Introduction
Colorectal cancer (CRC) is a common type of
malignant tumor and a frequent cause of cancer-related death
worldwide (1). Notably, CRC is the
fourth most frequent gastroenteric tumor and is associated with
high morbidity in China (2).
Although encouraging progress for early diagnosis and treatment of
CRC has been achieved in recent decades, the prognosis and 5-year
survival rate of patients with CRC is still unsatisfactory
(3). The tumorigenesis and
progression of CRC is a complex pathological process involving
numerous molecules and pathways, and the underlying mechanisms have
yet to be fully elucidated. Therefore, further insight into the
molecular mechanisms of CRC may help improve the diagnosis and
prognosis, and consequently identify novel therapeutic targets for
CRC.
Noncoding RNAs (ncRNAs) have been reported to serve
important biological roles in transcriptional regulation, RNA
modification, formation of chromosomes and cellular development
(4). Notably, ncRNAs are further
grouped into small ncRNAs (<200 bp) and long ncRNAs (lncRNAs;
>200 bp, up to 100 kb) based on transcript size, and lncRNAs
expression levels are indicative of those of active molecules and
lack significant open reading frames (5,6).
Previous studies have demonstrated that lncRNAs may serve important
roles in several biological processes, including cell
proliferation, differentiation and apoptosis (7,8). In
addition, it has been reported that dysregulation of lncRNAs may be
involved in the development of human cancer (9–15). The
lncRNA RPLP0P2 has been reported to exhibit significantly decreased
expression in lung adenocarcinoma (16). Chen et al (17) also observed that RPLP0P2 expression
was lower in lung adenocarcinoma, and low expression of RPLP0P2 was
associated with poor prognosis, as well as decreased proliferation
and adhesive ability, in tumor cells. Conversely, Li et al
(18) revealed that RPLP0P2 was
negatively associated with overall survival in gastric cancer,
suggesting that it may act as an oncogene in gastric cancer. These
studies indicated that the role of RPLP0P2 in cancer is related to
its tissue specificity.
Zhong et al (19) performed bioinformatics analysis on
RPLP0P2 in CRC and revealed that RPLP0P2 may represent an important
oncogene, which was negatively associated with overall survival.
This finding suggests that RPLP0P2 may have an important role in
CRC; however, the specific biological role and mechanism of RPLP0P2
remain unclear. The present study demonstrated that the lncRNA
RPLP0P2 may act as a candidate CRC biomarker according to lncRNA
gene expression data from The Cancer Genome Atlas (TCGA)
RNA-sequencing (RNA-Seq) datasets. These findings demonstrated that
the lncRNA RPLP0P2 may have an important role in CRC; therefore,
the present study aimed to further determine the biological role
and clinical significance of RPLP0P2 in CRC and to investigate the
underlying molecular mechanisms.
Materials and methods
Datasets and TCGA bioinformatics
TCGA data portal (https://tcga-data.nci.nih.gov/docs/publications/tcga/)
was used to locate microarray studies focusing on expression of
RPLP0P2 in CRC, and 50 matched cases of CRC (27 females and 23
males; age, 40–90 years; mean age 70.34±13.36 years) and paired
adjacent normal tissue, along with 647 cases of CRC (294 females
and 353 males; age, 31–90 years; mean age, 66.17±12.78 years) and
51 unpaired cases of normal tissue were collected, which were used
for RPLP0P2 expression analysis. The criterion for dividing the
expression of RPLP0P2 into high and low was the median value of
gene expression. Furthermore, 308 CRC cases (138 females and 170
males; age, 31–90 years; mean age, 67.29±12.79 years old) were
collected for survival analysis.
Cell culture
The human CRC cell lines (HCT116, HT29, SW480 and
RKO) were purchased from The Cell Bank of Type Culture Collection
of The Chinese Academy of Sciences. Cells were cultured in
RPMI-1640 medium supplemented with 10% fetal bovine serum
(Biological Industries) and were maintained at 37°C in an
atmosphere containing 5% CO2. Digestion and passage of
cells were performed until they reached 80% confluence.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from CRC cells using Total
RNA Isolation Reagent (cat. no. 3101-100; Shanghai Pufei Biotech
Co., Ltd.) in accordance with the manufacturer's instructions. The
M-MLV (Moloney Murine Leukemia Virus Reverse Transcriptase) kit
(cat. no. M1705; Promega Corporation) was used to reverse
transcribe total RNA according to the manufacturer's protocol. The
expression levels of RPLP0P2 and GAPDH were assessed in CRC cells
by qPCR using SYBR Master Mixture (cat. no. DRR041B; Takara Bio,
Inc.). The thermocycling conditions were as follows: 95°C for 30
sec, 95°C for 5 sec and 60°C for 30 sec, 40 cycles. Primer
sequences were as follows: RPLP0P2, forward
5′-TCAAGACGGGAGACAAAGTGG−3′ and reverse 5′-TCGATGATAGAATGGGGCACT−3′
(247 bp product); GAPDH, forward 5′-TGACTTCAACAGCGACACCCA−3′ and
reverse 5′-CACCCTGTTGCTGTAGCCAAA−3′ (121 bp product). All samples
were normalized to GAPDH, and all experiments were performed in
triplicate.
The mean value of each triplicate was used to
calculate relative lncRNA expression levels. 2−ΔΔCq
method was used to detect RNA level (20).
Lentivirus-mediated RPLP0P2 short
hairpin RNA (shRNA) vector infection
A shRNA lentivirus targeting RPLP0P2 was constructed
using the GV248 (hU6-MCS-Ubiquitin-EGFP-IRES-puromycin) vector
(Shanghai GeneChem Co., Ltd.). The shRNAs were synthesized and
inserted into GV248 lentivirus vector, and then verified by DNA
sequencing (Sanger). The shRNA sequences were as follows: shRNA-1,
forward
5′-CCGGCCCTGTAAAGTACCTGGAATACTCGAGTATTCCAGGTACTTTACAGGGTTTTTG-3′
and reverse
5′-AATTCAAAAACCCTGTAAAGTACCTGGAATACTCGAGTATTCCAGGTACTTTACAGGG-3′;
shRNA-2, forward
5′-CCGGCTGATCAAGACGGGAGACAAACTCGAGTTTGTCTCCCGTCTTGATCAGTTTTTG-3′
and reverse
5′-AATTCAAAAACTGATCAAGACGGGAGACAAACTCGAGTTTGTCTCCCGTCTTGATCAG-3′;
shRNA-3, forward
5′-CCGGGTCAGACGAGGATATGGGATTCTCGAGAATCCCATATCCTCGTCTGACTTTTTG-3′
and reverse
5′-AATTCAAAAAGTCAGACGAGGATATGGGATTCTCGAGAATCCCATATCCTCGTCTGAC−3′.
Lower case indicates protective bases; upper case indicates the
primer sequence.
As a negative control, a shRNA with a scrambled
sequence (TTCTCCGAACGTGTCACGT) was used. Subsequently, the
lentivirus-mediated RPLP0P2 shRNA vector was confirmed via
sequencing and used for subsequent experiments.
For infection, 1×105 RKO cells/per well
were seeded into 6-well plates and were incubated for 24 h at 37°C,
after which, medium containing the lentivirus (multiplicity of
infection, 10) was added, according to the manufacturer's
instructions. RKO cells were infected with the lentivirus for 72 h
at 37°C and infection efficiency was assessed. The approximate
fluorescence infection efficiency was estimated by comparing
bright-field and fluorescent images in the same field.
Subsequently, RT-qPCR was performed to detect knockdown
efficiency.
Detection of cell proliferation by
Cell Counting Kit-8 (CCK-8) and colony formation assay
RKO cells were infected with the RPLP0P2 shRNA
lentivirus for 3 days, and were then trypsinized and resuspended in
medium. For the CCK-8 assay, 2×104 cells/well were
seeded into 96-well plates. After 24 h, 10 µl CCK-8 solution
(Beyotime Institute of Biotechnology) was added to each well at 0,
1, 2 and 3 days, and the cells were incubated at 37°C for 2 h. The
absorbance at each time point was measured at 450 nm using a
microplate reader.
For the colony formation assay, 2 ml medium,
including RKO cells (500 cells), was added into each well of 6-well
culture plates, and three replicates for each group were performed.
Cells were further cultured at 37°C in an atmosphere containing 5%
CO2 for 8 days; the medium was changed every 3 days
while cell state was observed. After 8 days, a fluorescence
microscope was used to image the cell colonies. Briefly, 1 ml 4%
paraformaldehyde was used to fix the cells for 30–60 min at room
temperature and 1 ml crystal violet staining solution was added to
stain the cells for 10–20 min at room temperature. The cells were
then washed several times and air dried, and the number of colonies
containing >50 cells was counted.
Detection of cell migration by Celigo
imaging cytometry
Cells were seeded into 96-well plates at a
concentration of 50,000 cells/well (100 µl/well) and grown to 90%
confluence. Each group had three replicates and the cells were
cultured at 37°C in an atmosphere containing 5% CO2 for
24 h. Subsequently, the Celigo scratch instrument was used to
gently push upward to form scratches in the cell layer. Celigo
scanning (cat. no. VP408FH; V&P Scientific, Inc.) was performed
at 24 and 48 h according to the degree of healing, and the
migration rate was calculated. Celigo was used to to obtain the
cell area (S), migration area was calculated by
SXh-S0h (X=24 or 48), and migration rate was
calculated by (SXh-S0h)/area of scratch.
Cell migration and invasion
assays
A 24-well plate containing pore inserts (size, 8 µm)
with or without Matrigel-coated filters (Corning, Inc.) was used
for migration and invasion assay. In the upper chamber, 500 µl cell
suspension in serum-free medium (105 cells/well) was
added, and 750 µl medium containing 10% FBS was added into the
lower chamber; the cells were cultured at 37°C in an atmosphere
containing 5% CO2 for 72 h. Subsequently, the migratory
or invaded cells were stained with Giemsa for 3–5 min and air
dried. Migratory or invasive cells per field were imaged under a
light microscope and invasion rate was calculated. The number of
cells (N) in 5 fields was counted under the microscope, and the
control group was used as reference. The invasion rate was
calculated as Nexperimental group/Ncontrol
group.
Cell cycle assay
Cells were cultured in 6-cm dishes to ~80%
confluence and were digested and resuspended. After centrifugation
(400 × g, 10 min at 4°C), cells were washed with D-Hanks (pH,
7.2–7.4) and fixed in 75% ethanol for 1 h at 4°C. Cells were
stained with cell cycle dye, which was prepared using 40X propidium
iodide (2 mg/ml; cat. no. P4170; Sigma-Aldrich; Merck KGaA), 100X
RNase (10 mg/ml; cat. no. EN0531; Fermentas; Thermo Fisher
Scientific, Inc.) and 1X D-Hanks at a ratio of 25:10:1,000. After
incubation for 30 min at room temperature in the dark, samples were
analyzed with a flow cytometer (Guava easyCyte HT; EMD Millipore)
to detect DNA content. Guava InCyte software (3.1.1.0; EMD
Millipore) was used for analysis.
Apoptosis assay
Cells (5×105) were trypsinized, collected
and centrifuged at 400 × g for 5 min at 4°C. Subsequently,
pre-cooled D-Hanks (pH, 7.2–7.4) was used to wash the cells, and
they were resuspended in 200 µl 1X binding buffer (eBioscience;
Thermo Fisher Scientific, Inc.) and stained with 10 µl Annexin
V-APC (eBioscience; Thermo Fisher Scientific, Inc.) in the dark at
room temperature for 10–15 min. According to the number of cells,
400–800 µl binding buffer was added and samples were assessed using
a flow cytometry analyzer (Guava easyCyte HT; EMD Millipore). Guava
InCyte software (3.1.1.0; Millipore) was used for analysis.
Statistical analysis
Data are presented as the mean ± SD (n=3). All
results were analyzed using SPSS 16.0 (SPSS, Inc.). The expression
levels of RPLP0P2 in normal and cancer tissues were compared using
the Mann-Whitney U test and Wilcoxon signed-rank test. Survival
analysis with regard to RPLP0P2 expression levels in patients with
CRC was estimated by the Kaplan-Meier method accompanied by the
log-rank test to calculate differences between the curves.
Comparisons between experimental and control groups in colony
formation, wound-healing assay, Transwell assay, cell cycle
progression analysis and apoptosis analysis were performed by
paired-samples t-test. Differences in the effect of RPLP0P2 shRNA
on knockdown were compared using one-way ANOVA and Dunnett's test
was used for intra-group comparison. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression levels of RPLP0P2 are high
in CRC and are associated with prognosis
The present study analyzed the expression levels of
RPLP0P2 from lncRNA gene expression data in TCGA RNA-Seq datasets
and revealed that the expression levels of RPLP0P2 were higher in
CRC tissues compared with those in normal tissues (Fig. 1A). Paired samples of CRC also
revealed that RPLP0P2 expression levels were higher in CRC tissue
compared with paracancerous normal tissue (Fig. 1B). Further analysis revealed that
high levels of RPLP0P2 were associated with poor survival in
patients with CRC (Fig. 1C).
Infection efficiency of the RPLP0P2
shRNA lentivirus in RKO cells
In order to verify expression levels of RPLP0P2 in
CRC, levels of RPLP0P2 in CRC cell lines were assessed by RT-qPCR;
results showed that RPLP0P2 levels were higher in RKO cells
compared with other CRC cells (HCT116, HT29 and SW480; Fig. 1D). Subsequently, RKO cells were used
to assess the biological role of RPLP0P2 using a shRNA
lentivirus.
RKO CRC cells were infected with the RPLP0P2 shRNA
lentivirus for 72 h and then observed using a fluorescence
microscope. The results revealed that the cells appeared normal
after infection, and the infection efficiency of the shRNA
lentivirus was ~80% (Fig. 2A).
RT-qPCR also revealed that the expression levels of RPLP0P2 were
significantly downregulated by the RPLP0P2 shRNA lentivirus in all
groups, particularly in the shRNA-3 group (Fig. 2B). Therefore, RPLP0P2 shRNA-3 was
used in subsequent experiments. CCK-8 cell proliferation assay
revealed that shRNA-RPLP0P2 knockdown significantly decreased
proliferation in RKO cells (Fig.
2C).
RPLP0P2 shRNA lentivirus suppresses
colony formation in RKO cells
Following infection with the RPLP0P2 shRNA
lentivirus, the colony-forming ability of RKO cells was assessed.
The results revealed that RPLP0P2 knockdown significantly decreased
the number of cell colonies, whereas the control group exhibited a
normal ability to pack together and form colonies (Fig. 3), suggesting that RPLP0P2 may be
associated with the clonogenic capacity of RKO cells.
RPLP0P2 shRNA lentivirus suppresses
migration and invasion of RKO cells
The effects of RPLP0P2 on RKO cell migration and
invasion were assessed by wound-healing and Transwell assays. The
results of the wound-healing assay revealed that the wound area of
RPLP0P2 shRNA-infected cells was significantly larger compared with
that in the control group (Fig.
4A), indicating that RPLP0P2 shRNA inhibited cell migration.
The results of the Transwell assay demonstrated that the number of
migrated cells and invaded cells in the RPLP0P2 shRNA-infected
group was significantly decreased compared with in the control
group (Fig. 4B).
RPLP0P2 shRNA arrests RKO cells at S
stage
The present study also assessed changes in cell
cycle progression in response to RPLP0P2 knockdown with flow
cytometry. The results detected increased accumulation of RKO cells
in S phase in the RPLP0P2 knockdown group compared with in the
control group, which was evidenced by a significant increase in the
number of S phase cells (Fig. 5).
Consistently, this result was accompanied by a significant
reduction in the percentage of cells in G1 phase.
Meanwhile, there was no significant difference in G2/M
cell populations between the RPLP0P2 knockdown and control groups.
These results revealed that shRNA-mediated RPLP0P2 knockdown
arrested RKO cells from S stage to G2/M stage.
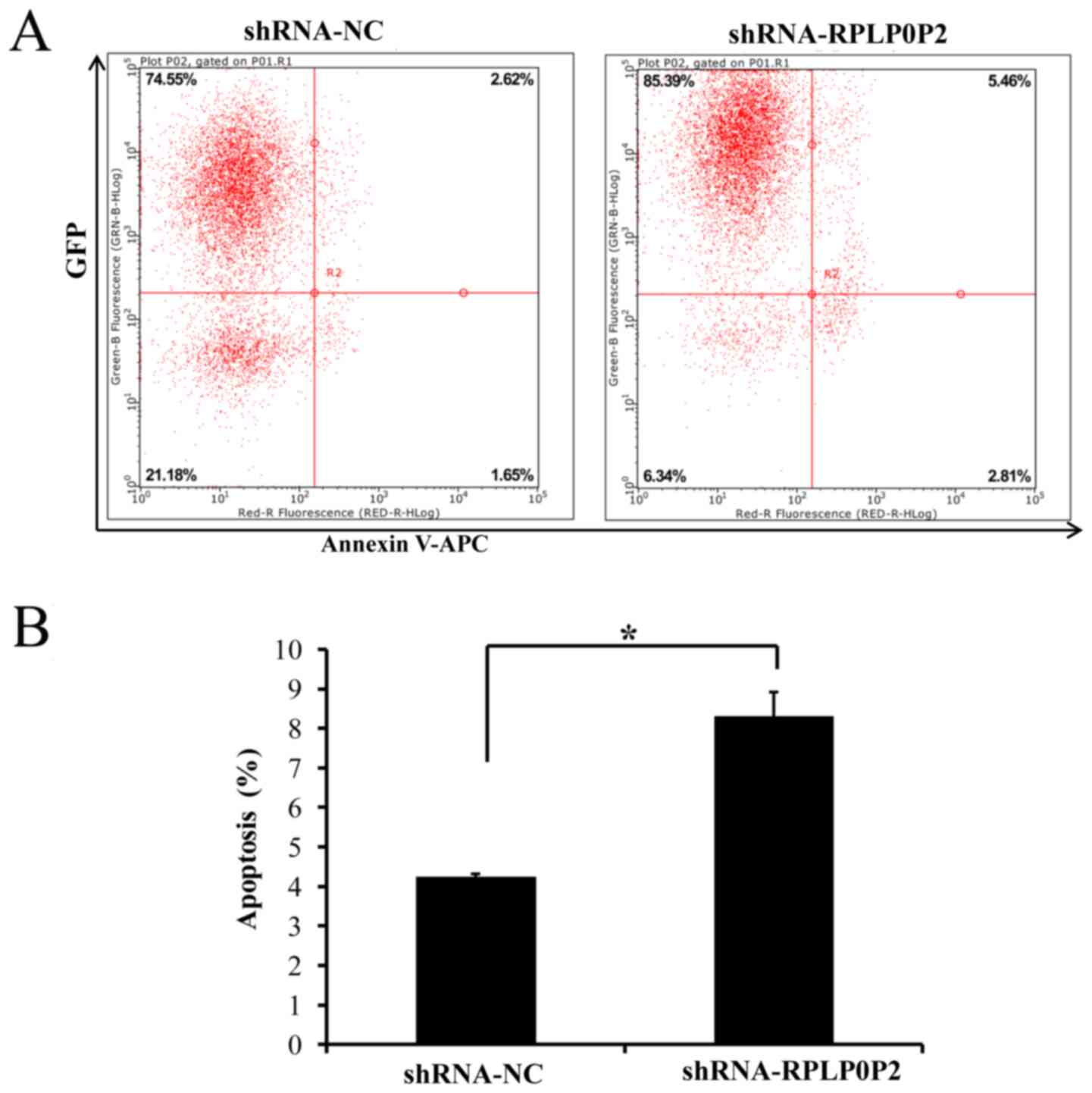
Infection with the RPLP0P2 shRNA
lentivirus promotes RKO cell apoptosis
Flow cytometry was also performed to assess
apoptosis of RKO cells infected with the RPLP0P2 shRNA lentivirus.
The results revealed that apoptosis was significantly increased in
RKO cells infected with the RPLP0P2 shRNA lentivirus compared with
in the control group (Fig. 6),
suggesting that RPLP0P2 inhibits the apoptosis of RKO cells.
Discussion
With their powerful proliferative ability and low
apoptotic rate, CRC cells are strongly resistant to chemotherapy,
radiation and other biological treatments, leading to malignant
tumor growth and recurrence after resection (21). Previous studies have revealed that
lncRNAs are increasingly known to be involved in numerous
biological processes, including acceleration of the development and
progression of cancer (22,23). It has also been suggested that
lncRNA RPLP0P2 levels may be significantly reduced in lung
adenocarcinoma (16).
Bioinformatics analysis of RPLP0P2 in CRC revealed that RPLP0P2 was
negatively associated with overall survival (19), suggesting that RPLP0P2 may serve a
key role in CRC.
The present study performed bioinformatics analysis
using publicly available TCGA data. The results revealed that the
expression levels of RPLP0P2 were higher in CRC tissues compared
with in normal tissues. Further analysis verified that the
expression levels of RPLP0P2 were also high in RKO cells. These
results indicated that RPLP0P2 may be abnormally expressed in CRC
and may act as an oncogene. RPLP0P2 was also previously reported to
be negatively associated with overall survival in gastric cancer
(18). However, Xu et al
(16) reported that RPLP0P2
expression was significantly lower in lung adenocarcinoma compared
with in adjacent tissues. These studies indicated that tissue
specificity may be one of the most important properties for
RPLP0P2.
In the present study, RPLP0P2 knockdown using a
shRNA lentivirus suppressed proliferation and colony formation in
RKO cells, thus suggesting that RPLP0P2 may be associated with the
proliferative capacity of CRC. shRNA-mediated RPLP0P2 knockdown
also induced cell cycle arrest of RKO cells in S stage, which
confirmed that the RPLP0P2 expression levels were associated with
cell proliferation. Chen et al (17) also reported that low expression of
lncRNA RPLP0P2 decreased cell proliferation and adhesive ability in
lung adenocarcinoma. This finding is in agreement with the present
experimental results, which revealed that RPLP0P2 knockdown induced
cell accumulation in S phase of the cell cycle, suggesting that
involvement of RPLP0P2 in CRC may be related to cells arrested from
S phase to G2/M phase.
The present study demonstrated that RPLP0P2 was
associated with the migratory and invasive ability of CRC cells.
Furthermore, flow cytometric analysis revealed that RPLP0P2 may
inhibit apoptosis of CRC cells, suggesting that RPLP0P2 may promote
CRC development via inhibition of apoptosis and metastasis.
However, whether and how RPLP0P2 is implicated in CRC requires
further study. It has been reported that lncRNA-associated
competing endogenous RNA networks, for example, lncRNA-microRNA
(miRNA/miR)-mRNA networks, may participate in the progression of
CRC (24). Expression of EZH2 has
been reported to be associated with the risk of lung cancer
(25), and a co-methylation network
revealed that RPLP0P2 was co-methylated with EZH2, suggesting
involvement of RPLP0P2 in the progression of lung cancer (26). Further analysis revealed that the
lncRNA-miRNA-mRNA network miR-29c-3p-RPLP0P2-EZH2 was significantly
associated with the prognosis of lung adenocarcinoma (26). These reports may provide further
understanding of the role of RPLP0P2 in CRC pathogenesis and
prognosis.
In conclusion, the present study revealed that the
expression levels of RPLP0P2 were associated with the development
of CRC through promotion of proliferation and metastasis, and
inhibition of apoptosis. These findings suggested that RPLP0P2 may
function as an oncogene in CRC, as well as a therapeutic target in
CRC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HY, ST and YS conceptualized and designed the study.
HY and YM analyzed the data and wrote the manuscript. YM performed
the experiments. YS and HY confirmed the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
McGuire S: World cancer report 2014.
Geneva, Switzerland: World health organization, international
agency for research on cancer, WHO Press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gu MJ, Huang QC, Bao CZ, Li YJ, Li XQ, Ye
D, Ye ZH, Chen K and Wang JB: Attributable causes of colorectal
cancer in China. BMC Cancer. 18:382018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sana J, Faltejskova P, Svoboda M and Slaby
O: Novel classes of non-coding RNAs and cancer. J Transl Med.
10:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gong Z, Zhang S, Zhang W, Huang H, Li Q,
Deng H, Ma J, Zhou M, Xiang J, Wu M, et al: Long non-coding RNAs in
cancer. Sci China Life Sci. 55:1120–1124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang JP, Tang YY, Fan CM, Guo C, Zhou YH,
Li Z, Li XL, Li Y, Li GY, Xiong W, et al: The role of exosomal
non-coding RNAs in cancer metastasis. Oncotarget. 9:12487–12502.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gong Z, Zhang S, Zeng Z, Wu H, Yang Q,
Xiong F, Shi L, Yang J, Zhang W, Zhou Y, et al: LOC401317, a
p53-regulated long non-coding RNA, inhibits cell proliferation and
induces apoptosis in the nasopharyngeal carcinoma cell line HNE2.
PLoS One. 9:e1106742014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang L, Tang Y, Xiong F, He Y, Wei F,
Zhang S, Guo C, Xiang B, Zhou M, Xie N, et al: LncRNAs regulate
cancer metastasis via binding to functional proteins. Oncotarget.
9:1426–1443. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fan C, Tang Y, Wang J, Xiong F, Guo C,
Wang Y, Zhang S, Gong Z, Wei F, Yang L, et al: Role of long
non-coding RNAs in glucose metabolism in cancer. Mol Cancer.
16:1302017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gong Z, Yang Q, Zeng Z, Zhang W, Li X, Zu
X, Deng H, Chen P, Liao Q, Xiang B, et al: An integrative
transcriptomic analysis reveals p53 regulated miRNA, mRNA, and
lncRNA networks in nasopharyngeal carcinoma. Tumour Biol.
37:3683–3695. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He Y, Jing Y, Wei F, Tang Y, Yang L, Luo
J, Yang P, Ni Q, Pang J, Liao Q, et al: Long non-coding RNA PVT1
predicts poor prognosis and induces radioresistance by regulating
DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell
Death Dis. 9:2352018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang
B, Zhou M, Li X, Xiong W, Li G, et al: The long noncoding RNA
MALAT-1 is a novel biomarker in various cancers: A meta-analysis
based on the GEO database and literature. J Cancer. 7:991–1001.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang W, Huang C, Gong Z, Zhao Y, Tang K,
Li X, Fan S, Shi L, Li X, Zhang P, et al: Expression of LINC00312,
a long intergenic non-coding RNA, is negatively correlated with
tumor size but positively correlated with lymph node metastasis in
nasopharyngeal carcinoma. J Mol Histol. 44:545–554. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y,
Fan C, Li X, Li G, Li Y, et al: Circular RNAs (circRNAs) in cancer.
Cancer Lett. 425:134–142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang
W, Chen J, Yu F, Zhou T and Wang Y: Long noncoding RNA expression
profiles of lung adenocarcinoma ascertained by microarray analysis.
PLoS One. 9:e1040442014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen J, Hu L, Chen J, Wu F, Hu D, Xu G,
Zhu P and Wang Y: Low expression lncRNA RPLP0P2 is associated with
poor prognosis and decreased cell proliferation and adhesion
ability in lung adenocarcinoma. Oncol Rep. 36:1665–1671. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li CY, Liang GY, Yao WZ, Sui J, Shen X,
Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, et al: Integrated
analysis of long non-coding RNA competing interactions reveals the
potential role in progression of human gastric cancer. Int J Oncol.
48:1965–1976. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhong ME, Chen Y, Zhang G, Xu L, Ge W and
Wu B: LncRNA H19 regulates PI3K-Akt signal pathway by functioning
as a ceRNA and predicts poor prognosis in colorectal cancer:
Integrative analysis of dysregulated ncRNA-associated ceRNA
network. Cancer Cell Int. 19:1482019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Selzner M, Bielawska A, Morse MA, Rüdiger
HA, Sindram D, Hannun YA and Clavien PA: Induction of apoptotic
cell death and prevention of tumor growth by ceramide analogues in
metastatic human colon cancer. Cancer Res. 61:1233–1240.
2001.PubMed/NCBI
|
|
22
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Khachane AN and Harrison PM: Mining
mammalian transcript data for functional long non-coding RNAs. PLoS
One. 5:e103162010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jalali S, Bhartiya D, Lalwani MK,
Sivasubbu S and Scaria V: Systematic transcriptome wide analysis of
lncRNA-miRNA interactions. PLoS One. 8:e538232013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoon KA, Gil HJ, Han J, Park J and Lee JS:
Genetic polymorphisms in the polycomb group gene EZH2 and the risk
of lung cancer. J Thorac Oncol. 5:10–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wei Y, Chang Z, Wu C, Zhu Y, Li K and Xu
Y: Identification of potential cancer-related pseudogenes in lung
adenocarcinoma based on ceRNA hypothesis. Oncotarget.
8:59036–59047. 2017. View Article : Google Scholar : PubMed/NCBI
|