The nuclei were stained with
DAPI (2 µg/ml) at room temperature for 5 min. After washing twice
with PBS, images were captured from three fields of view using a
fluorescence microscope (magnification, ×200).
Western blotting
Total protein was extracted from H9c2 cardiomyocytes
using lysis buffer (Beyotime Institute of Biotechnology) containing
a protease inhibitor and phosphatase inhibitor. After being
quantified using a BCA kit (Beyotime Institute of Biotechnology),
equal amounts of protein (50 µg) were separated via 12% SDS-PAGE
and transferred to PVDF membranes. The membranes were blocked with
5% non-fat milk at 37°C for 2 h. Subsequently, the membranes were
incubated overnight at 4°C with primary antibodies targeted
against: Bcl-2 (Abcam; cat. no. ab32124; 1:1,000), Bax (Abcam; cat.
no. ab32503; 1:10,000), cleaved (c)-caspase-9 (Abcam; cat. no.
ab32539; 1:5,000), caspase-9 (Abcam; cat. no. ab184786; 1:1,000),
c-caspase-3 (Abcam; cat. no. ab32042; 1:500), caspase-3 (Abcam;
cat. no. ab13847; 1:500), glucose-regulated protein 78 kDa (GRP78;
Abcam; cat. no. ab21685; 1:1,000), phosphorylated
(p)-inositol-requiring enzyme-1 (IRE1)α (Abcam; cat. no. ab124945;
1:1,000), IRE1α (Abcam; cat. no. ab37073; 1:1,000), Xbp1s
(ProteinTech Group, Inc.; cat. no. 24868-1-AP; 1:1,000), unspliced
Xbp1 (Xbp1u; ProteinTech Group, Inc.; cat. no. 25997-1-AP;
1:1,000), activating transcription factor (ATF)6 (ProteinTech
Group, Inc.; cat. no. 24169-1-AP; 1:2,000), ATF4 (ProteinTech
Group, Inc.; cat. no. 10835-1-AP; 1:1,000), Beclin 1 (ProteinTech
Group, Inc.; cat. no. 11306-1-AP; 1:10,000), microtubule associated
protein 1 light chain 3α (LC3)II/I (ProteinTech Group, Inc.; cat.
no. 14600-1-AP; 1:2,000), p62 (Abcam; cat. no. ab56416; 1:1,000)
and GAPDH (Abcam; cat. no. ab8245; 1:10,000). Following primary
incubation, the membranes were incubated with horseradish
peroxidase-conjugated goat anti-rabbit/mouse IgG secondary
antibodies at room temperature for 2 h. Protein bands were
visualized using an enhanced chemiluminescence reagent (Thermo
Fisher Scientific, Inc.) and detection system (Amersham; Cytiva).
Protein expression levels were semi-quantified using Image-Pro Plus
software version 6.0 (Media Cybernetics, Inc.) with GAPDH as the
loading control.
RT-qPCR
Total RNA was extracted from H9C2 cardiomyocytes
using an RNA isolation kit (Total RNA Extraction Reagent; Vazyme
Biotech Co., Ltd.). Total RNA was reverse transcribed into cDNA
using a reverse transcriptase (Vazyme Biotech Co., Ltd.).
Subsequently, qPCR was performed using the CFX384 Real-Time System
C1000 Thermocycler (Bio-Rad Laboratories, Inc.) and SYBR-Green
ROX-mix (Vazyme Biotech Co., Ltd.). The following primers were used
for qPCR: Dancr forward, 5′-CTCGGATAGAAGCGCAGGTT-3′ and reverse,
5′-AGGCAAGCGGGGTCATTAAA-3′; miR-6324 forward,
5′-ATAGCTGGGGTCAAGGTGCT-3′ and reverse, 5′-CTTGCTGTCTGGCCTACTGA-3′;
GAPDH forward, 5′-TTGTGCAGTGCCAGCCTC-3′ and reverse,
5′-GGTAACCAGGCGTCCGATAC-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The following
thermocycling conditions were used for qPCR: Initial denaturation
at 95°C for 30 sec then 40 cycles of 95°C for 5 sec and 60°C for 15
sec, followed by default of melt curve. miRNA and mRNA expression
levels were quantified using the 2−∆∆Cq method (37) and normalized to the internal
reference genes U6 and GAPDH, respectively.
Dual-luciferase reporter assay
The wild-type (WT) and mutated (MUT) Dancr fragments
containing the miR-6324 binding sites were synthesized and inserted
into the pmirGLO luciferase vector (Promega Corporation) to
generate WT-pmirGLO and MUT-pmirGLO, respectively. For the promoter
analysis, miR-6324 mimic or mimic NC were cloned into the pGL3
luciferase reporter (Promega Corporation). Cells were
co-transfected with WT-pmirGLO or MUT-pmirGLO vectors and miR-6324
mimic or mimic NC. At 48 h post-transfection, luciferase activities
were measured using a Dual-Luciferase reporter assay system
(Promega Corporation) as previously described (38).
Statistical analysis
Data are presented as the mean ± standard deviation
of from at least three independent experiments. Comparisons between
two groups were analyzed using the Students t-test. Comparisons
among multiple groups were analyzed using one-way ANOVA followed by
Tukeys post hoc test. Statistical analyses were performed using
GraphPad Prism software (version 5.0; GraphPad Software, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
lncRNA Dancr is downregulated upon Tm
stimulation
The present study aimed to determine whether Dancr
could regulate cardiomyocytes that underwent ERS. Following
stimulation with different concentrations of the ERS agonist Tm
(0.1, 0.5, 2.5 or 12.5 µM), Dancr expression levels and cell
viability in H9C2 cells were measured. Compared with the control
group, Tm decreased Dancr mRNA expression levels (Fig. 1A) and cell viability (Fig. 1B) in a concentration-dependent
manner, suggesting a modulatory effect of Dancr on cardiomyocyte
ERS. Based on the finding that 2.5 µM Tm resulted in a >50%
reduction in Dancr expression but maintained cell viability at
≥50%, 2.5 µM Tm was selected for subsequent experiments (39).
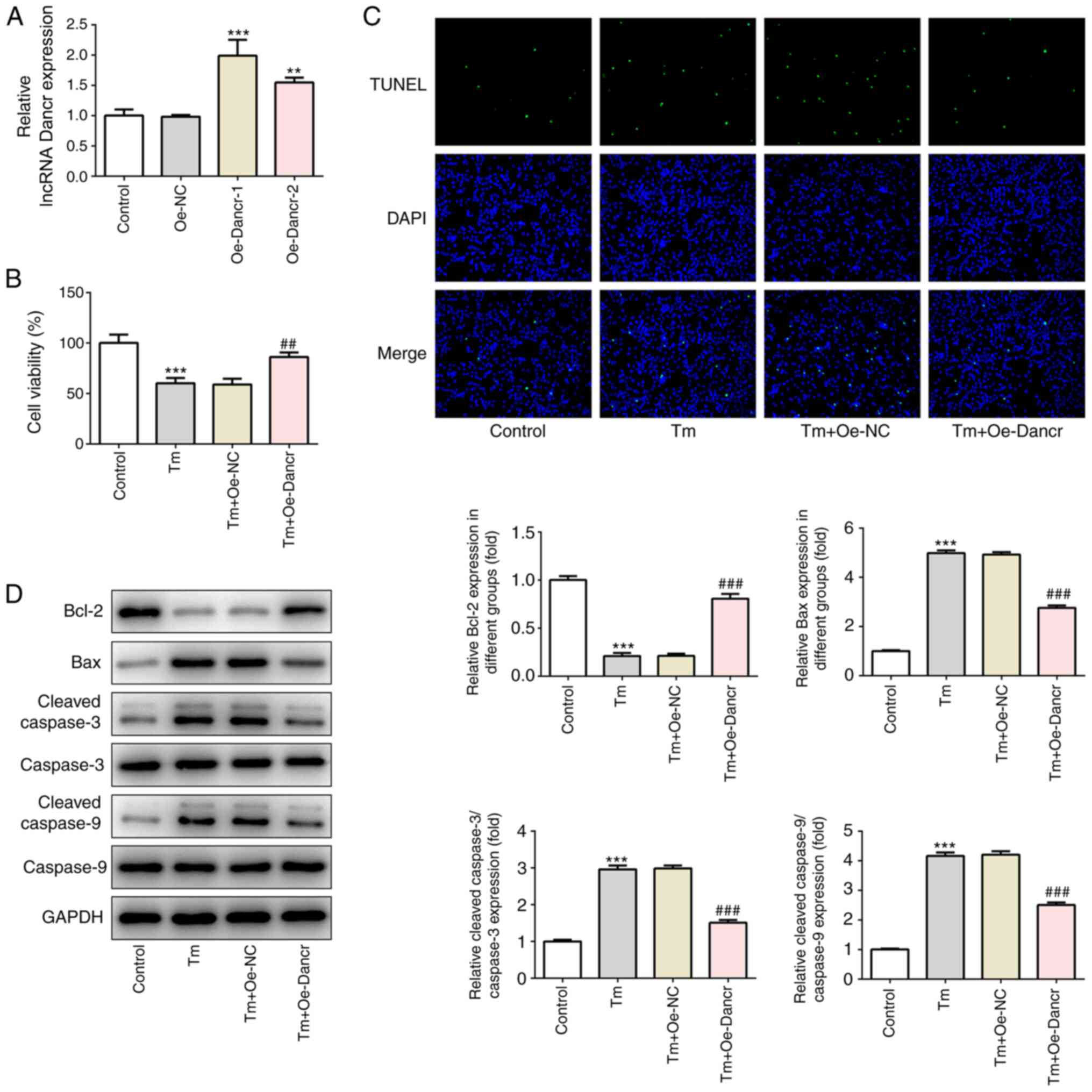
Dancr overexpression enhances cell
viability and decreases cell apoptosis in Tm-stimulated H9C2
cardiomyocytes
To determine the specific role of Dancr in
ERS-induced cardiomyocyte injury, Dancr was overexpressed in H9C2
cells. The results demonstrated the successful overexpression of
Dancr (Fig. 2A). Oe-Dancr-1 was
selected to establish Dancr-overexpression H9C2 cells in subsequent
experiments based on its higher efficacy compared with Oe-Dancr-2.
Moreover, compared with the control group, 2.5 µM Tm treatment
significantly decreased cell viability, markedly increased the
ratio of apoptotic cells, significantly decreased the expression
levels of the antiapoptosis protein Bcl-2, and significantly
increased the expression levels of the proapoptosis proteins Bax
and c-caspase-3/9 (Fig. 2B-D).
However, following Tm stimulation, compared with Oe-NC-transfected
cells, Oe-Dancr-transfected cells displayed significantly higher
cell viability, a notably lower ratio of cell apoptosis,
significantly increased expression levels of Bcl-2, and
significantly decreased expression levels of Bax and
c-caspase-3/9.
Dancr overexpression enhances ERS and
autophagy in Tm-stimulated H9C2 cardiomyocytes
The present study analyzed the expression levels of
proteins associated with ERS and autophagy. Compared with the
control group, Tm treatment resulted in significantly increased
expression levels of GRP78, p-IRE1α, Xbp1s, IRE1α, ATF6 and ATF4,
significantly decreased expression levels of Xbp1u and no
significant alterations to the p-IRE1α/IRE1α ratio, indicating the
induction of ERS (Fig. 3A).
Compared with the Tm + Oe-NC group, the Tm + Oe-Dancr group
displayed significantly enhanced p-IRE1α, p-IRE1α/IRE1α and Xbp1s
expression levels and significantly decreased Xbp1u expression
levels, but there were no significant alterations to the expression
levels of IRE1α, ATF6 or ATF4, suggesting that Dancr selectively
promoted the phosphorylation of IRE1α and activated the IRE1α
branch in the UPR. Moreover, compared with the control group, Tm
treatment induced autophagy, as evidenced by significantly
increased expression levels of Beclin 1 and LC3II/I, and
significantly decreased expression levels of p62 (Fig. 3B). Compared with the Tm + Oe-NC
group, the Tm + Oe-Dancr group displayed increased levels of
autophagy, as demonstrated by significantly increased Beclin 1 and
LC3II/I expression levels, and significantly lower p62 expression
levels.
 | Figure 3.Effects of Dancr overexpression on
ERS and autophagy. Western blotting was performed to measure the
expression levels of (A) ERS-related proteins, including GRP78,
p-IRE1α, IRE1α, p-IRE1α/IRE1α, ATF6, ATF4, Xbp1s and Xbp1u and (B)
autophagy-related proteins, including Beclin1, LC3II/I and p62.
**P<0.01 and ***P<0.001 vs. control; ##P<0.01
and ###P<0.001 vs. Tm + Oe-NC. Dancr, discrimination
antagonizing non-protein coding RNA; ERS, endoplasmic reticulum
stress; GRP78, glucose-regulated protein 78 kDa; p, phosphorylated;
IRE1, inositol-requiring enzyme-1; ATF, activating transcription
factor; Xbp1s, spliced X-box-binding protein 1; Xbp1u, unspliced
Xbp1; LC3, microtubule associated protein 1 light chain 3α; Tm,
Tunicamycin; Oe, overexpression; NC, negative control. |
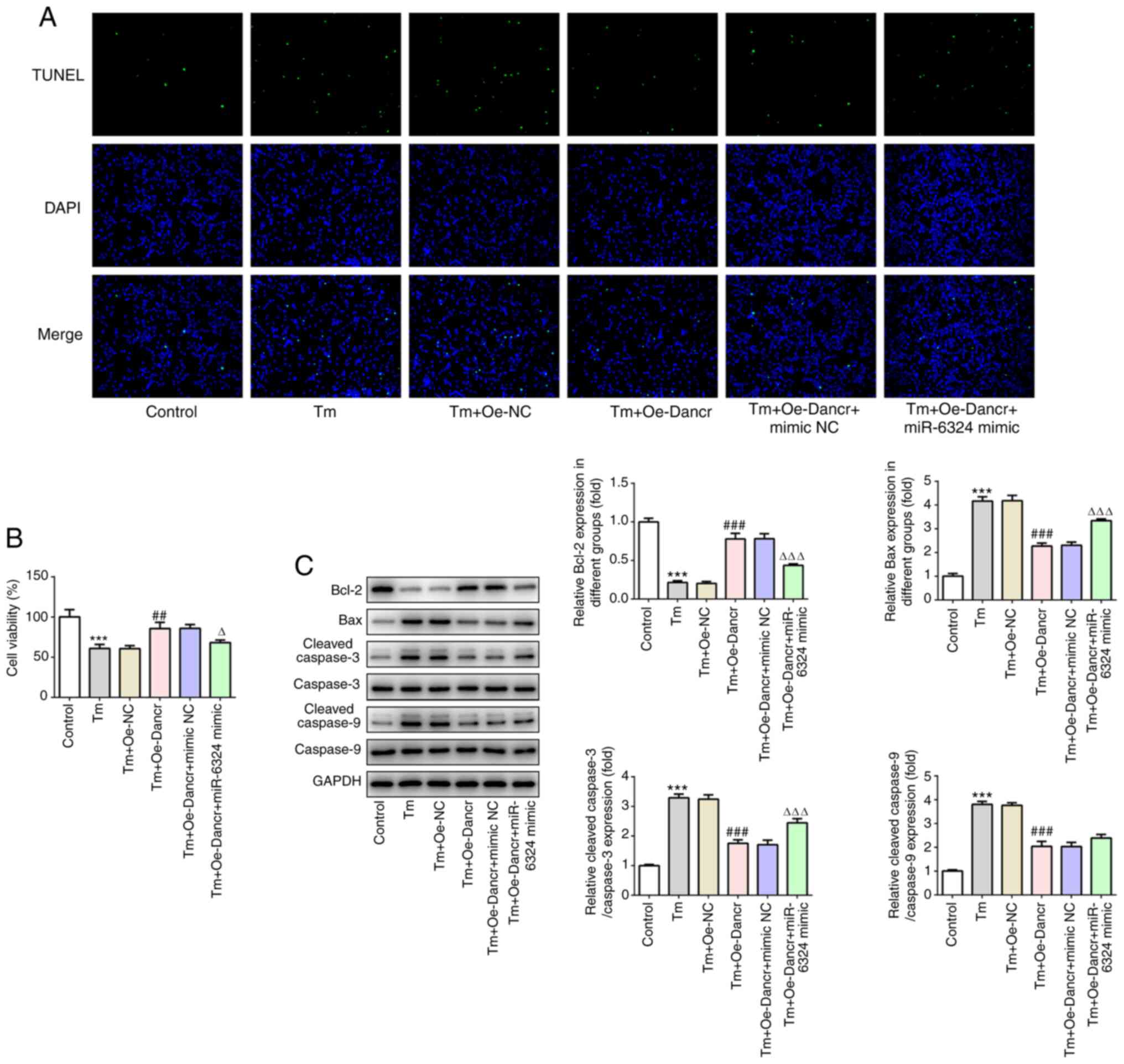
miR-6324 mimic partially reverses the
effects of Dancr overexpression on Tm-induced apoptosis, ERS and
autophagy
miR-6324 was predicted to bind to Dancr (Fig. 4A). To verify whether miR-6324 was
the target of Dancr, the present study constructed two miR-6324
mimics. The results verified the successful transfection of the
miR-6324 mimics (Fig. 4B). The
direct interaction between Dancr and miR-6324 was assessed by
performing a Dual-luciferase reporter assay (Fig. 4C). Moreover, Dancr overexpression
significantly downregulated miR-6324 expression levels compared
with Oe-NC (Fig. 4D). The results
suggested that miR-6324 might participate in the actions of Dancr
on ERS-induced cardiomyocyte injury.
To investigate the modulatory effects of miR-6324 on
Dancr-mediated H9C2 ERS, Tm-treated and Dancr-overexpression H9C2
cells were transfected with miR-6324 mimic or mimic-NC. Compared
with the Tm + Oe-Dancr + mimic NC group, the Tm + Oe-Dancr +
miR-6324 mimic group displayed significantly decreased cell
viability and markedly increased numbers of apoptotic cells
(Fig. 5A and B). Furthermore,
compared with the Tm + Oe-Dancr + mimic NC group, the Tm + Oe-Dancr
+ miR-6324 mimic group displayed significantly decreased Bcl-2
expression levels, and significantly increased Bax and
c-caspase-3/9 expression levels (Fig.
5C).
The expression levels of proteins associated with
ERS and autophagy were measured (Fig.
6). Oe-Dancr-mediated upregulation of GRP78, p-IRE1α,
p-IRE1α/IRE1α, Xbp1s, Beclin 1 and LC3II/I expression levels was
significantly reversed by miR-6324 mimic. Similarly, miR-6324 mimic
partially recovered the expression levels of Xbp1u and p62, which
were decreased by Tm and further decreased by Dancr overexpression
combined with Tm treatment. The aforementioned results indicated
that miR-6324 mimic partially abolished Dancr
overexpression-mediated effects on Tm-induced apoptosis, ERS and
autophagy.
 | Figure 6.miR-6324 mimic reverses Dancr
overexpression-mediated effects on Tm-induced ERS and autophagy.
Western blotting was performed to measure the expression levels of
(A) ERS-related proteins, including GRP78, p-IRE1α, IRE1α,
p-IRE1α/IRE1α, ATF6, ATF4, Xbp1s and Xbp1u, (B) autophagy-related
proteins, including Beclin1, LC3II/I and p62. **P<0.01 and
***P<0.001 vs. control; ##P<0.01 and
###P<0.001 vs. Tm + Oe-NC; ∆∆P<0.01 and
∆∆∆P<0.001 vs. Tm + Oe-Dancr + mimic NC. miR,
microRNA; Dancr, discrimination antagonizing non-protein coding
RNA; Tm, Tunicamycin; ERS, endoplasmic reticulum stress; GRP78,
glucose-regulated protein 78 kDa; p, phosphorylated; IRE1,
inositol-requiring enzyme-1; ATF, activating transcription factor;
Xbp1s, spliced X-box-binding protein 1; Xbp1u, unspliced Xbp1; LC3,
microtubule associated protein 1 light chain 3α; Oe,
overexpression; NC, negative control. |
Discussion
lncRNA Dancr has been reported to protect H9c2
cardiomyocytes against hypoxia-induced damage (18). The present study indicated that
Dancr protected H9C2 cells against ERS-induced apoptosis in
vitro via the selective activation of the IRE1α/XBP1 signaling
pathway in the UPR, which suggested that Dancr provided
cytoprotection in response to ERS.
When the ER senses a wide variety of perturbations,
including UPR, the adaptive process of ERS occurs to maintain ER
homeostasis and mitigate or eliminate the stress (22). There are three primary ER
transmembrane stress sensors that initiate the UPR, including IRE1,
protein kinase RNA-like endoplasmic reticulum kinase (PERK) and
ATF6, are maintained in an inactive state via binding to their
luminal domains with the ER chaperone GRP78. However, in the
presence of ERS, GRP78 is released from these complexes and
recruited to misfolded proteins, leading to the activation of three
distinct UPR branches (40). For
instance, the IRE1 branch possesses endoribonuclease activity that
splices mRNAs encoding Xbp1u to form mature Xbp1/Xbp1s mRNA, which
is then translated to a potent transcription factor that controls
the genes encoding proteins that target misfolded proteins for
ubiquitination and ER-associated degradation (ERAD). Meanwhile,
ATF6 cooperates with IRE1α for the induction of Xbp1 transcription.
The proteins balance the unfolded protein/chaperone system to
provide ER homeostasis. If the cell fails to recover from ERS, the
UPR represses the adaptive response and triggers apoptosis.
Notably, PERK stimulates the expression of the proapoptotic
transcription factor C/EBP homologous protein via the mobilization
of ATF4 (41). Thus, the ERS serves
a dual role, transmitting both adaptive and apoptotic signals. In
the present study, compared with the control group, Tm decreased
cell viability and induced cell apoptosis, which could be explained
by the occurrence of excessive ERS, as evidence by increased
expression levels of GRP78, p-IRE1α, IRE1α, ATF6, ATF4 and Xbp1s,
and decreased expression levels of Xbp1u. As previously described,
if ERS is too severe or prolonged, the UPR initiates an apoptotic
response (42).
Autophagy is essential for maintaining protein
homeostasis and is an evolutionarily conserved protein degradation
signaling pathway that removes damaged or expired proteins and
organelles by chelating in autophagosomes, which subsequently
undergo lysosomal degradation (43). Previous studies have demonstrated
that the ERS response can activate the autophagy-lysosome signaling
pathway, which serves a major role in the cardiac stress response
(44,45). Autophagy functions as a cellular
stress signaling pathway and can assist with the degradation of
proteins to recover ER homeostasis (44). In the present study, compared with
the control group, Tm treatment increased the expression levels of
Beclin 1 and LC3II/I, and decreased the expression levels of p62,
indicating the induction of autophagy in response to ERS.
lncRNAs have been identified as novel targets for
the treatment of cardiovascular diseases. For example, lncRNA
CDKN2B antisense RNA 1 has been identified as the most powerful
predictor of atherosclerosis (46,47).
Chi et al (47) revealed
that the lncRNA myocardial infarction associated transcript 1 led
to a decrease in the expression of inflammatory factors via
inhibition of the NF-κB signaling pathway, thereby decreasing
myocardial cell apoptosis and inflammatory cell infiltration to
decrease AMI damage. To the best of our knowledge, the present
study suggested the role of Dancr in cardiomyocyte ERS for the
first time. Compared with the Tm + Oe-NC group, Dancr
overexpression increased cell viability by decreasing cell
apoptosis and promoting autophagy in Tm-treated H9C2
cardiomyocytes. Furthermore, compared with the Tm + Oe-NC group,
Dancr overexpression enhanced p-IRE1α, p-IRE1α/IRE1α and Xbp1s
expression levels, and decreased Xbp1u expression levels, but
displayed no significant effects on IRE1α, ATF6 and ATF4 expression
levels in Tm-treated H9C2 cardiomyocytes, suggesting that Dancr
selectively activated the IRE1α branch in the UPR, thus promoting
autophagy and ERAD, and ultimately alleviating ERS. It has been
reported that enhancing autophagy could protect cardiomyocytes from
ERS and apoptosis (48). However,
whether the antiapoptotic effect of Dancr is dependent on the
autophagy signaling pathway, the ERS signaling pathway or other
mediators requires further investigation.
lncRNAs regulate the occurrence and development of
human diseases, including cardiovascular diseases, primarily via
sponging miRNAs (11). Our previous
unpublished data demonstrated that miR-6324 was upregulated in MI
model rats. In the present study, online analysis and luciferase
reporter assays confirmed that miR-6324 interacted with lncRNA
Dancr. Therefore, the present study hypothesized that Dancr
displayed cardioprotective effects via sponging miR-6324 and
inhibiting its expression. To verify the hypothesis, the present
study co-transfected Tm-treated H9C2 cells with Oe-Dancr and
miR-6324 mimic. miR-6324 mimic partially reversed Dancr
overexpression-mediated effects on cell viability, cell apoptosis,
ERS and autophagy. The results indicated that miR-6324 may serve as
a downstream target miRNA of Dancr to rescue cell viability,
inhibit apoptosis and promote autophagy, thereby relieving
cardiomyocyte ERS. However, apoptosis and autophagy are
double-edged swords, thus consistent with previous studies
(20,24,25),
the present study identified Dancr as a potential target to reduce
apoptosis and ERS, while enhancing autophagy in Tm-induced H9C2
cardiomyocytes. The enhanced cell viability indicated the
protective effect of Dancr, but in vivo studies are required
to verify the results of the present study. Moreover, as miR-6324
mimic did not completely reverse the effects of Dancr, other
downstream targets of Dancr may exist and should be investigated in
future studies.
In summary, the present study provided evidence that
lncRNA Dancr sponged miR-6324, selectively activated the IRE1α/Xbp1
signaling pathway of autophagy, repressed apoptosis and enhanced
autophagy, leading to amelioration of ERS. The actions of Dancr
identified in the present study suggested its potential for the
treatment of cardiovascular diseases.
Acknowledgements
Not applicable.
Funding
This study was supported by the Lanzhou Talent
Project for Innovation and Entrepreneurship (grant no. 2015-RC-12)
and the Health Science and Technology Development Project of
Lanzhou (grant no. 2019-002).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors contributions
YHD, DXX, JL and JX conceived and designed the
study. JL, JX, YZW, YRG, LW and GWD acquired and analyzed the data.
JL and JX drafted the manuscript and figures. YHD and DXX revised
the manuscript for critically important intellectual content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Moran AE, Roth GA, Narula J and Mensah GA:
1990-2010 global cardiovascular disease atlas. Glob Heart. 9:3–16.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mehta LS, Beckie TM, DeVon HA, Grines CL,
Krumholz HM, Johnson MN, Lindley KJ, Vaccarino V, Wang TY, Watson
KE, et al American Heart Association Cardiovascular Disease in
Women and Special Populations Committee of the Council on Clinical
Cardiology, Council on Epidemiology and Preventionm Council on
Cardiovascular and Stroke Nursing, Council on Quality of Care and
Outcomes Research, : Acute Myocardial Infarction in Women: A
Scientific Statement From the American Heart Association.
Circulation. 133:916–947. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reed GW, Rossi JE and Cannon CP: Acute
myocardial infarction. Lancet. 389:197–210. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tamis-Holland JE, Jneid H, Reynolds HR,
Agewall S, Brilakis ES, Brown TM, Lerman A, Cushman M, Kumbhani DJ,
Arslanian-Engoren C, et al American Heart Association
Interventional Cardiovascular Care Committee of the Council on
Clinical Cardiology; Council on Cardiovascular and Stroke Nursing;
Council on Epidemiology and Prevention; and Council on Quality of
Care and Outcomes Research, : Contemporary Diagnosis and Management
of Patients With Myocardial Infarction in the Absence of
Obstructive Coronary Artery Disease: A Scientific Statement From
the American Heart Association. Circulation. 139:e891–e908. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Heusch G and Gersh BJ: The pathophysiology
of acute myocardial infarction and strategies of protection beyond
reperfusion: A continual challenge. Eur Heart J. 38:774–784.
2017.PubMed/NCBI
|
|
6
|
Asaria P, Elliott P, Douglass M, Obermeyer
Z, Soljak M, Majeed A and Ezzati M: Acute myocardial infarction
hospital admissions and deaths in England: A national follow-back
and follow-forward record-linkage study. Lancet Public Health.
2:e191–e201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dall C, Khan M, Chen CA and Angelos MG:
Oxygen cycling to improve survival of stem cells for myocardial
repair: A review. Life Sci. 153:124–131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jarroux J, Morillon A and Pinskaya M:
History, Discovery, and Classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Deniz E and Erman B: Long noncoding RNA
(lincRNA), a new paradigm in gene expression control. Funct Integr
Genomics. 17:135–143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao Z, Sun W, Guo Z, Zhang J, Yu H and
Liu B: Mechanisms of lncRNA/microRNA interactions in angiogenesis.
Life Sci. 254:1169002020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sallam T, Sandhu J and Tontonoz P: Long
noncoding RNA discovery in cardiovascular disease: Decoding Form to
Function. Circ Res. 122:155–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ballantyne MD, McDonald RA and Baker AH:
lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol
Ther. 99:494–501. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thin KZ, Liu X, Feng X, Raveendran S and
Tu JC: LncRNA-DANCR: A valuable cancer related long non-coding RNA
for human cancers. Pathol Res Pract. 214:801–805. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yuan SX, Wang J, Yang F, Tao QF, Zhang J,
Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, et al: Long noncoding RNA
DANCR increases stemness features of hepatocellular carcinoma by
derepression of CTNNB1. Hepatology. 63:499–511. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo L, Gu J, Hou S, Liu D, Zhou M, Hua T,
Zhang J, Ge Z and Xu J: Long non-coding RNA DANCR promotes the
progression of non-small-cell lung cancer by inhibiting p21
expression. OncoTargets Ther. 12:135–146. 2018. View Article : Google Scholar
|
|
16
|
Bai Y, Zhang G, Chu H, Li P and Li J: The
positive feedback loop of lncRNA DANCR/miR-138/Sox4 facilitates
malignancy in non-small cell lung cancer. Am J Cancer Res.
9:270–284. 2019.PubMed/NCBI
|
|
17
|
Wang Y, Zeng X, Wang N, Zhao W, Zhang X,
Teng S, Zhang Y and Lu Z: Long noncoding RNA DANCR, working as a
competitive endogenous RNA, promotes ROCK1-mediated proliferation
and metastasis via decoying of miR-335-5p and miR-1972 in
osteosarcoma. Mol Cancer. 17:892018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiu L, Zhao Q, Dai L, Zhu A, Xu X, Zhao S
and Chen J: Long non-coding RNA DANCR alleviates hypoxia-caused
H9c2 cells damage through up regulation of HIF-1α. Artif Cells
Nanomed Biotechnol. 48:533–541. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang M, Tang M, Wu Q, Wang Z, Chen Z,
Ding H, Hu X, Lv X, Zhao S, Sun J, et al: LncRNA DANCR attenuates
brain microvascular endothelial cell damage induced by
oxygen-glucose deprivation through regulating of miR-33a-5p/XBP1s.
Aging (Albany NY). 12:1778–1791. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zuo S, Kong D, Wang C, Liu J, Wang Y, Wan
Q, Yan S, Zhang J, Tang J, Zhang Q, et al: CRTH2 promotes
endoplasmic reticulum stress-induced cardiomyocyte apoptosis
through m-calpain. EMBO Mol Med. 10:e82372018.simplehttps://doi.org/10.15252/emmm.201708237
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Xu L, Gillette TG, Jiang X and
Wang ZV: The unfolded protein response in ischemic heart disease. J
Mol Cell Cardiol. 117:19–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan M, Shu S, Guo C, Tang C and Dong Z:
Endoplasmic reticulum stress in ischemic and nephrotoxic acute
kidney injury. Ann Med. 50:381–390. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yamamoto K and Ichikawa S: Tunicamycin:
Chemical synthesis and biosynthesis. J Antibiot (Tokyo).
72:924–933. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang S, Wang Z, Fan Q, Guo J, Galli G, Du
G, Wang X and Xiao W: Ginkgolide K protects the heart against
endoplasmic reticulum stress injury by activating the
inositol-requiring enzyme 1α/X box-binding protein-1 pathway. Br J
Pharmacol. 173:2402–2418. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fernández A, Ordóñez R, Reiter RJ,
González-Gallego J and Mauriz JL: Melatonin and endoplasmic
reticulum stress: Relation to autophagy and apoptosis. J Pineal
Res. 59:292–307. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu J, Huang CX, Rao PP, Cao GQ, Zhang Y,
Zhou JP, Zhu LY, Liu MX and Zhang GG: MicroRNA-155 inhibition
attenuates endoplasmic reticulum stress-induced cardiomyocyte
apoptosis following myocardial infarction via reducing macrophage
inflammation. Eur J Pharmacol. 857:1724492019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Zhao J, Geng J, Chen F, Wei Z, Liu
C, Zhang X, Li Q, Zhang J, Gao L, et al: Long non-coding RNA MEG3
knockdown attenuates endoplasmic reticulum stress-mediated
apoptosis by targeting p53 following myocardial infarction. J Cell
Mol Med. 23:8369–8380. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao N, Mi L, Zhang X, Xu M, Yu H, Liu Z,
Liu X, Guan G, Gao W and Wang J: Enhanced MiR-711 transcription by
PPARγ induces endoplasmic reticulum stress-mediated apoptosis
targeting calnexin in rat cardiomyocytes after myocardial
infarction. J Mol Cell Cardiol. 118:36–45. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu Y, Wu L, Chen A, Xu C and Feng Q:
Protective effects of olive leaf extract on acrolein-exacerbated
myocardial infarction via an endoplasmic reticulum stress pathway.
Int J Mol Sci. 19:4932018.simplehttps://doi.org/10.3390/ijms19020493 View Article : Google Scholar
|
|
30
|
Cătană CS, Atanasov AG and Berindan-Neagoe
I: Natural products with anti-aging potential: Affected targets and
molecular mechanisms. Biotechnol Adv. 36:1649–1656. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong Z, Chu G, Sima Y and Chen G: Djhsp90s
are crucial regulators during planarian regeneration and tissue
homeostasis. Biochem Biophys Res Commun. 498:723–728. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang L, Li Y, Ning N, Wang J, Yan Z, Zhang
S, Jiao X, Wang X and Liu H: Decreased autophagy induced by
β1-adrenoceptor autoantibodies contributes to cardiomyocyte
apoptosis. Cell Death Dis. 9:4062018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aisa Z, Liao GC, Shen XL, Chen J, Li L and
Jiang SB: Effect of autophagy on myocardial infarction and its
mechanism. Eur Rev Med Pharmacol Sci. 21:3705–3713. 2017.PubMed/NCBI
|
|
34
|
Kanamori H, Takemura G, Goto K, Maruyama
R, Ono K, Nagao K, Tsujimoto A, Ogino A, Takeyama T, Kawaguchi T,
et al: Autophagy limits acute myocardial infarction induced by
permanent coronary artery occlusion. Am J Physiol Heart Circ
Physiol. 300:H2261–H2271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu D, Zhang K and Hu P: The role of
autophagy in acute myocardial infarction. Front Pharmacol.
10:5512019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu Y, Sun G, Luo Y, Wang M, Chen R, Zhang
J, Ai Q, Xing N and Sun X: Cardioprotective effects of
Notoginsenoside R1 against ischemia/reperfusion injuries by
regulating oxidative stress- and endoplasmic reticulum stress-
related signaling pathways. Sci Rep. 6:217302016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liao B, Dong S, Xu Z, Gao F, Zhang S and
Liang R: LncRNA Kcnq1ot1 renders cardiomyocytes apoptosis in acute
myocardial infarction model by up-regulating Tead1. Life Sci.
256:1178112020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lai L, Liu Y, Liu Y, Zhang N, Cao S, Zhang
X and Wu D: Role of endoplasmic reticulum oxidase 1α in H9C2
cardiomyocytes following hypoxia/reoxygenation injury. Mol Med Rep.
22:1420–1428. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Walter P and Ron D: The unfolded protein
response: From stress pathway to homeostatic regulation. Science.
334:1081–1086. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Oyadomari S and Mori M: Roles of
CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ.
11:381–389. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Durante W: Targeting endoplasmic reticulum
stress in hypoxia-induced cardiac injury. Vascul Pharmacol. 83:1–3.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Levy JMM, Towers CG and Thorburn A:
Targeting autophagy in cancer. Nat Rev Cancer. 17:528–542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lin R, Su Z, Tan X, Su Y, Chen Y, Shu X,
Liang S, Wang J and Xie S: Effect of endoplasmic reticulum stress
and autophagy in the regulation of post-infarct cardiac eepair.
Arch Med Res. 49:576–582. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wiersma M, Meijering RAM, Qi XY, Zhang D,
Liu T, Hoogstra-Berends F, Sibon OCM, Henning RH, Nattel S and
Brundel BJJM: Endoplasmic reticulum stress is associated with
autophagy and cardiomyocyte remodeling in experimental and human
atrial fibrillation. J Am Heart Assoc. 6:e0064582017.doi:
10.1161/JAHA.117.006458. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Holdt LM and Teupser D: Long noncoding RNA
ANRIL: Lnc-ing genetic variation at the chromosome 9p21 locus to
molecular mechanisms of atherosclerosis. Front Cardiovasc Med.
5:1452018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chi JS, Li JZ, Jia JJ, Zhang T, Liu XM and
Yi L: Long non-coding RNA ANRIL in gene regulation and its duality
in atherosclerosis. J Huazhong Univ Sci Technolog Med Sci.
37:816–822. 2017.doi: 10.1007/s11596-017-1812-y. PubMed/NCBI
|
|
48
|
Chang JC, Hu WF, Lee WS, Lin JH, Ting PC,
Chang HR, Shieh KR, Chen TI and Yang KT: Intermittent hypoxia
induces autophagy to protect cardiomyocytes from endoplasmic
reticulum stress and apoptosis. Front Physiol. 10:9952019.
View Article : Google Scholar : PubMed/NCBI
|