Introduction
Colorectal cancer (CRC) is one of the most common
types of cancer and has high incidence and mortality rates
(1,2). Despite advancements in the clinical
treatments for patients with CRC, the overall survival (OS) rate
has not improved due to recurrence and metastasis (3). Therefore, it is urgently necessary to
identify new biomarkers for the malignant progression of CRCs.
Long non-coding RNAs (lncRNAs) are RNAs >200
nucleotides in length that do not have protein-coding capacity
(4). Numerous studies have reported
that lncRNAs are involved in the regulation of biological behaviors
of tumors, including proliferation, apoptosis and metastasis
(5,6). For example, lncRNA Down syndrome cell
adhesion molecule antisense RNA 1 (DSCAM-AS1) facilitates
osteosarcoma cell metastasis by sponging microRNA (miR)-101-3p
(7). In addition, it has been
reported that the lncRNA wee1-like protein kinase 2 antisense RNA 1
(WEE2-AS1) suppresses breast cancer cell apoptosis by modulating
transducer of ERBB2, 1 (TOB1) and sponging miR-32-5p (8), whereas LINC01535 promotes the
development of esophageal squamous cell cancer by regulating the
JAK/STAT3 pathway (9). The present
study aimed to investigate the role of lncRNAs in CRC progression.
Previous studies have revealed that feline leukemia virus subgroup
C receptor 1 antisense RNA 1 (FLVCR1-AS1) serves an oncogenic role
in the development of several types of tumors. For instance, it has
been demonstrated that lncRNA FLVCR1-AS1 promotes glioma by
modulating the miR-4731-5p/E2F transcription factor 2 (E2F2) axis
(10), and contributes to the
malignant progression of ovarian cancer by sponging miR-513
(11). Furthermore, it has been
shown that silencing lncRNA FLVCR1-AS1 represses the proliferation
of lung cancer cells by regulating the Wnt/β-catenin pathway
(12). Nevertheless, the mechanism
of FLVCR1-AS1 in CRC remains unclear.
Numerous studies have indicated that lncRNAs exert
their functions via the targeting of miRNAs (13–15).
miRNAs have been reported to have tumor suppressive functions in
multiple human cancers. For instance, studies have shown that
miRNA-339-5p inhibits gastric cancer development via the
upregulation of alkB homolog 1 (ALKBH1) (16), miR-95-3p regulates CRC cell
proliferation via the downregulation of hepatoma-derived growth
factor (HDGF) (17), and miR-30a-3p
represses renal cancer cell metastasis via the regulation of
autophagy-related protein 12 (ATG12) expression (18). Furthermore, several studies have
shown that miR-381 is associated with the development of different
types of cancer, including cervical (19), gastric (20) and prostate cancer (21). A previous research has reported that
miR-381 acted as a tumor suppressor in CRC by targeting Twist1
(22). However, the regulatory
mechanism of miR-381 in CRC needs to be further elucidated.
The current study aimed to evaluate the FLVCR1-AS1
expression in CRC, and to investigate the biological role of
FLVCR1-AS1 in modulating the malignant phenotypes of CRC cells
in vitro.
Materials and methods
The Cancer Genome Atlas (TCGA)
analysis
The expression of FLVCR1-AS1 and CRC clinical data
were downloaded from the TCGA database (https://tcga-data.nci.nih.gov/tcga/). Kaplan-Meier
analysis and the log-rank test were used to analyze survival
curves. Cut-off values were determined using mean expression level
of FLVCR1-AS1.
Tissue specimens
A total of 26 pairs of CRC tissues and adjacent
non-tumor tissues were obtained from patients with CRC (14 males
and 12 females) with a mean age of 52 years (range, 34–82 years)
between April 2017 and September 2018 at Ruijin Hospital Affiliated
to Shanghai Jiaotong University School of Medicine. Written
informed consent was obtained from all subjects and the study was
approved by the Ethics Committee of Ruijin Hospital Affiliated to
Shanghai Jiaotong University School of Medicine. The specimens were
immediately cryopreserved in liquid nitrogen after surgery.
Cell lines and cell culture
Four human CRC cell lines, namely Caco-2, SW480,
LoVo and SW1116, the NCM460 normal colonic epithelial cell line and
293T cells were purchased from the American Type Culture
Collection. All cells were cultured in Dulbecco's modified Eagle's
medium (DMEM) containing 10% fetal bovine serum (both Gibco; Thermo
Fisher Scientific, Inc.) at 37°C in a humified atmosphere
containing 5% CO2.
Cell transfection
Short hairpin RNA (shRNA) targeting FLVCR1-AS1
(shFLVCR1-AS1) and its corresponding control shRNA (shNC), miR-381
mimics, 5′-UACAGUACUGUGAUAACUGAA-3′; NC mimics,
5′UUUGUACUACACAAAAGUACUG-3′; miR-381 inhibitor,
5′-UUCAGUUAUCACAGUACUGUA-3′; NC inhibitor,
5′-UCACAACCUCCUAGAAAGAGUAGA-3′. The full-length of FLVCR1-AS1 and
RAP2A were subcloned into pcDNA3.1 to overexpress FLVCR1-AS and
RAP2A levels, with empty pcDNA3.1 serving as control. The
overexpression plasmid and pcDNA3.1 were bought from Shanghai
GenePharma Co., Ltd. shRNA (100 nm), miRNA (50 nM) or plasmid (1
µg/ml) was transfected into cells using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequent
experiments were performed at 48 h post-transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues and cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
Subsequently, total RNA was reverse transcribed into cDNA using the
PrimeScript RT reagent kit (Takara Bio, Inc.) at 37°C for 15 min.
RT-qPCR was performed on an ABI 7500 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with SYBR-Green PCR
Master Mix kit (Takara Bio, Inc.). The PCR amplification reaction
was carried out using cDNA as template with the following
conditions: 95°C for 10 min, and 40 cycles of 95°C for 15 sec, 62°C
for 30 sec, and 72°C for 30 sec. Primers were as follows:
FLVCR1-AS1 forward, 5′-GAAAGCTGCAACATGCTCCC-3′ and reverse,
5′-TCCATGTCGTCCCAGTTGGT-3′; miR-381 forward,
5′-GCAGGGCTTCTGAGCTCCTTAA-3′ and reverse,
5′-CAAATTCGTGAAGCGTTCCATAT-3′; RAP2A forward,
5′-ACACGTCTGGAGGAGACAGC-3′ and reverse, 5′-GAGAGGTTGTGCCGGATAGA-3′;
GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse,
5′-TCCACCACCCTGTTGCTGTA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The relative expression of
genes was calculated using the 2−ΔΔCq method (23), and glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) and U6 were used as internal references.
Cell proliferation
Cell viability was evaluated using a Cell Counting
Kit 8 (CCK-8; Dojindo Molecular Technologies, Inc.) assay. Briefly,
2×104 cells/well were seeded into 96-well plates and 48
h following transfection, each well was supplemented with 10 µl
CCK-8 reagent. Subsequently, the optical density value at 450 nm
was measured using a microplate reader (Thermo Fisher Scientific,
Inc.).
TUNEL assay
An in situ Cell Death Detection kit (cat. no.
11684817910; Roche Diagnostics GmbH) was used to analyze cell
apoptosis. Caco-2 and SW480 cells were washed with PBS, and fixed
in 4% paraformaldehyde solution for 1 h at 4°C. The cells were
permeabilized in a solution containing 0.1% Triton X-100 for 2 min
and cultured with TUNEL reaction mixture (Roche Diagnostics GmbH)
for 1 h at 37°C in the dark. The TUNEL-stained coverslips were then
washed with PBS and then counterstained with DAPI (1:20 dilution;
Beyotime Institute of Biotechnology) for 15 min at room
temperature. A fluorescence microscope (magnification, ×100;
Olympus Corporation) was utilized to observe TUNEL-positive cells
in at least 5 fields of view.
Bioinformatics analysis and
dual-luciferase reporter assay
The starBase database (http://starbase.sysu.edu.cn/) were used for predicting
the binding sites between miR-381 and FLVCR1-AS1 or RAP2A. The
wild-type (wt) or mutant (mut) FLVCR1-AS1/RAP2A was inserted into a
pGL3.0 vector (Shanghai GenePharma Co., Ltd.). Subsequently, 293T
cells were transfected with one of the above vectors and either
miR-381 mimics or NC mimics using Lipofectamine 2000. Following
incubation for 48 h, luciferase activity was measured using the
Dual-Luciferase Reporter Assay System (Promega Corporation).
Firefly luciferase activity was normalized to Renilla
luciferase gene activity.
Pull-down assay
Biotin-labeled wt miR-381 (bio-miR-381-wt), mut
miR-381 (bio-miR-381-mut) and miR-NC were obtained from GenePharma
Co., Ltd. The biotinylated RNAs were transfected into Caco-2 and
SW480 cells using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), and the cell lysates were
incubated with streptavidin magnetic beads (Invitrogen; Thermo
Fisher Scientific, Inc.) overnight. Following RNA extraction, the
abundance of FLVCR1-AS1 was detected by RT-qPCR.
Transwell assay
Cell migration and invasion abilities were assessed
using Transwell chambers (8.0 µm pore size; BD Biosciences). For
cell migration, 1×105 transfected cells and 200 µl
serum-free DMEM were added to the upper chamber with an uncoated
membrane. Then, complete DMEM was added to the lower chamber.
Following seeding for 48 h, cells in the upper chamber were
removed, and those in the lower chamber were stained with 0.1%
crystal violet (Sigma-Aldrich; Merck KGaA). For cell invasion, the
membranes were precoated with Matrigel at 37°C for 2 h, and the
other steps were the same as those carried out in the migration
assay. Cell numbers were counted under a light microscope.
(magnification, ×200; Zeiss GmbH).
Western blot assay
Total protein extracts were isolated from
transfected cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology) and protein concentrations were determined using a
bicinchoninic acid protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Subsequently, 10 µg protein/lane was separated
by 10% SDS-PAGE (Bio-Rad Laboratories, Inc.). The proteins were
then electrotransferred onto a PVDF membrane. Following blocking
with non-fat milk for 2 h at room temperature, membranes were first
incubated with primary antibodies against RAP2A (1:1,000; cat. no.
ab101369; Abcam) and GAPDH (1:1,000; cat. no. ab9485; Abcam) at 4°C
overnight and then with corresponding secondary antibodies
(1:1,000; goat anti-rabbit IgG, cat. no. ab205718; Abcam) for 1 h
at room temperature. Finally, blots were visualized with an
enhanced chemiluminescent detection system (EMD Millipore). Protein
expression was evaluated using Image-Pro® Plus software
(version 6.0; Media Cybernetics, Inc.).
Statistical analysis
SPSS 19.0 (IBM Corp.) software was used to perform
the statistical analyses, and results are expressed as the mean ±
standard deviation. Each experiment was performed at least three
times. Clinicopathological characteristics were evaluated using the
χ2 test. Paired or unpaired Student's t-tests, or
one-way ANOVA followed by Tukey's post hoc test were used to
compare the groups. The survival curve was generated using the
Kaplan-Meier method and log-rank test with the ‘survival’ R package
(version 3.4.1; http://bioconductor.org/packages/survivalr/). The
patients were divided into a high and a low expression group
according to the mean gene expression. The correlation between gene
expression levels was analyzed using Pearson's correlation
coefficient. P<0.05 was considered to indicate a statistically
significant difference.
Results
FLVCR1-AS1 is upregulated in CRC
Firstly, the expression of FLVCR1-AS1 in CRC tissues
was examined, and the results indicated that the FLVCR1-AS1
expression levels were significantly elevated in CRC tissues
compared with the adjacent normal tissue (Fig. 1A). Consistent with this, FLVCR1-AS1
was found to be upregulated in CRC cells (Caco-2, SW480, LoVo and
SW111) compared with normal colonic epithelial cells (Fig. 1B). Analysis of the expression of
FLVCR1-AS1 according to different clinicopathological
characteristics of the patients with CRC indicated that high levels
of FLVCR1-AS1 were positively associated with TNM stage, distant
metastasis and lymph node metastasis (Table I). Notably, the expression of
FLVCR1-AS1 was significantly higher in CRC tissues of advanced TNM
stages compared with earlier ones (Fig.
1C). Furthermore, Kaplan-Meier analysis showed that CRC
patients with high levels of FLVCR1-AS1 exhibited a shorter
survival time than those with low FLVCR1-AS1 levels (Fig. 1D). These findings suggest that
FLVCR1-AS1 is involved in CRC tumorigenesis.
 | Table I.Association between
clinicopathological factors and FLVCR1-AS1 expression in patients
with colorectal cancer. |
Table I.
Association between
clinicopathological factors and FLVCR1-AS1 expression in patients
with colorectal cancer.
|
|
| lncRNA FLVCR1-AS1
expression, n |
|
|---|
|
|
|
|
|
|---|
| Parameters | Total | Low | High | P-value |
|---|
| Sex |
|
|
| 0.603 |
|
Female | 12 | 6 | 6 |
|
|
Male | 14 | 7 | 7 |
|
| Age (years) |
|
|
| 0.552 |
|
≤60 | 15 | 8 | 7 |
|
|
>60 | 11 | 5 | 6 |
|
| TNM stage |
|
|
| 0.024 |
|
I–II | 12 | 7 | 5 |
|
|
III–IV | 14 | 6 | 8 |
|
| Tumor size
(cm) |
|
|
| 0.817 |
| ≤2 | 11 | 6 | 5 |
|
|
>2 | 15 | 7 | 8 |
|
| Lymph node
metastasis |
|
|
| 0.016 |
|
Positive | 13 | 4 | 9 |
|
|
Negative | 13 | 9 | 4 |
|
| Distant
metastasis |
|
|
| 0.033 |
|
Positive | 8 | 3 | 5 |
|
|
Negative | 18 | 10 | 8 |
|
FLVCR1-AS1 modulates CRC cell
proliferation, metastasis and apoptosis
The biological role of FLVCR1-AS1 in CRC was further
explored. The expression of FLVCR1-AS1 was significantly decreased
in Caco-2 and SW480 cells following transfection with shFLVCR1-AS1
compared with shNC (Fig. 2A). CCK-8
and Transwell assays demonstrated that the knockdown of FLVCR1-AS1
attenuated CRC cell viability, migration and invasion (Fig. 2B-D). Furthermore, TUNEL assay
revealed that FLVCR1-AS1 silencing induced Caco-2 and SW480 cell
apoptosis (Fig. 2E). Subsequently,
RT-qPCR analysis showed that FLVCR1-AS1 was efficiently
overexpressed following transfection with an FLVCR1-AS1
overexpression plasmid (Fig. 2F).
CCK-8 and Transwell assays indicated that transfection with
pcDNA3.1/FLVCR1-AS1 increased the viability, migration and invasion
of Caco-2 and SW480 cells (Fig.
2G-I). In addition, TUNEL assay revealed that FLVCR1-AS1
overexpression markedly attenuated CRC cell apoptosis (Fig. 2J). The aforementioned data indicate
that FLVCR1-AS1 promotes CRC progression.
 | Figure 2.FLVCR1-AS1 modulates the
proliferation, metastasis and apoptosis of colorectal cancer cells.
(A) RT-qPCR was used to confirm the reduction of FLVCR1-AS1
expression in Caco-2 and SW480 cells transfected with shFLVCR1-AS1
compared with shNC. (B) CCK-8 and Transwell (magnification, ×200)
(C) migration and (D) invasion assays were performed to evaluate
the proliferation, migration and invasion abilities of Caco-2 and
SW480 cells transfected with shFLVCR1-AS1 and shNC. (E) TUNEL assay
(magnification, ×100) was used to estimate the apoptosis of Caco-2
and SW480 cells after FLVCR1-AS1 knockdown. (F) RT-qPCR was used to
determine the expression of FLVCR1-AS1 in Caco-2 and SW480 cells
transfected with pcDNA3.1/FLVCR1-AS1 and pcDNA3.1. (G) CCK-8 and
Transwell (magnification, ×200) (H) migration and (I) invasion
assays were performed to evaluate the proliferation, migration and
invasion abilities of Caco-2 and SW480 cells transfected with
pcDNA3.1/FLVCR1-AS1 and pcDNA3.1. (J) TUNEL assay (magnification,
×100) was used to estimate the apoptosis of Caco-2 and SW480 cells
transfected with pcDNA3.1/FLVCR1-AS1 and pcDNA3.1. *P<0.05.
FLVCR1-AS1, feline leukemia virus subgroup C receptor 1 antisense
RNA 1; RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; sh, short hairpin; NC, negative control; CCK-8, Cell
Counting Kit 8; OD450, optical density at 450 nm. |
FLVCR1-AS1 acts as a competing
endogenous RNA (ceRNA) for miR-381 in CRC
To explore the molecular mechanism of FLVCR1-AS1 in
CRC, its downstream targets were identified. Using the starBase
online tool, the binding sequence between miR-381 and FLVCR1-AS1
was predicted (Fig. 3A). RT-qPCR
analysis showed that miR-381 expression was successfully elevated
in Caco-2 and SW480 cells following transfection with miR-381
mimics compared with NC mimics (Fig.
3B). A luciferase reporter assay indicated that the luciferase
activity of FLVCR1-AS1-wt was significantly inhibited by miR-381
mimics. However, miR-381 mimics had no effect on the luciferase
activity of FLVCR1-AS1-mut in 293T cells (Fig. 3C). Moreover, the RNA pull-down assay
demonstrated that the FLVCR1-AS1 expression levels were elevated in
Caco-2 and SW480 cells following transfection with bio-miR-381-wt
(Fig. 3D). Furthermore, RT-qPCR
analysis revealed that the expression of miR-381 was downregulated
in CRC tissues compared with adjacent normal tissues (Fig. 3E), and the knockdown of FLVCR1-AS1
increased miR-381 expression in Caco-2 and SW480 cells (Fig. 3F). Following confirmation by RT-qPCR
analysis that miR-381 was successfully knocked down in Caco-2 and
SW480 cells following transfection with miR-381 inhibitor (Fig. 3G), whether FLVCR1-AS1 promotes CRC
progression by modulating miR-381 was further investigated. CRC
cells were transfected with shNC, shFLVCR1-AS1 and shFLVCR1-AS1 +
miR-318 inhibitor. CCK-8 and Transwell assays of the transfected
cells demonstrated that the miR-381 inhibitor attenuated the
inhibitory effect of FLVCR1-AS1 knockdown on CRC cell viability,
migration and invasion (Fig. 3H-J).
The aforementioned findings suggest that FLVCR1-AS1 may act as a
molecular sponge for miR-381 in CRC.
 | Figure 3.FLVCR1-AS1 functions as a competing
endogenous RNA for miR-381 in CRC. (A) The binding sequence between
FLVCR1-AS1 and miR-381 was predicted using starBase. (B) RT-qPCR
results show that the level of miR-381 was successfully increased
in Caco-2 and SW480 cells transfected with miR-381 mimics. (C)
Luciferase reporter assay results showing the luciferase activity
of FLVCR1-AS1-wt and FLVCR1-AS1-mut in 293T cells transfected with
NC mimics or miR-381 mimics. *P<0.05. (D) RNA pull-down assay
was used to verify the interaction between FLVCR1-AS1 and miR-381
in Caco-2 and SW480 cells transfected with bio-miR-381-wt or
bio-miR-381-mut. *P<0.05 vs. Bio-miR-NC. (E) RT-qPCR assay
results show that the level of miR-381 is lower in CRC tissues
compared with normal tissues. (F) RT-qPCR assay results show that
the expression of miR-381 is increased in cells transfected with
shFLVCR1-AS1. (G) RT-qPCR results show that the level of miR-381 in
Caco-2 and SW480 cells was successfully reduced by transfection
with miR-381 inhibitor. (H) Cell Counting Kit-8, (I) Transwell
migration assay (magnification, ×200) and (J) invasion assay
results demonstrating the proliferation, migration and invasion of
Caco-2 and SW480 cells transfected with shNC, shFLVCR1-AS1 and
shFLVCR1-AS1+ miR-381 inhibitor. *P<0.05. FLVCR1-AS1, feline
leukemia virus subgroup C receptor 1 antisense RNA 1; miR,
microRNA; CRC, colorectal cancer; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; bio, biotin;
wt, wild-type; mut, mutant; sh, short hairpin; NC, negative
control; OD450, optical density at 450 nm. |
RAP2A is directly targeted by
miR-381
Bioinformatics analysis was conducted using starBase
software to predict the downstream targets of miR-381. The analysis
revealed that RAP2A includes a potential binding site for miR-381
(Fig. 4A). A luciferase reporter
assay demonstrated that miR-381 mimics inhibit the luciferase
activity of RAP2A-wt but have no effect on the luciferase activity
of RAP2A-mut (Fig. 4B). In
addition, RT-qPCR results showed that RAP2A was highly expressed in
CRC tissues compared with adjacent normal tissue (Fig. 4C). In addition, RT-qPCR and western
blot assays indicated that overexpression of miR-381 significantly
reduced RAP2A expression in Caco-2 and SW480 cells (Fig. 4D). Subsequently, the association
between RAP2A and FLVCR1-AS1 was evaluated. The results indicated
that FLVCR1-AS1 knockdown downregulated RAP2A at the mRNA and
protein levels (Fig. 4E). In
addition, transfection with pcDNA3.1/FLVCR1-AS1 attenuated the
miR-381-mediated inhibition of RAP2A expression (Fig. 4F). Additionally, Pearson's
correlation analysis showed that the expression of miR-381 was
negatively correlated with that of FLVCR1-AS1 and RAP2A in CRC
tissues (Fig. 4G). The
aforementioned data suggest that FLVCR1-AS1 increases RAP2A
expression via the repression of miR-381 in CRC.
FLVCR1-AS1 promotes CRC progression by
upregulating RAP2A
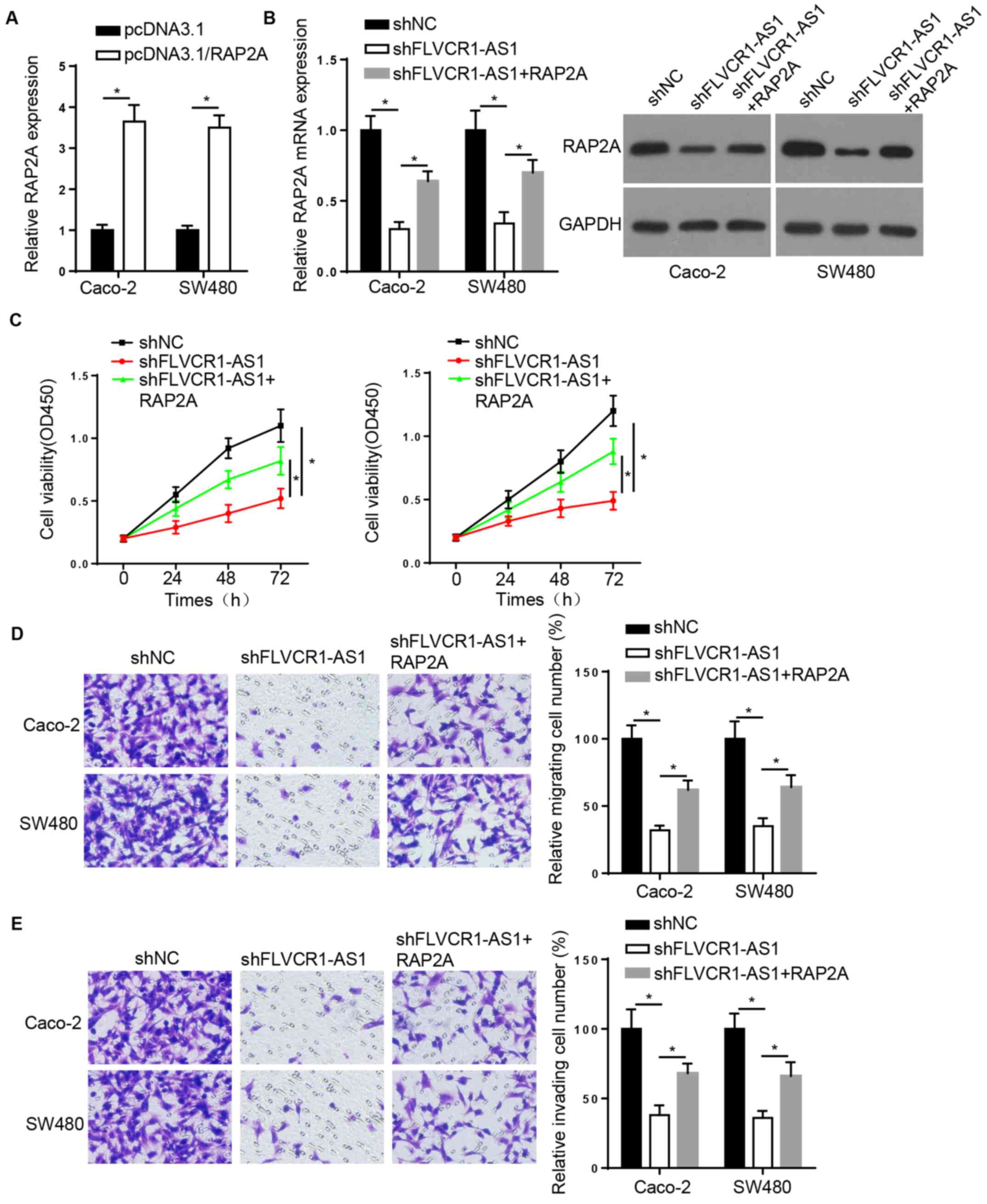
To investigate whether the molecular functions of
FLVCR1-AS1 are mediated by the upregulation of RAP2A in CRC, the
Caco-2 and SW480 cells were transfected with shNC, shFLVCR1-AS1 or
shFLVCR1-AS1 + RAP2A, and RT-qPCR and western blot assays were then
carried out. First, it was confirmed that RAP2A expression was
successfully increased by transfection with RAP2A overexpression
vector (Fig. 5A). The RT-qPCR and
western blotting results demonstrated that RAP2A overexpression
attenuated the FLVCR1-AS1 knockdown-mediated reduction of RAP2A
expression (Fig. 5B). Similarly,
CCK-8 and Transwell assays confirmed that RAP2A overexpression
attenuated the effect of FLVCR1-AS1 downregulation on Caco-2 and
SW480 cell proliferation, migration, and invasion (Fig. 5C-E). These findings indicate that
FLVCR1-AS1 promotes the development of CRC by modulating the
expression of RAP2A.
Discussion
The present study demonstrated that lncRNA
FLVCR1-AS1 was elevated in CRC tissues compared with adjacent
normal tissues, and the upregulated expression of FLVCR1-AS1 was
associated with poor prognosis in patients with CRC, thus implying
its essential role in CRC progression. Previous studies have shown
that the abnormal expression of FLVCR1-AS1 is associated with the
tumorigenesis of multiple cancers (24,25).
For example, FLVCR1-AS1 was reported to be upregulated in lung
cancer and hepatocellular carcinoma, in which it promoted cell
metastasis (26) and tumor growth
(27), respectively. Therefore, we
hypothesized that FLVCR1-AS1 could be involved in the development
of CRC. The findings of the current study reveal that the
downregulation of FLVCR1-AS1 attenuated CRC cell viability,
migration and invasion, thus suggesting that FLVCR1-AS1 serves an
oncogenic role in CRC.
There is considerable evidence that lncRNAs act as
molecular sponges to modulate miRNAs (28). For example, it has been reported
that lncRNA associated with poor prognosis of hepatocellular
carcinoma (AWPPH) sponges miRNA-204 to promote the development of
non-small cell lung cancer (29).
In addition, lncRNA olfactory receptor 3A4 (OR3A4) has been shown
to accelerate osteosarcoma cell proliferation via the targeting of
miR-1207-5p (30). Furthermore, the
miRNA-20a-5p/tripartite motif containing 32 (TRIM32) axis has been
shown to be associated with the apoptosis of breast cancer cells
following hepatocyte nuclear factor 1-α antisense RNA 1 (HNF1A-AS1)
silencing (31). It has also been
revealed that FLVCR1-AS1 acts as a ceRNA that is involved in the
pathogenesis and development of several types of cancer. Bao et
al (24) demonstrated that
lncRNA FLVCR1-AS1 acts as a sponge for miR-573 and thereby
regulates cholangiocarcinoma cell proliferation. Liu et al
(25) suggested that lncRNA
FLVCR1-AS1 acts as an oncogene in gastric cancer via regulation of
the miR-155/c-Myc pathway. The present study identified miR-381 as
a novel target of FLVCR1-AS1, and transfection with miR-381
inhibitor attenuated the suppressive effect of FLVCR1-AS1 silencing
on CRC cell viability and metastasis. These findings indicate that
miR-381 exerts an antitumor effect on CRC.
Furthermore, RAP2A was identified as a direct target
of miR-381 in the present study. Several studies have reported that
RAP2A may be used as a therapeutic biomarker and acts as an
oncogene in various cancers. For example, Zhang et al
(32) revealed that the
overexpression of RAP2A promoted gastric cancer progression, and
another study demonstrated that it accelerated the metastasis and
growth of hepatocellular carcinoma cells (33). In addition, RAP2A has been
identified to be contribute to the progression of tumors via the
regulation of the phosphorylation of AKT. For instance, one study
showed that RAP2A promoted the migration and invasion of renal
cancer cells by increasing phosphorylated (p)-AKT levels (34), while another demonstrated that the
overexpression of RAP2A increased cancer cell migration, invasion
and metastasis by the upregulation of p-AKT (35). Consistent with previous studies, the
current study revealed that the expression of RAP2A was markedly
increased in CRC tissues, and its expression was regulated by the
FLVCR1-AS1/miR-381 axis. Moreover, RAP2A overexpression reversed
the inhibitory effect of FLVCR1-AS1 silencing on CRC cell
viability, apoptosis, migration and invasion, thus supporting the
oncogenic role of RAP2A in CRC.
In conclusion, the present study suggests that
FLVCR1-AS1 acts as a ceRNA to increase RAP2A expression by sponging
miR-381, thereby facilitating CRC tumorigenesis. However, certain
limitations remain to be addressed in future studies. In
particular, the biological roles of FLVCR1-AS1 in CRC in
vivo, and the downstream effectors of RAP2A require further
investigation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YH, XW and LH designed the present study. EM and BS
performed the experiments. LH and BS analyzed the data and prepared
the figures. YH and XW drafted the initial manuscript. EM and LH
reviewed and revised the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Ruijin Hospital Affiliated to Shanghai Jiaotong
University School of Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ahnen DJ, Wade SW, Jones WF, Sifri R,
Mendoza Silveiras J, Greenamyer J, Guiffre S, Axilbund J, Spiegel A
and You YN: The increasing incidence of young-onset colorectal
cancer: A call to action. Mayo Clin Proc. 89:216–224. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ladabaum U, Dominitz JA, Kahi C and Schoen
RE: Strategies for colorectal cancer screening. Gastroenterology.
158:418–432. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Uszczynska-Ratajczak B, Lagarde J,
Frankish A, Guigó R and Johnson R: Towards a complete map of the
human long non-coding RNA transcriptome. Nat Rev Genet. 19:535–548.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Davalos V and Esteller M: Disruption of
long noncoding RNAs targets cancer hallmark pathways in lung
tumorigenesis. Cancer Res. 79:3028–3030. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu F, Sui Y, Wang Y, Xu T, Fan L and Zhu
H: Long noncoding RNA SNHG7, a molecular sponge for microRNA-485,
promotes the aggressive behavior of cervical cancer by regulating
PAK4. Onco Targets Ther. 13:685–699. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang S, Ding L, Gao F and Fan H: Long
non-coding RNA DSCAM-AS1 upregulates USP47 expression through
sponging miR-101-3p to accelerate osteosarcoma progression. Biochem
Cell Biol. 98:600–611. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang R, Huang Z, Qian C, Wang M, Zheng Y,
Jiang R and Yu C: LncRNA WEE2-AS1 promotes proliferation and
inhibits apoptosis in triple negative breast cancer cells via
regulating miR-32-5p/TOB1 axis. Biochem Biophys Res Commun.
526:1005–1012. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fang Y, Zhang S, Yin J, Shen YX, Wang H,
Chen XS and Tang H: LINC01535 promotes proliferation and inhibits
apoptosis in esophageal squamous cell cancer by activating the
JAK/STAT3 pathway. Eur Rev Med Pharmacol Sci. 24:3694–3700.
2020.PubMed/NCBI
|
|
10
|
Yan Z, Zhang W, Xiong Y, Wang Y and Li Z:
Long noncoding RNA FLVCR1-AS1 aggravates biological behaviors of
glioma cells via targeting miR-4731-5p/E2F2 axis. Biochem Biophys
Res Commun. 521:716–720. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan H, Li H, Silva MA, Guan Y, Yang L, Zhu
L, Zhang Z, Li G and Ren C: LncRNA FLVCR1-AS1 mediates miR-513/YAP1
signaling to promote cell progression, migration, invasion and EMT
process in ovarian cancer. J Exp Clin Cancer Res. 38:3562019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin H, Shangguan Z, Zhu M, Bao L, Zhang Q
and Pan S: lncRNA FLVCR1-AS1 silencing inhibits lung cancer cell
proliferation, migration, and invasion by inhibiting the activity
of the Wnt/β-catenin signaling pathway. J Cell Biochem.
120:10625–10632. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo H, Xu C, Le W, Ge B and Wang T: lncRNA
CASC11 promotes cancer cell proliferation in bladder cancer through
miRNA-150. J Cell Biochem. 120:13487–13493. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peng CL, Zhao XJ, Wei CC and Wu JW: LncRNA
HOTAIR promotes colon cancer development by down-regulating
miRNA-34a. Eur Rev Med Pharmacol Sci. 23:5752–5761. 2019.PubMed/NCBI
|
|
15
|
Lai XJ and Cheng HF: LncRNA colon
cancer-associated transcript 1 (CCAT1) promotes proliferation and
metastasis of ovarian cancer via miR-1290. Eur Rev Med Pharmacol
Sci. 22:322–328. 2018.PubMed/NCBI
|
|
16
|
Wang C, Huang Y, Zhang J and Fang Y:
MiRNA-339-5p suppresses the malignant development of gastric cancer
via targeting ALKBH1. Exp Mol Pathol. 115:1044492020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hong YG, Huang ZP, Liu QZ, E JF, Gao XH,
Xin C, Zhang W, Li P and Hao LQ: MicroRNA-95-3p inhibits cell
proliferation and metastasis in colorectal carcinoma by HDGF.
Biomed J. 43:163–173. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Y, Zhou J, Wu X, Huang J, Chen W, Liu
D, Zhang J, Huang Y and Xue W: MiR-30a-3p inhibits renal cancer
cell invasion and metastasis through targeting ATG12. Transl Androl
Urol. 9:646–653. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shang A, Zhou C, Bian G, Chen W, Lu W,
Wang W and Li D: miR-381-3p restrains cervical cancer progression
by downregulating FGF7. J Cell Biochem. 120:778–789. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie Y, Qi J, Zhu C, Zhao D and Liao G:
MiR-381 functions as a tumor suppressor in gastric cancer by
targeting ROCK2. Int J Clin Exp Pathol. 12:164–172. 2019.PubMed/NCBI
|
|
21
|
Hu J, Wu X, Yang C, Rashid K, Ma C, Hu M,
Ding Q and Jiang H: Anticancer effect of icaritin on prostate
cancer via regulating miR-381-3p and its target gene UBE2C. Cancer
Med. 8:7833–7845. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He X, Wei Y, Wang Y, Liu L, Wang W and Li
N: MiR-381 functions as a tumor suppressor in colorectal cancer by
targeting Twist1. Onco Targets Ther. 9:1231–1239. 2016.PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bao W, Cao F, Ni S, Yang J, Li H, Su Z and
Zhao B: lncRNA FLVCR1-AS1 regulates cell proliferation, migration
and invasion by sponging miR-485-5p in human cholangiocarcinoma.
Oncol Lett. 18:2240–2247. 2019.PubMed/NCBI
|
|
25
|
Liu Y, Guo G, Zhong Z, Sun L, Liao L, Wang
X, Cao Q and Chen H: Long non-coding RNA FLVCR1-AS1 sponges miR-155
to promote the tumorigenesis of gastric cancer by targeting c-Myc.
Am J Transl Res. 11:793–805. 2019.PubMed/NCBI
|
|
26
|
Gao X, Zhao S, Yang X, Zang S and Yuan X:
Long non-coding RNA FLVCR1-AS1 contributes to the proliferation and
invasion of lung cancer by sponging miR-573 to upregulate the
expression of E2F transcription factor 3. Biochem Biophys Res
Commun. 505:931–938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang L, Cheng H, Yue Y, Li S, Zhang D and
He R: TUG1 knockdown ameliorates atherosclerosis via up-regulating
the expression of miR-133a target gene FGF1. Cardiovasc Pathol.
33:6–15. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Riaz F and Li D: Non-coding RNA associated
competitive endogenous RNA regulatory network: Novel therapeutic
approach in liver fibrosis. Curr Gene Ther. 19:305–317. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu D, Qin BY, Qi XG, Hong LL, Zhong HB and
Huang JY: LncRNA AWPPH accelerates the progression of non-small
cell lung cancer by sponging miRNA-204 to upregulate CDK6. Eur Rev
Med Pharmacol Sci. 24:4281–4287. 2020.PubMed/NCBI
|
|
30
|
Wang X, Chen K and Zhao Z: LncRNA OR3A4
regulated the growth of osteosarcoma cells by modulating the
miR-1207-5p/G6PD Signaling. Onco Targets Ther. 13:3117–3128. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meng Q, Wang L, Lv Y, Wu J and Shi W:
Deletion of HNF1A-AS1 suppresses the malignant phenotypes of breast
cancer cells in vitro and in vivo through targeting
miRNA-20a-5p/TRIM32 axis. Cancer Biother Radiopharm. Apr
22–2020.(Epub ahead of print). doi: 10.1089/cbr.2019.3168.
View Article : Google Scholar
|
|
32
|
Zhang J, Wei Y, Min J, Wang Y, Yin L, Cao
G and Shen H: Knockdown of RAP2A gene expression suppresses
cisplatin resistance in gastric cancer cells. Oncol Lett.
19:350–358. 2020.
|
|
33
|
Zheng X, Zhao W, Ji P, Zhang K, Jin J,
Feng M, Wang F, Zheng S and Wang X: High expression of Rap2A is
associated with poor prognosis of patients with hepatocellular
carcinoma. Int J Clin Exp Pathol. 10:9607–9613. 2017.PubMed/NCBI
|
|
34
|
Wu JX, Du WQ, Wang XC, Wei LL, Huo FC, Pan
YJ, Wu XJ and Pei DS: Rap2a serves as a potential prognostic
indicator of renal cell carcinoma and promotes its migration and
invasion through up-regulating p-Akt. Sci Rep. 7:66232017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu JX, Zhang DG, Zheng JN and Pei DS:
Rap2a is a novel target gene of p53 and regulates cancer cell
migration and invasion. Cell Signal. 27:1198–1207. 2015. View Article : Google Scholar : PubMed/NCBI
|