Introduction
Lung cancer is a leading cause of death from cancer
worldwide with an annual death rate of ~1,000,000 individuals and a
low 5-year survival rate of ~19% diagnosed between 2009 and 2015 in
the United States, although efforts are increasingly being made to
improve these outcomes (1–3). Lung adenocarcinoma (LUAD) is a common
type of non-small cell lung cancer (NSCLC) that accounts for ~90%
of all lung cancer cases (2).
Unfortunately, no completely effective therapeutic methods for the
treatment of lung cancer are currently available.
In clinical practice, platinum-based combined
chemotherapy is used as the essential pharmacotherapy for patients
with lung cancer who need adjuvant chemotherapy or for patients
whose cancer is in an advanced, nonoperative state (4). However, chemotherapy failure and a
poor prognosis that impairs the quality of life of patients with
lung cancer occur frequently due to acquired chemotherapy
resistance (4). Thus, studying the
mechanism by which chemoresistance develops is urgently required to
identify effective therapeutic targets and strategies.
Glycolysis is a glucose metabolism pathway with a
high amount of glucose utilization and a low level of oxygen
consumption, which is essential for cancer cells to achieve a high
rate of proliferation and to avoid apoptosis (5–7).
Glycolysis is involved in cancer progression as well as
chemoresistance (5,6). Similarly to solid cancers, LUAD
exhibits a metabolic switch toward glycolysis (8,9).
Accordingly, the dynamic expression of certain molecules involved
in glycolysis, such as glucose transporters (GLUT), sodium/glucose
cotransporters and hexokinase, has been observed in lung cancer
(10–12). In addition, previous studies have
reported an association between glycolysis and cancer cell survival
and between cisplatin (DDP) sensitivity and poor prognosis of
patients with lung cancer, suggesting that glycolysis predicts
patient overall survival and chemotherapy failure (8,13–15).
However, the mechanism between glycolysis and DDP sensitivity is
poorly understood at present.
PR domain zinc finger protein (PRDM) family proteins
have been demonstrated to regulate cell differentiation, especially
in embryonic stem cells (16–18).
Recently, PRDM14, a PR-domain-containing transcriptional regulator,
has been reported to be differentially expressed in various types
of cancer such as NSCLC and breast cancer and to be involved in
cancer cell differentiation as well as cancer stemness, migration
and metastasis (19–21). However, studies on whether PRDM14
affects chemoresistance and glycolysis in LUAD cells are
limited.
The present study aimed to determine the functions
of knockdown or overexpression of PRDM14 in the chemosensitivity
and glycolysis of LUAD cells. The results may offer an insightful
perspective on the function of PRDM14 in glycolysis-mediated
chemoresistance of LUAD.
Materials and methods
Clinical subjects and samples
A total of 40 patients with LUAD (20 DDP-sensitive
and 20 DDP-resistant), 22 female and 18 male, aged between 28 and
67 years (median, 48 years), whose first-line treatment was DDP
were recruited at The Seventh People's Hospital (Shanghai, China)
between March 2014 and October 2017. Using the proportion of
changes in tumor volume after treatment, the patients were grouped
into four subgroups: i) Complete response (CR; no tumor); ii)
partial response (PR; tumor shrinkage by >50%); iii) stable
disease (tumor shrinkage by <50% or tumor enlargement by
<25%); and iv) progressive disease (PD; tumor enlargement by
>25%). Patients in the CR and PR groups were defined as
DDP-sensitive, and those in the SD and PD groups were defined as
DDP-resistant. A total of 20 normal adjacent non-tumor lung
tissues, which were resected within at ≥5 cm of the tumor margin
when the patients (10 DDP-sensitive and 10 DDP-resistant) underwent
surgery, were used as the control samples. All tissues were
immediately frozen with liquid nitrogen until further experiments.
In addition, a total of 12 primary LUAD cell samples were isolated
from 12 patients with LUAD recruited at The Seventh People's
Hospital (Shanghai, China) between March 2014 and October 2017, 7
female and 5 male, aged between 35 and 61 years (median, 42 years),
as previously described (22) and
were classified on the basis of median PRDM14 mRNA expression
levels relative to GAPDH mRNA expression levels determined by
reverse transcription quantitative (RT-q)PCR into low and high
PRDM14 level groups (median value, 3.16). Cases that received
preoperative radiochemotherapy before surgical resection were
excluded. Written informed consent was obtained from all patients.
The study was approved by the Ethics Committee of The Seventh
People's Hospital (approval no. 2014-002).
Cell lines and culture
The cell lines used in the present study were
obtained from JRDUN Biotechnology (Shanghai) Co., Ltd., including
two human LUAD cell lines, A549 and A549/DDP, and a human bronchial
epidermal 16HBE cell line, which was used as the control. The cells
were cultured in the basal culture medium comprising RPMI-1640
medium (HyClone; Cytiva) mixed with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2 in a
cell incubator with a humidified atmosphere.
Vector construction of PRDM14
overexpression and short hairpin (sh)RNA interference
To overexpress PRDM14, the specific coding sequence
was synthesized and cloned into pLVX-Puro plasmids (Clontech
Laboratories, Inc.). To knock down PRDM14, the specific shRNA
sequences targeting PRDM14 (shRNA-1 5′-CCTCATGCAGACGGTGTTT-3′;
shRNA-2, 5′-GGATATTCCTGTGAGCCTT-3′; and shRNA-3,
5′-GCATACTCCGCACACACAT-3′) or scramble shRNA
(5′-CATTCCGCAGTGGTGCATT-3′) were cloned into linearized pLKO.1
plasmids (Addgene, Inc.). To produce transducer plasmids, the
recombinant plasmids (1,000 ng) were transfected along with the
packaging plasmids psPAX2 (100 ng) and pMD2G (900 ng; Addgene,
Inc.) and amplified in 293T cells (ATCC; ACS-4500) plated in a
9-well plate (1×105 cells/well) for 6 h at 37°C. The
transfection procedures used Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Subsequently, 48 h after transfection,
the recombinant lentivirus in the cell supernatant was collected by
centrifugation at 5,000 × g for 5 min and the purification and
titration of recombinant lentivirus was performed as previously
described (23). A549 or A549/DDP
cells were plated in a 6-well plate (5×105 cells/well)
and infected with the recombinant lentivirus-transducing units at
an MOI of 20 in the presence of 8 µg/ml polybrene (Sigma-Aldrich;
Merck KGaA) for 24 h at 37°C. Stable cells were selected by
puromycin (3 µg/ml; Thermo Fisher Scientific) for four more days.
pLKO.1-scrambled shRNA (shNC) and blank pLVX-Puro (vector) were
used as negative controls.
Cell Counting Kit-8 (CCK-8) cell
viability assay
The primary LUAD, A549 and A549/DDP cells were
plated in a 96-well plate (3×103 cells/well) and
cultured overnight at 37°C. The following day, primary LUAD cells
were exposed to 10 µM DDP (Selleck Chemicals) for 48 h. A549 cells
were transduced with the PRDM14 overexpression vector (oePRDM14)
and treated with 10 or 20 µM DDP or 20 µM antiglycolytic agent
3-Bromopyruvate (3-BrPA; MedKoo Biosciences, Inc.) for 48 h at
37°C. A549/DDP cells were transfected with shPRDM14 and treated
with 10 or 20 µM DDP for 48 h at 37°C. Following treatment, 10 µl
CCK-8 solution (Signalway Antibody LLC) was added to each well for
1 h according to the manufacturer's instructions. The optical
density (OD) value of each well was read on a microplate reader at
450 nm to calculate relative cell viability inhibition rate as
follows: Viability inhibition rate = (ODvehicle + vector
- ODDDP + vector)/ODvehicle + vector
×100%.
Apoptosis analysis following DDP
treatment by flow cytometry
A549 and A549/DDP cells were cultured in a 6-well
plate (3×105 cells/well) until they reached 50%
confluency. Subsequently, the cells were subjected to transduction
or drug treatment. Cells were collected at 48 h post-treatment. The
staining procedure included a 15-min incubation at 4°C with 5 µl
annexin-V-FITC, followed by a 15-min incubation with 5 µl propidium
iodide (PI; all from Beyotime Institute of Biotechnology). An
Accuri™ C6 flow cytometer (BD Biosciences) using CellQuest Pro
software, version 3.3 (Becton, Dickinson and Company) was used to
examine early [(annexin-V-FITC)+/PI-] and late
[(annexin-V-FITC)+/PI+] apoptosis.
Measurement of glucose uptake
A549 and A549/DDP cells (5×105
cells/well) were cultured in a 6-well plate, kept in continuous
cell culture for 24 h at 37°C and subjected to transduction or drug
treatment. At 48 h post-treatment, low-glucose DMEM (Hyclone;
Cytiva) was added to each well. Following 3-h incubation, the
Krebs-Ringer Bicarbonate Buffer (Guangzhou Weibo Biological
Technology Co Ltd.) supplemented with 2% bovine serum albumin
(Beijing Solarbio Science & Technology Co., Ltd.) was used to
replace the culture medium and wash the cells. Subsequently,
glucose-free DMEM supplemented with 100 µM
2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxyglucose
(2-NBDG; Cayman Chemical Company) was used to culture cells for an
additional 45 min, and cell glucose uptake analysis was performed
using a flow cytometer.
Measurement of lactate
A549 and A549/DDP cells were seeded in a 6-well
plate (5×105 cells/well), cultured for 24 h and
subjected to transfection or drug treatment for 48 h. The medium
was collected for the analysis of lactate concentration using a
Lactic Acid Assay kit (Nanjing Jiancheng Bioengineering Institute)
according to the manufacturer's instructions.
RTq-PCR
Total RNA was isolated from the tissue homogenate
and cell lines using TRIzol® reagent (Thermo Fisher
Scientific, Inc.). The RNA was subjected to reverse transcription
into cDNA by using a PrimeScript™ kit (Takara Biotechnology Co.,
Ltd.). The cDNA synthesis conditions were 37°C for 60 min, followed
by 85°C for 5 min and 4°C for 5 min. The PCR amplification of cDNA
product was performed using the SYBR® Green PCR Master
Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) according
to the manufacturer's instructions. The primer sequences were as
follows: PRDM14 forward, 5′-GACTCACGCCTGTAATCC-3′ and reverse,
5′-GTCTCCTGTGCTCAAACC-3′; and GAPDH forward,
5′-AATCCCATCACCATCTTC-3′ and reverse, 5′-AGGCTGTTGTCATACTTC-3′. The
fold-changes of the PRDM14 mRNA levels were calculated by the
2−ΔΔCq method (24).
GAPDH mRNA was used as the internal control. The PCR cycling
conditions were 95°C for 10 min followed by 40 cycles at 95°C for
15 sec and 60°C for 45 sec followed by a final extension step of
95°C for 15 sec, 60°C for 1 min, 95°C for 15 sec and 60°C for 15
sec.
Western blotting
Total protein was extracted from the LUAD tissue
homogenate or cell lines using RIPA lysis buffer supplemented with
a mixture of protease inhibitors (Sigma-Aldrich; Merck KGaA). Total
protein concentration in each tissue sample was measured using a
Lowry protein assay kit (Bio-Rad Laboratories, Inc.). Equivalent
quantities (25 µg) of protein was separated by 10 or 15% SDS-PAGE,
transferred to a nitrocellulose membrane (EMD Millipore), blocked
in 5% fat-free milk overnight at 4°C and incubated with primary
antibodies against PRDM14 (1:500; cat. no. ab187881), γ-H2A histone
family member X (γ-H2AX; 1:500; cat. no. ab26350), cleaved
caspase-3 (1:500; cat. no. ab2302), pro-caspase-3 (1:500; cat. no.
ab32150), cleaved poly(ADP-Ribose) polymerase 1 (PARP1; 1:500; cat.
no. ab32064), PARP1 (1:500; ab191217) and GLUT1 (1:1,000; cat. no.
ab32551) (all from Abcam) overnight at 4°C, and then washed three
times with Tris-buffered saline with 0.1% Tween-20 (Amresco, LLC).
The membranes were subsequently incubated with anti-rabbit
horseradish peroxidase-conjugated immunoglobulin G secondary
antibody (1:1,000; cat. no. A0208; Beyotime Institute of
Biotechnology;) for 1 h at 37°C. GAPDH (1:2,000.; cat. no. 5174;
Cell Signaling Technology, Inc.) served as the internal control.
Finally, the specific signals were visualized using an enhanced
chemiluminescence system (Bio-Rad Laboratories, Inc.) and
quantified by densitometry (Quantity One software, version 4.62;
Bio-Rad Laboratories, Inc.).
Xenograft establishment with stable
cell lines and DDP treatment
A total of 80 BALB/c nude mice (male; age, 6–8
weeks; weight, 20–25 g) obtained from Shanghai SLAC Laboratory
Animal Co., Ltd. were randomly divided into two groups (n=40 per
group), and their flank regions were subcutaneously injected with
5×106 A549/DDP cells transfected with shPRDM14 or shNC.
The animals were maintained at a constant temperature of 25°C with
~50% humidity under a regular 12:12-h light/dark cycle with food
and water available ad libitum.
To examine the xenograft sensitivity to DDP, each
group was evenly divided into two further subgroups receiving an
intraperitoneal injection of 5 mg/kg DDP or an equal volume of
vehicle once a week for 3 weeks (n=20 per group). The xenograft
tumors were measured 12 days after the cells were injected, and the
tumor volumes were calculated. Tumor xenografts of mice in each
group (n=6 per group) were dissected 33 days after cell injection
for visual evaluation of morphology and were weighed. Tumor volume
was calculated twice a week using the following equation:
Volume=(length × width2)/2. The remaining mice (n=14 per
group) were used to analyze survival rates for 90 days after cell
injection.
The animals were sacrificed by an intraperitoneal
injection of sodium pentobarbital (30 mg/kg; Vetoquinol UK, Ltd.)
followed by cervical dislocation on days 33 and 90 or when they
reached certain humane endpoints (tumor diameter >2 cm or 20%
body weight loss). Any procedure performed on mice followed the
guidelines of the Ethics Committee of Institutional Animal Care and
Use Committee of The Seventh People's Hospital (approval no.
2019-078).
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analyses were performed by GraphPad Prism 8.0.2
(GraphPad Software, Inc.). Comparisons between two groups were
performed using unpaired Student's t-test and among multiple groups
using one- or two-way ANOVA followed by Bonferroni or Tukey's
multiple comparisons test when the data were compared among
multiple groups or with a single control group, respectively. Mouse
survival was analyzed by the Kaplan-Meier method and log-rank test.
Bonferroni correction was used to adjust P-values for multiple
measures. Cell experiments were performed in triplicate. P<0.05
was considered to indicate a statistically significant
difference.
Results
PRDM14 expression levels are
upregulated in patients with DDP-resistant LUAD and A549/DDP
cells
RT-qPCR results demonstrated that patients in the
DDP-resistant group expressed higher levels of PRDM14 compared with
patients in the DDP-sensitive group (Fig. 1A). To further determine the
association between PRDM14 level and the sensitivity of LUAD cells
to DDP, 12 primary LUAD cell lines, collected from 12 patients with
LUAD, were classified into low and high PRDM14 expression groups
(Fig. 1B) and treated with DDP.
Following treatment, low cell viability inhibition was observed in
the high PRDM14 expression group compared with that in the low
PRDM14 group (Fig. 1C).
Additionally, in the A549/DDP cells, the expression levels of
PRDM14 were higher level compared with those in the progenitor A549
cells (Fig. 1D and E). These data
suggested a potential relationship between the chemosensitivity of
LUAD cells to DDP and PRDM14.
PRDM14 regulates A549 and A549/DDP
cell sensitivity to DDP in vitro
To determine the regulatory effects of PRDM14 on the
chemosensitivity of LUAD cells to DDP, PRDM14 was overexpressed in
A549 cells (Fig. 2A), and cell
viability and the percentage of apoptotic cells sensitive to DDP
treatment were measured. In the vector-transfected control cells,
10 and 20 µM DDP inhibited cell viability and promoted the
apoptotic rate, whereas in cells overexpressing PRDM14, these
changes were significantly attenuated (Fig. 2B and C). In A549 cells, the cell
viability inhibition rate in response to DDP in the oePRDM14 group
(10 µM, 10.65±1.99%; 20 µM, 19.93±0.26%) was significantly lower
compared with that in the vector group (10 µM, 15.67±0.99%; 20 µM,
27.97±0.83%; Fig. 2B). By contrast,
in A549/DDP cells, PRDM14 silencing by shRNA targeting PRDM14
(Fig. 2D) enhanced the decrease in
cell viability and the increase in the percentage of apoptotic
cells in response to 10 and 20 µM DDP (Fig. 2E and F). In A549/DDP cells, the cell
viability inhibition rate response to DDP in the shPRDM14 group (10
µM, 24.16±2.39%; 20 µM, 41.67±3.39%) was significantly higher
compared with that in the shNC group (10 µM, 17.98±1.49%; 20 µM,
29.3±1.38; Fig. 2E).
Chemosensitivity is associated with not only the
activation of apoptosis, but also with DNA damage (25). Therefore, the expression of γ-H2AX,
a marker of DNA damage, was also determined. In the
vector-transfected cells, the two doses of DDP promoted an increase
in the expression levels of γ-H2AX compared with those in the
vehicle group, whereas in cells overexpressing PRDM14, this
increase was significantly attenuated (Fig. 2G). By contrast, in A549/DDP cells,
PRDM14 silencing enhanced the increase in the γ-H2AX expression
levels in response to the two doses of DDP compared with that in
the vehicle group (Fig. 2G). These
results suggested that PRDM14 overexpression inhibited the DDP
sensitivity of A549 cells, whereas PRDM14 silencing enhanced that
of A549/DDP cells.
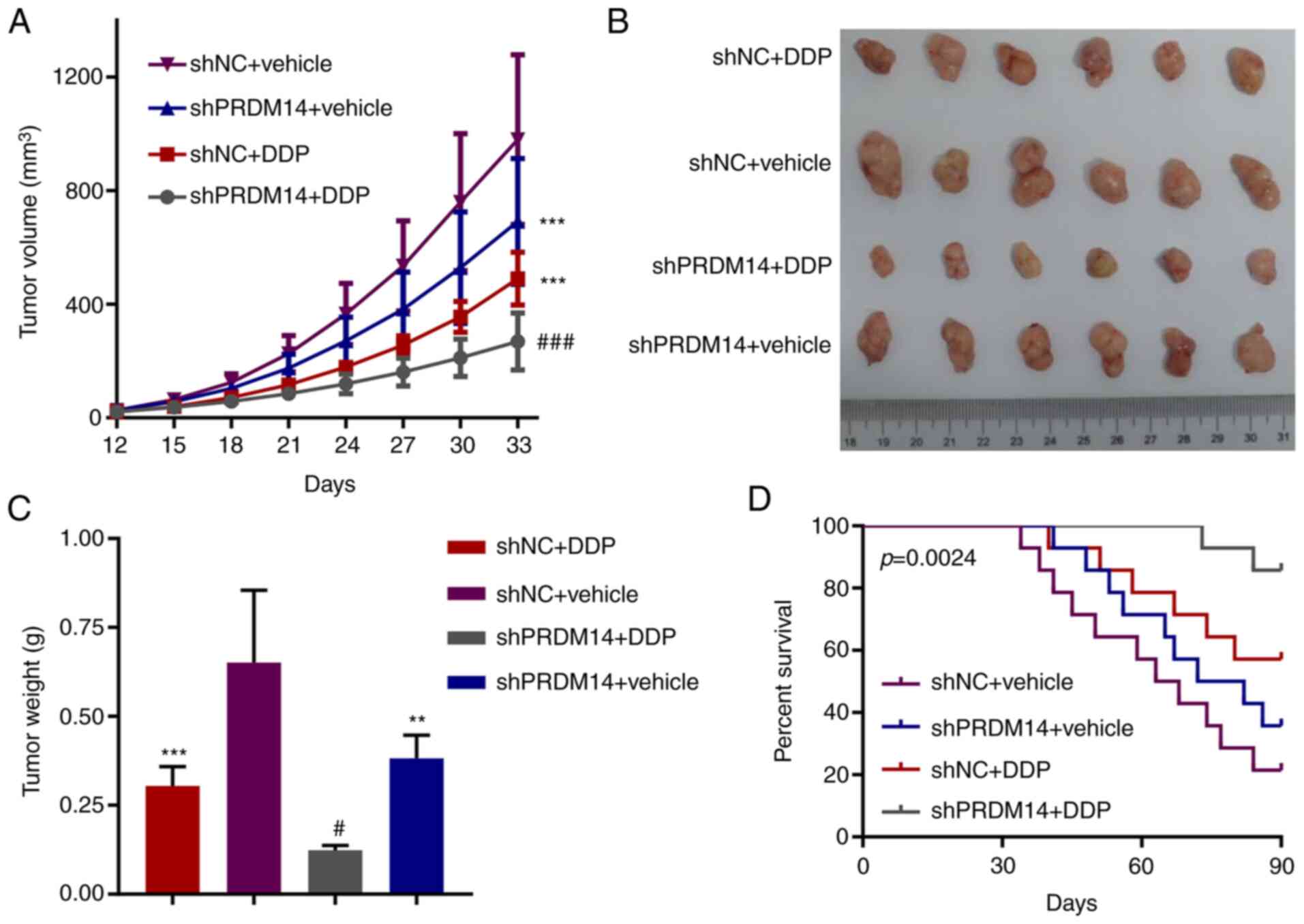
Knockdown of PRDM14 restrains
DDP-resistant tumor growth in vivo
A model using mice bearing a DDP-resistant tumor
with A549/DDP cells transfected with shPRDM14 or shNC was
established and treated with DDP treatment to evaluate the role of
PRDM14 knockdown in the tumor sensitivity to DDP. As presented in
Fig. 3A-C, PRDM14 knockdown alone
or DDP alone suppressed tumor growth, whereas PRDM14 knockdown
combined with DDP treatment resulted in the lowest tumor volume and
weight at 33 days among all groups, suggesting that PRDM14
knockdown enhanced the chemosensitivity of A549/DDP cells to DDP.
In addition, PRDM14 knockdown resulted in the highest survival rate
of tumor-bearing mice receiving DDP treatment among all groups
(Fig. 3D).
PRDM14 silencing inhibits glucose
uptake and lactate release in A549/DDP cells
To assess the involvement of PRDM14 in glycolysis in
LUAD cells, PRDM14 was silenced by shPRDM14 transfection. The
results of glycolysis analysis demonstrated that, compared with the
negative control shRNA, PRDM14 silencing reduced the glucose uptake
(Fig. 4A and B) and lactate
production (Fig. 4C). Additionally,
the levels of GLUT1, a protein essential for glucose transport
during glycolysis (26), were also
repressed by PRDM14 silencing compared with those in the shNC group
(Fig. 4D). These results suggested
that PRDM14 silencing inhibited the glycolysis of A549/DDP
cells.
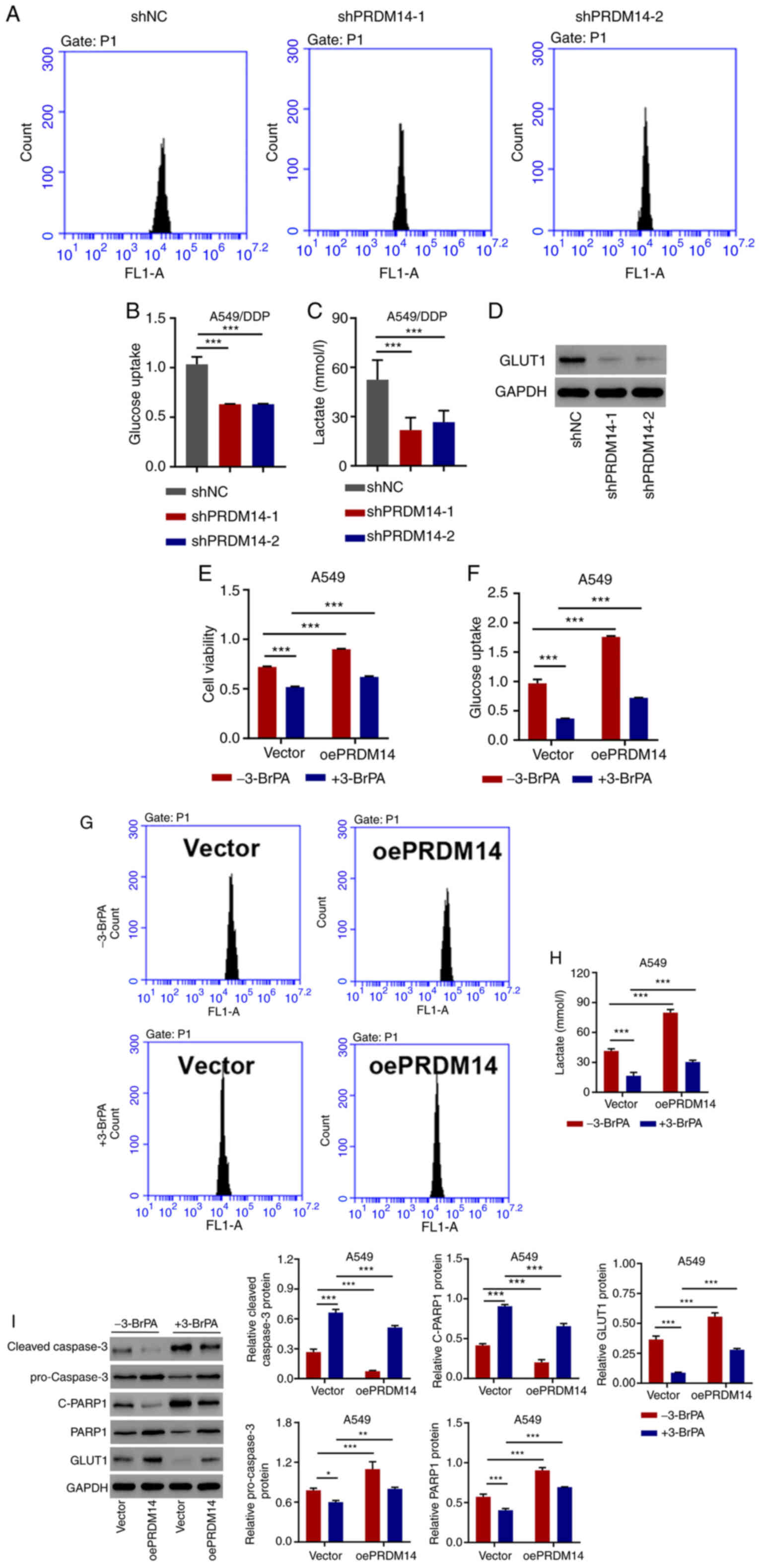 | Figure 4.PRDM14 regulates glucose uptake and
lactate release in A549/DDP cells and alleviates the effects of
3-BrPA in A549 cells. (A and B) Glucose uptake, (C) lactate release
and (D) GLUT1 expression in A549/DDP cells transfected with
shPRDM14 or shNC. (E) Cell viability, (F and G) glucose uptake and
(H) lactate release, and (I) C-caspase-3, pro-caspase-3, C-PARP1,
PARP1 and GLUT1 expression levels in A549 cells transduced with
oePRDM14 or blank vector in the absence or presence of the
antiglycolytic agent 3-BrPA. *P<0.05, **P<0.01 and
***P<0.001. DDP, cisplatin; oe, overexpression; sh, short
hairpin; NC, negative control; GLUT, glucose transporter; C-,
cleaved; PARP1, poly(ADP-ribose) polymerase 1; 3-BrPA,
3-bromopyruvate; PRDM14, PR domain zinc finger protein 14. |
PRDM14 alleviates the antiglycolytic
and pro-apoptotic effects of 3-BrPA in A549 cells
To determine the role of PRDM14 in cell viability
and glycolysis, A549 cells were treated with the PRDM14
overexpression vector and 3-BrPA, which is a potent glycolysis
inhibitor and a specific anticancer agent for lung tumorigenesis
(27,28). The antiglycolytic agent 3-BrPA alone
exhibited a significant inhibitory effect on cell viability,
glucose uptake, lactate release and the expression levels of
pro-caspase-3, PARP1 and GLUT1, but increased the expression levels
of cleaved caspase-3 and cleaved PARP1 compared with the untreated
group. PRDM14 overexpression alone exerted the opposite effects;
PRDM14 overexpression combined with 3-BrPA reversed the inhibitory
effects of 3-BrPA in A549 cells (Fig.
4E-I), suggesting that PRDM14 overexpression alleviated the
antiglycolytic and pro-apoptotic effects of 3-BrPA.
Discussion
DDP-based combined chemotherapy is an essential
strategy for lung cancer treatment, which fails to improve patient
survival rates due to chemotherapy resistance (4). Identifying the key molecules or
signaling pathways that control chemoresistance is an urgent
research target. The results of the present study demonstrated that
PRDM14, a protein highly expressed in DDP-resistant patients and in
LUAD cells, negatively controlled the sensitivity of LUAD cells to
DDP by promoting glycolysis, suggesting that PRDM14 may be a target
for chemotherapy in patients with LUAD.
PRDM14 exhibits a low expression level in
adjacent-normal tissues in adults, but is upregulated in various
types of cancer, whereas aberrant PRDM14 expression controls the
cancer cell phenotype and acts as a driver of oncogenic processes,
such as in colorectal (29),
pancreatic (30) and breast
(21) cancer. Similarly, the
results of the present study demonstrated that PRDM14 was
upregulated in patients with DDP-resistant LUAD and A549/DDP cells,
and enhanced the resistance of LUAD cells to DDP. Since limited
studies have reported the function of PRDM14 in the chemoresistance
of LUAD cells, the present study focused on PRDM14 and explored its
underlying mechanism in this process.
A previous study has demonstrated that PRDM14
regulates the cell migration and invasion in human NSCLC (20). The metabolic priorities of cancer
cells differ from those of normal cells, thus providing a new
therapeutic window; the Warburg effect, also termed glycolysis, is
characterized by high glucose uptake and lactate release, and is
considered to be a hallmark of prostate cancer (31). This metabolic adaptation benefits
cancer cells in surviving through hypoxic conditions, which are
common in tumors, and supports their anabolic requirements
(32). Elaborating the central role
of PRDM14 in the glycolysis of DDP-resistant lung cancer cells is
an important outcome of the present study. Aerobic glycolysis is
actively promoted by cancer cells to sustain the metabolic
requirements for tumor progression as well as in cells escaping
damage from chemotherapeutic agents (33,34),
which has been demonstrated in the chemoresistance of lung cancer
(9,35). Notably, the results of the present
study demonstrated that PRDM14 silencing inhibited glucose uptake
and lactate release in A549/DDP cells and that PRDM14
overexpression restored the glycolysis of A549 cells under the
inhibition by antiglycolytic agent 3-BrPA, indicating that PRDM14
may positively promote the chemoresistance of LUAD cells by
increasing glycolysis.
Despite an increasing number of studies focusing on
the role of PRDM14 in tumor progression and chemoresistance, the
understanding of the targets of PRDM14 is still limited. Previous
studies have attempted to determine the interaction between PRDM14
and gene associated with cell proliferation, apoptosis and
glycolysis, which are crucial cancer cell processes during
chemoresistance (36–38). PRDM14 may affect the proliferation
of 293T cells by inducing cell cycle arrest at G1/S phase (36), target apoptosis regulators
phorbol-12-myristate-13-acetate-induced protein 1 [PMAIP1, also
known as NOXA) and Bcl-2 binding component 3 (BBC3, also known as
PUMA)] in the apoptosis evasion of HPV-positive cancer cells
(37), and interact with heat shock
protein 90α and glucose-regulated protein 78, two molecules
associated with chemotherapy resistance (38).
The present study analyzed the regulatory effects of
PRDM14 on the expression of caspase-3, which is a pro-apoptotic
gene (39), and GLUT1, which is a
protein mediating glucose transport, metabolism and chemoresistance
of NSCLC cells (26). The
activation of caspase-3 and PARP1 and the downregulation of GLUT1
induced by 3-BrPA was reversed by PRDM14 overexpression in A549
cells, suggesting that caspase-3 and GLUT1 may be downstream
targets for PRDM14. However, whether PRDM14 regulates caspase-3 and
GLUT1 via its transcriptional activity directly or other indirectly
mechanisms require further validation in future studies.
There were several limitations to the present study
that need to be considered when interpreting these results. First,
although this cohort quite accurately reflected the normal span of
patients in a clinic, the sample size was quite small. Second,
clinical parameter information necessary to assess a correlation
between PRDM14 expression and LUAD progression needs to be further
validated. Finally, the pathways should be clarified in further
investigation, especially those involved in the apoptosis,
glycolysis and chemoresistance-related signals.
In conclusion, the results of the present study
suggested the key role of PRDM14 in the chemoresistance of LUAD
cells and demonstrated the regulatory effects of PRDM14 on
apoptosis and glycolysis in LUAD cells resistant to DDP. The
results of the present study provide new insights and support the
application of PRDM14 as a chemotherapy target for LUAD.
Acknowledgements
Not applicable.
Funding
This work was funded by Natural Science Foundation
of China (grant no. 81904044), Talents Training Program of Seventh
People's Hospital of Shanghai University of TCM (grant no.
XX2017-06), Shanghai Health Commission Youth Project (grant no.
20174Y0044), Key Disciplines Construction Project of the Municipal
Health Commission, Pudong New Area (grant no. PWZxk2017-06) and the
Budget Project of Shanghai University of Traditional Chinese
Medicine (grant no. 2019LK045).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SH, XM and DY performed the experiments. NZ and GW
acquired, analyzed and interpreted the data. MW and WX designed the
study and drafted the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Seventh People's Hospital (approval no. 2014-002),
and written informed consent was obtained from each patient.
Procedures performed on mice followed the guidelines of the Ethics
Committee of Institutional Animal Care and Use Committee of The
Seventh People's Hospital (approval no. 2019-078).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pan X, Chen Y, Shen Y and Tantai J:
Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in
A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis.
10:4292019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stankovic B, Bjørhovde HAK, Skarshaug R,
Aamodt H, Frafjord A, Müller E, Hammarström C, Beraki K, Bækkevold
ES, Woldbæk PR, et al: Immune cell composition in human non-small
cell lung cancer. Front Immunol. 9:31012018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rossi A and Di Maio M: Platinum-based
chemotherapy in advanced non-small-cell lung cancer: Optimal number
of treatment cycles. Expert Rev Anticancer Ther. 16:653–660. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li FL, Liu JP, Bao RX, Yan GQ, Feng X, Xu
YP, Sun YP, Yan W, Ling ZQ, Xiong Y, et al: Acetylation accumulates
PFKFB3 in cytoplasm to promote glycolysis and protects cells from
cisplatin-induced apoptosis. Nat Commun. 9:5082018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bose S and Le A: Glucose metabolism in
cancer. Adv Exp Med Biol. 1063:3–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim JH, Nam B, Choi YJ, Kim SY, Lee JE,
Sung KJ, Kim WS, Choi CM, Chang EJ, Koh JS, et al: Enhanced
glycolysis supports cell survival in EGFR-Mutant lung
adenocarcinoma by inhibiting autophagy-mediated EGFR degradation.
Cancer Res. 78:4482–4496. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang W, Bouchard G, Yu A, Shafiq M,
Jamali M, Shrager JB, Ayers K, Bakr S, Gentles AJ, Diehn M, et al:
GFPT2-expressing cancer-associated fibroblasts mediate metabolic
reprogramming in human lung adenocarcinoma. Cancer Res.
78:3445–3457. 2018.PubMed/NCBI
|
|
10
|
Higashi K, Yamagishi T, Ueda Y, Ishigaki
Y, Shimasaki M, Nakamura Y, Oguchi M, Takegami T, Sagawa M and
Tonami H: Correlation of HIF-1α/HIF-2α expression with FDG uptake
in lung adenocarcinoma. Ann Nucl Med. 30:708–715. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huber SM, Misovic M, Mayer C, Rodemann HP
and Dittmann K: EGFR-mediated stimulation of sodium/glucose
cotransport promotes survival of irradiated human A549 lung
adenocarcinoma cells. Radiother Oncol. 103:373–379. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ciribilli Y, Singh P, Inga A and Borlak J:
c-Myc targeted regulators of cell metabolism in a transgenic mouse
model of papillary lung adenocarcinoma. Oncotarget. 7:65514–65539.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong T, Cui L, Wang H, Wang H and Han N:
Knockdown of KLF5 suppresses hypoxia-induced resistance to
cisplatin in NSCLC cells by regulating HIF-1α-dependent glycolysis
through inactivation of the PI3K/Akt/mTOR pathway. J Transl Med.
16:1642018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu J, Lu F, Gong Y, Zhao C, Pan Q,
Ballantyne S, Zhao X, Tian S and Chen H: High expression of
synthesis of cytochrome c oxidase 2 and TP53-induced glycolysis and
apoptosis regulator can predict poor prognosis in human lung
adenocarcinoma. Hum Pathol. 77:54–62. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang L, Zhang Z and Yu Z: Identification
of a novel glycolysis-related gene signature for predicting
metastasis and survival in patients with lung adenocarcinoma. J
Transl Med. 17:4232019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tan SX, Hu RC, Xia Q, Tan YL, Liu JJ, Gan
GX and Wang LL: The methylation profiles of PRDM promoters in
non-small cell lung cancer. Onco Targets Ther. 11:2991–3002. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fog CK, Galli GG and Lund AH: PRDM
proteins: Important players in differentiation and disease.
Bioessays. 34:50–60. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hohenauer T and Moore AW: The prdm family:
Expanding roles in stem cells and development. Development.
139:2267–2282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Taniguchi H and Imai K: PRDM14, a zinc
finger protein, regulates cancer stemness. Methods Mol Biol.
1867:3–13. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bi HX, Shi HB, Zhang T and Cui G: PRDM14
promotes the migration of human non-small cell lung cancer through
extracellular matrix degradation in vitro. Chin Med J (Engl).
128:373–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Taniguchi H and Imai K: Silencing PRDM14
via oligonucleotide therapeutics suppresses tumorigenicity and
metastasis of breast cancer. Methods Mol Biol. 1974:233–243. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tuthill MH, Montinaro A, Zinngrebe J,
Prieske K, Draber P, Prieske S, Newsom-Davis T, von Karstedt S,
Graves J and Walczak H: TRAIL-R2-specific antibodies and
recombinant TRAIL can synergise to kill cancer cells. Oncogene.
34:2138–2144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiong S, Zheng Y, Jiang P, Liu R, Liu X
and Chu Y: MicroRNA-7 inhibits the growth of human non-small cell
lung cancer A549 cells through targeting BCL-2. Int J Biol Sci.
7:805–814. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sakurai Y, Ichinoe M, Yoshida K, Nakazato
Y, Saito S, Satoh M, Nakada N, Sanoyama I, Umezawa A, Numata Y, et
al: Inactivation of REV7 enhances chemosensitivity and overcomes
acquired chemoresistance in testicular germ cell tumors. Cancer
Lett. 489:100–110. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Suzuki S, Okada M, Takeda H, Kuramoto K,
Sanomachi T, Togashi K, Seino S, Yamamoto M, Yoshioka T and
Kitanaka C: Involvement of GLUT1-mediated glucose transport and
metabolism in gefitinib resistance of non-small-cell lung cancer
cells. Oncotarget. 9:32667–32679. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan T, Sun G, Sun X, Zhao L, Zhong R and
Peng Y: Tumor energy metabolism and potential of 3-bromopyruvate as
an inhibitor of aerobic glycolysis: Implications in tumor
treatment. Cancers (Basel). 11:3172019. View Article : Google Scholar
|
|
28
|
Zhang Q, Pan J, North PE, Yang S, Lubet
RA, Wang Y and You M: Aerosolized 3-bromopyruvate inhibits lung
tumorigenesis without causing liver toxicity. Cancer Prev Res
(Phila). 5:717–725. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Igarashi H, Taniguchi H, Nosho K, Ishigami
K, Koide H, Mitsuhashi K, Okita K, Takemasa I, Imai K and Nakase H:
PRDM14 promotes malignant phenotype and correlates with poor
prognosis in colorectal cancer. Clin Transl Oncol. 22:1126–1137.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moriya C, Taniguchi H, Miyata K, Nishiyama
N, Kataoka K and Imai K: Inhibition of PRDM14 expression in
pancreatic cancer suppresses cancer stem-like properties and liver
metastasis in mice. Carcinogenesis. 38:638–648. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang T, Ye X, Yan D, Deng C, Li Z and
Tian B: FAM46B promotes apoptosis and inhibits glycolysis of
prostate cancer through inhibition of the MYC-LDHA axis. Onco
Targets Ther. 13:8771–8782. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heydarzadeh S, Moshtaghie AA, Daneshpoor M
and Hedayati M: Regulators of glucose uptake in thyroid cancer cell
lines. Cell Commun Signal. 18:832020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chakraborty PK, Mustafi SB, Xiong X,
Dwivedi SKD, Nesin V, Saha S, Zhang M, Dhanasekaran D, Jayaraman M,
Mannel R, et al: MICU1 drives glycolysis and chemoresistance in
ovarian cancer. Nat Commun. 8:146342017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ganapathy-Kanniappan S and Geschwind JF:
Tumor glycolysis as a target for cancer therapy: Progress and
prospects. Mol Cancer. 12:1522013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kikuchi R, Iwai Y, Tsuji T, Watanabe Y,
Koyama N, Yamaguchi K, Nakamura H and Aoshiba K: Hypercapnic tumor
microenvironment confers chemoresistance to lung cancer cells by
reprogramming mitochondrial metabolism in vitro. Free Radic Biol
Med. 134:200–214. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu Y, Wan Z, Zhang X, Zhong X, Rui L and
Li Z: PRDM14 inhibits 293T cell proliferation by influencing the
G1/S phase transition. Gene. 595:180–186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Snellenberg S, Cillessen SA, Van Criekinge
W, Bosch L, Meijer CJLM, Snijders PJF and Steenbergen RDM:
Methylation-mediated repression of PRDM14 contributes to apoptosis
evasion in HPV-positive cancers. Carcinogenesis. 35:2611–2618.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moriya C, Taniguchi H, Nagatoishi S,
Igarashi H, Tsumoto K and Imai K: PRDM14 directly interacts with
heat shock proteins HSP90α and glucose-regulated protein 78. Cancer
Sci. 109:373–383. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Feng Y, Hu J, Xie D, Qin J, Zhong Y, Li X,
Xiao W, Wu J, Tao D, Zhang M, et al: Subcellular localization of
caspase-3 activation correlates with changes in apoptotic
morphology in MOLT-4 leukemia cells exposed to X-ray irradiation.
Int J Oncol. 27:699–704. 2005.PubMed/NCBI
|