Introduction
Gliomas are the commonest type of primary brain
tumors with high morbidity, mortality and recurrence in addition to
being one of the challenging disorders to treat among all central
nervous system diseases (1).
According to histopathological features, the World Health
Organization classifies gliomas into four grades. The latest
classification stated that low-grade gliomas (LGG) can be
completely cured if effective treatment is administered (2). Glioblastoma multiforme (GBM), the most
malignant glioma, is difficult to be cured using surgery or
chemotherapy (2,3). Patients with glioma also face the
problem of chemotherapy drug resistance clinically. Due to high
drug resistance, several chemotherapeutic drugs traditionally used
for treating glioma, including temozolomide, are often not
effective (4). Therefore,
development of more efficient drugs with novel mechanisms of action
is required.
Schmidt and Lademann (5) first isolated corilagin in 1951;
Kakiuchi et al (6)
discovered in 1985 that corilagin exhibits reverse transcriptase
activity against RNA tumor viruses. Over the past decades, a number
of studies have reported various pharmacological activities of
corilagin, including anti-tumor activity (7), anti-oxidation activity (8), liver and lung protection (9,10) and
anti-inflammatory activity (11).
Other studies have noted that corilagin can inhibit various tumors
such as gastric (12), liver
(13), breast (14) and ovarian cancers (15). Corilagin not only directly inhibits
tumor cell proliferation, but also indirectly exerts anti-tumor
effects by enhancing the effect of traditional chemotherapy drugs
(16). A recent study reported that
corilagin can induce tumor cell apoptosis through mitochondrial
damage (14). According to the
potential application value of corilagin for the treatment of
glioma, the present study identified that corilagin could induce
apoptosis of glioma cells.
Autophagy, also known as type II cell death, is
regulated by autophagy-related genes (17). It is the cellular process in which
damaged organelles and macromolecules are transported to lysosomes
for degradation to maintain the stability of the intracellular
environment (18). Autophagy
usually occurs at a low level but increases rapidly when adenosine
triphosphate energy is depleted, reactive oxygen species (ROS) are
released and mitochondrial membrane channels are opened (19). Microtubule-associated protein light
chain 3 (LC3) is recognized as a universal marker for autophagosome
detection. It has two molecular forms, LC3I and LC3II. LC3II is a
crucial molecule in the formation of autophagosomes and is widely
used to detect the activation of autophagy. SQSTM1/p62 is a
connexin that guides the degradation of ubiquitinated proteins into
autophagosomes, which can be degraded along with autophagic flow;
p62 accumulates when autophagic flow is disordered (20). The present study observed that the
rate of conversion of LC3I into LC3II increases and the amount of
p62 protein decreases following corilagin activation in U251 glioma
cells, suggesting that corilagin induces autophagy.
Nuclear factor erythroid 2 like 2 (NFE2L2 or NRF2)
is a protective factor against cellular stress response (21). Studies (21–23)
have demonstrated that NRF2 activates various gene transcriptional
factors to protect cells against oxidative stress and endogenous
and exogenous stimulation; therefore, NRF2 is considered a
protective transcription factor for cells (24). Evidence has indicated that NRF2
inhibits cell apoptosis (25,26).
In addition, NRF2 is demonstrated to be associated with metastasis
and resistance mechanisms of cholangiocarcinoma (27). As a result of these advantages,
cancer cells with sustained NRF2 activation often develop ‘NRF2
addiction’ and display a malignant phenotype, leading to a poor
prognosis among patients with cancer (28). Previous studies have demonstrated
that U251 cells exhibit aberrant activation of NRF2 and knockdown
of NRF2 inhibits the proliferation and growth of U251 cells in a
mouse xenograft model (28,29); therefore the present study termed
U251 cells NRF2-addicted. Inhibition of NRF2 is a promising
therapeutic strategy for NRF2-addicted cancers and the development
of NRF2 inhibitors is urgent (28).
Under normal conditions, Kelch-like epichlorohydrin-related protein
1 (Keap1) binds to NRF2 protein and the complex is degraded rapidly
by the 26S proteasome, with the NRF2 protein escaping capture by
the Keap1 accumulated in the nucleus (30). This process can be regulated by the
autophagy activity in cells. As a substrate for autophagy,
SQSTM1/p62 binds to Keap1 and causes NRF2 and Keap1 to separate
(30). Subsequently, NRF2 enters
the nucleus, activates the transcription of various antioxidant
genes and inhibits the expression of oxidative-stress-related
proteins (30). The present study
preliminarily confirmed that NRF2 is highly expressed in glioma
cancer. In addition, corilagin stimulated the U251 cell line, which
increased autophagy activity, enhanced p62 degradation and reduced
NRF2 protein expression; The present study concluded that the
promotion of autophagy by corilagin induced the downregulation of
NRF2 expression.
Materials and methods
Reagents and antibodies
Corilagin (98% purity) was purchased from Shanghai
Yuanye Bio-Technology Co., Ltd., dissolved with phosphate buffer
saline (Beijing Solarbio Science & Technology Co., Ltd.) to
prepare stock solution (4 mg/ml) and stored at 4°C. Dulbecco's
modified Eagle's medium high glucose (DMEM) medium, RPMI-1640
Medium and Earle's Balanced Salt Solution (EBSS) were obtained from
HyClone (Cytiva). Fetal bovine serum (FBS) were purchased from
HyClone (Cytiva), penicillin and streptomycin were purchased from
Beijing Solarbio Science & Technology Co., Ltd. The trypsin
solution was obtained from Gibco (Thermo Fisher Scientific, Inc.).
RIPA Lysis Buffer, phenylmethylsulfonyl fluoride, BCA protein assay
kit and 5X loading buffer were purchased from Beyotime Institute of
Biotechnology. β-actin (cat. no. AC026, monoclonal, rabbit
anti-human) antibody was obtained from ABclonal Biotech Co., Ltd.
NRF2 (cat. no. SC-365949, monoclonal, mouse anti-human) antibody
was obtained from Santa Cruz Biotechnology, Inc. Bcl-2 (cat. no.
4223, monoclonal, rabbit anti-human) and LC3 (cat. no. 12741,
monoclonal, rabbit anti-human) antibody were purchased from Cell
Signaling Technology, Inc. P62 (cat. no. BA2849, polyclonal, rabbit
anti-human) antibody was obtained from Wuhan Boster Biological
Technology, Ltd. ECL chemiluminescence solution and 0.2 µm
thickness PVDF membrane were purchased from EMD Millipore. The
primers of NRF2 and Bcl-2 were obtained from Beijing Liuhe BGI.
Horseradish peroxidase-labeled secondary antibody (anti-rabbit IgG:
cat. no. AS014; anti-mouse IgG: AS003) were purchased from ABclonal
Biotech Co., Ltd. Coralite488-labeled secondary antibody (cat. no.
SA00013-2) was purchased from ProteinTech Group, Inc.
Collection of patient samples
The present study was approved by the Ethics
Committee of Affiliated Hospital of Jining Medical University
(Jining, China; approval no. 2019C008). Each participant (or their
legal representative) provided written informed consent. The 15
glioma tissues were obtained from patients with glioma following
surgical resection at Affiliated Hospital of Jining Medical
University. The inclusion criteria were patients diagnosed with
glioma and treated with surgical resection. In addition, none of
the patients received chemotherapy prior to surgery. The exclusion
criteria were patients with unconfirmed pathology and/or patients
with incomplete data records. The nine non-glioma specimens were
obtained from patients with cerebral hemorrhage (CH) and Traumatic
Brain Injury (TBI) following surgical resection at Affiliated
Hospital of Jining Medical University. All tissues were promptly
snap-frozen in liquid nitrogen following surgical resection and
stored at −80°C refrigerator until further use.
Cell culture
The U251 cell line was obtained from the Shandong
Provincial Key Laboratory of Stem Cells and Neuro-oncology. U251
cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml
penicillin and 100 µg/ml streptomycin. The U251 cells were cultured
in a humidified atmosphere at 37°C under 5% CO2.
According to the Cellosaurus reference website (https://web.expasy.org/cellosaurus/CVCL_2219), the
U251 cell line used was U251 MG and was authenticated using short
tandem repeat analysis. U251 cells were seeded at 1×105
per well into a 6-well plate and stimulated by corilagin at
different concentrations of 0, 25, 50, 100 µg/ml in a 37°C
incubator for 48 h beginning at the second day. EBSS was added to
control group cells to make positive control group in 37°C
incubator for 2 h. Cells were collected for reverse
transcription-quantitative (RT-q) PCR, western blot analysis,
immunofluorescence and Hoechst 33258 staining.
RNA extraction and RT-qPCR
Cells were harvested for RNA extraction when at
80–90% confluence. The total RNA was isolated from U251 cells with
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. Total RNA (1.5 µg) was
prepared for reverse transcription using a FastQuant RT kit
(Tiangen Biotech Co., Ltd.) to obtain the cDNA. The cDNA and
SYBR-Green mixture (CoWin Biosciences) were used following the
manufacturer's protocol for RT-qPCR. The machine used was from
Bio-Rad Laboratories, Inc. and the RT-qPCR program was as follows:
95°C 10 min, 95°C 10 sec and 60°C 30 sec (45 cycles), 95°C 1 min,
55°C 1 min, 55°C 10 sec, 95°C 5 sec. The 2−ΔΔCq method
was used for data analysis (31)
and the relative expression was normalized to β-actin. The primers
sequences used for the experiment were: β-actin, forward,
5′-GAAGTGTGACGTGGACATCC-3′ and reverse, 5′-CCGATCCACACGGAGTACTT-3′;
NRF2, forward, 5′-GAGAGCCCAGTCTTCATTGC-3′ and reverse,
5′-TGCTCAATGTCCTGTTGCAT-3′; Bcl-2, forward,
5′-GGTGGGGTCATGTGTGTGG-3′ and reverse,
5′-CGGTTCAGGTACTCAGTCATCC-3′. At least three independent assays
were performed in triplicate.
Western blot analysis
Following corilagin stimulation, U251 cells were
harvested for lysis using RIPA lysate buffer. Lysates were
incubated on ice for >30 min and centrifuged at 12,000 × g for
15 min at 4°C. The supernatant was transferred to a new Eppendorf
tube and the precipitate discarded. Protein concentration was
detected by a BCA determination kit. Proteins (50 µg) were
separated by 12% SDS-PAGE and transferred onto a PVDF membrane.
Membranes were blocked with 5% skimmed milk for 1 h at room
temperature and incubated with primary antibodies at a dilution of
1:1,000 overnight at 4°C. The membranes were incubated with a
horseradish peroxidase (HRP)-conjugated secondary (1:5,000
dilution) at 37°C for 1 h. Blots were detected using ECL
chemiluminescent reagents. The relative protein expression of genes
was quantified by ImageJ v.1.46r software (National Institutes of
Health) using β-actin as control, Data are shown as mean ± standard
deviation from at least three independent experiments. P<0.05
was considered to indicate a statistically significant
difference.
Hoechst 33258 staining
Following corilagin stimulation, U251 cells were
washed with PBS twice at room temperature. The cells were fixed
with methanol for 10 min and incubated with Hoechst 33258 staining
reagent for 10 min at room temperature. A confocal microscope
(Zeiss LSM800; Zeiss AG) was used to detect the apoptosis levels at
×100 magnification and the apoptotic cells appeared as dense bright
blue. The experiments were carried out three times,
individually.
Immunofluorescence staining
Following corilagin stimulation, U251 cells were
washed with PBS three times and fixed with 4% paraformaldehyde for
10 min at room temperature. Cell membrane was penetrated using PBS
containing 0.1% Triton X-100 and blocked with 10% FBS at room
temperature. Cells were incubated with LC3B primary antibody (1:200
dilution) at room temperature for 2 h. The cells were washed three
times and incubated with coralite488-labeled secondary antibody
(1:100) at room temperature for 30 min. Following washing with PBS
for three times, the cells were incubated with DAPI for 10 min at
room temperature. Finally, the slides were sealed with
anti-fluorescent quencher to delay fluorescence quenching. All
experiments were performed in triplicate and repeated at least
three times.
Cell transfection
U251 cells were seeded in 6-well plates
(2×105/well) until reached 30–40%, before transfection,
the transfection reagent Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), 10 nM negative
control (NC) or 10 nM small interfering (si)RNAs were mixed and
incubated for 20 min, and then added to the wells with serum-free
medium. The NC and siRNAs were synthesized by Shanghai GenePharma
Co., Ltd. Sequence of NC was: 5′-UUCUCCGAACGUGUCACGUTT-3′; sequence
of siRNAs were: siNrf2-1, 5′-GGUUGAGACUACCAUGGUUTT-3′; siNrf2-2,
5′-GACAGAAGUUGACAAUUAUTT-3′; siNrf2-3, 5′-CCAGAACACUCAGUGGAAUTT-3′.
Following transfection for 72 h, cells were harvested for further
study.
GEPIA dataset
GEPIA is an interactive web server for analyzing the
RNA sequencing expression data that includes 9,736 tumors and 8,587
normal samples from The Cancer Genome Atlas and the Genotype-Tissue
Expression projects, using the output of a standard processing
pipeline for RNA sequencing data. GEPIA provides customizable
functions such as tumor/normal differential expression analysis,
profiling according to cancer types or pathological stages, patient
survival analysis, similar gene detection, correlation analysis,
and dimensionality reduction analysis. GEPIA dynamically plots
expression profiles of a given gene according to user-defined
sample selections and methods. The results can be presented in box
plots. In addition, GEPIA plots gene expression by pathological
stages based on the TCGA clinical annotation. GEPIA performs
survival analysis based on gene expression levels. This function
allows users to select their custom cancer types for survival
analysis. For example, to examine the survival curves of an input
gene in glioma, we select both GBM and LGG for the survival
analyses. GEPIA uses log-rank test for the hypothesis evaluation.
The hazard ratio and the 95% confidence interval information can
also be included in the survival plot (32).
Statistical analysis
All data were expressed as mean ± standard deviation
and analyzed by one-way ANOVA followed by Bonferroni's post hoc
test using GraphPad Prism v.5 (GraphPad Software, Inc.). The error
bars were derived from at least three independent experiments.
P<0.05 was considered to indicate a statistically significant
difference.
Results
NRF2 is overexpressed in glioma
tissues and is negatively correlated with patient survival
The nine non-glioma specimens were taken from
patients with CH or TBI requiring intracranial decompression and
the 15 tumor tissues were taken from patients with glioma. The
basic characteristics of these 24 brain tissues are listed in
Tables I and II. Total proteins were extracted the from
these tissues and it was discovered by using western blotting that
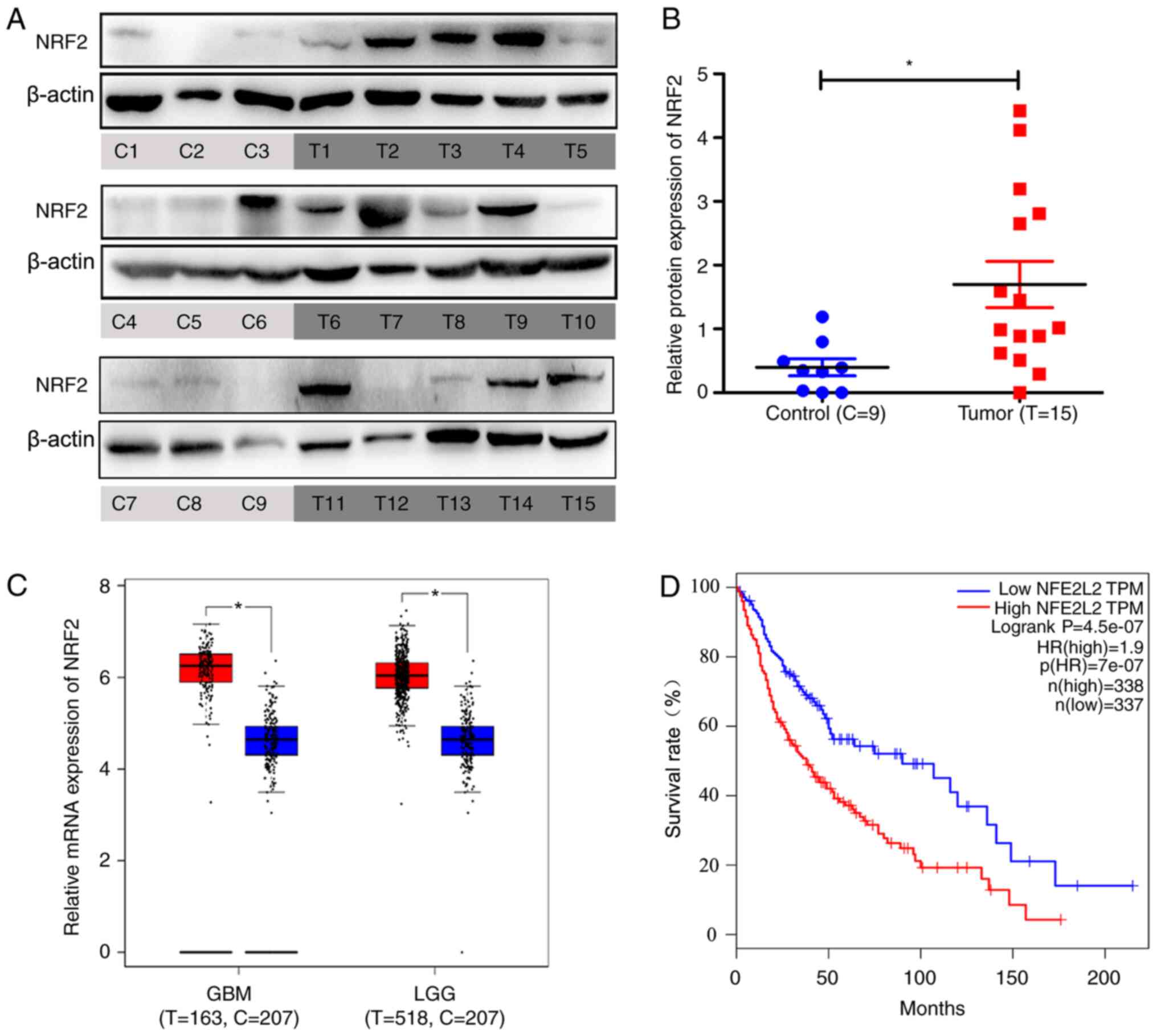
NRF2 was overexpressed in glioma tissues compared with non-glioma
specimens (Fig. 1A and B).
Information from a large sample database (Gene Expression Profiling
Interactive Analysis, GEPIA) revealed that NRF2 was overexpressed
in GBM and LGG (Fig. 1C). NRF2 was
negatively correlated with patient survival using GEPIA. The low
NRF2 expression group demonstrated higher percent survival rates
compared with the high NRF2 expression group (Fig. 1D).
 | Table I.Characteristics of non-glioma patient
samples. |
Table I.
Characteristics of non-glioma patient
samples.
| Number | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 |
|---|
| Sex | M | F | M | M | F | M | M | F | F |
| Age (years) | 40 | 43 | 69 | 44 | 27 | 51 | 31 | 44 | 55 |
| Source | CH | TBI | CH | CH | CH | CH | TBI | CH | CH |
 | Table II.Clinicopathological characteristics
of glioma patient samples. |
Table II.
Clinicopathological characteristics
of glioma patient samples.
| Number | T1 | T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 | T10 | T11 | T12 | T13 | T14 | T15 |
|---|
| Sex | F | F | M | F | M | F | F | M | F | F | M | F | M | M | M |
| Age (years) | 67 | 72 | 67 | 40 | 60 | 50 | 67 | 40 | 60 | 68 | 44 | 53 | 39 | 68 | 70 |
| WHO Grade | IV | IV | III | IV | IV | IV | IV | III | III–IV | III | III | IV | IV | IV | IV |
Corilagin downregulates the expression
of NRF2 in the U251 glioma cell line
The present study demonstrated that corilagin, an
anti-glioma drug, possessed a clear ability to inhibit the
proliferation of glioma cells (33). To determine whether it inhibits NRF2
express ion to suppress glioma cells, the regulation effect exerted
by corilagin on NRF2 was investigated. Following the use of
corilagin to stimulate U251 cells, the protein expression of NRF2
was detected though western blotting. The protein expression of
NRF2 was weakened in the corilagin treatment group compared with
the control group (Fig. 2A and B).
Following stimulation of U251 cells by using 100 µg/ml corilagin,
the protein expression of NRF2 was detected though
immunofluorescence. The fluorescence intensity of NRF2 was weakened
in the corilagin treatment group (Fig.
2C).
Corilagin induces apoptosis of the
U251 glioma cell line
To determine whether corilagin exerts other effects
on U251 glioma cells, the following experiments were performed.
Different concentrations of corilagin (0, 25, 50 and 100 µg/ml)
were used to stimulate cells for 48 h and the level of cell
apoptosis was detected using Hoechst 33258 staining. The results
demonstrated that cells appeared bright blue with the intensity of
the color increasing as concentration increased (Fig. 3A). This indicated that corilagin
induced apoptosis of the U251 glioma cells. To further explore the
induced-apoptosis, the results were verified at the molecular
level. The corilagin-stimulated cells were harvested and the
protein expression of Bcl-2 was detected using western blotting.
The result indicated that the protein expression of Bcl-2 was
reduced by the increased concentration of corilagin (Fig. 3B). The mRNA expression of Bcl-2 was
detected using RT-qPCR and the results indicated that the mRNA
expression of Bcl-2 was also reduced by the increasing
concentration of corilagin (Fig.
3C).
Corilagin induces the autophagy of the
U251 glioma cell line
Whether corilagin regulated cell autophagy was
investigated. EBSS is a positive inducer of autophagy. Following
stimulation of U251 cells using EBSS and 100 µg/ml corilagin, the
cell autophagy-related protein LC3II was detected though
immunofluorescence. The level of LC3II increased significantly in
the EBSS and corilagin treatment groups (Fig. 4A) by immunofluorescence, whereas the
expression of P62 decreased compared with the control group, as
observed using western blotting (Fig.
4B). Although the decrease of P62 appears slight, no less than
three experiments prove that the decrease is statistically
significant. Therefore, the level of autophagy in the corilagin
treatment group was higher compared with the control group. The
relative protein expression levels of P62 and the ratio of LC3II to
LC3I are illustrated in Fig. 4C. In
addition, the mRNA level of P62 was reduced in the
corilagin-treated group (Fig. 4D)
confirming that the level of autophagy in the Corilagin treatment
group was also greater than that in the control group.
NRF2 regulates apoptosis and
autophagy
To demonstrate whether corilagin induced apoptosis
and autophagy via inhibiting NRF2, small interfering (si)RNAs were
used to knock down the NRF2 expression in U251 cell line. In the
present study, three siRNAs sequences of NRF2 were designed, but
only one of these siRNA sequences significantly inhibited the
expression of NRF2. It was discovered that as the expression of
NRF2 was suppressed, the expression of P62 and Bcl-2 were also
suppressed and the ratio of LC3IIto LC3Iis increased (Fig. 5A and B). This result is consistent
with the effect of corilagin on stimulating cells. As mentioned
above, corilagin clearly inhibited NRF2 expression, so it was
hypothesized that the inhibitory effect of corilagin on cells was
at least partly dependent on NRF2 inhibition.
Discussion
The present study demonstrated that corilagin
induced apoptosis and autophagy of the NRF2-addicted U251 glioma
cell line and reduced NRF2 expression. The significance of the
target gene NRF2 in the anti-glioma role of corilagin was
identified. The present study aimed to discover a new function of
corilagin as a novel antitumor drug and its key target. It also
provided a new theoretical basis for tumor cell
chemoresistance.
Overcoming chemotherapy resistance is a challenging
problem in glioma treatment. Temozolomide is expensive and has poor
clinical efficacy (34); the
development of new anticancer drugs is thus an urgent matter. A
number of studies have demonstrated numerous pharmacological
properties of corilagin in vivo and in vitro. In
vivo experiments have revealed that corilagin inhibits growth
of tumors, such as hepatocellular carcinoma and ovarian cancer, in
an athymic implant tumor model (35,36).
Due to the blood brain barrier (BBB), animal experiments are
crucial for researching anti-glioma chemotherapy drugs. Corilagin
has three phenolic rings for π-π stacking interactions with some
amino acid residues and exhibits excellent amyloid-β (Aβ) oligomer
selectivity with a high binding affinity (37,38).
The primary influence driving Alzheimer's disease (AD) pathogenesis
is accumulation of Aβ in the brain, therefore, corilagin
demonstrates the potential to alleviate AD (37–39).
Although the evidence (37–39) support the view that corilagin can
pass through the BBB, animal experiments are required to support
the conclusion of the present study. In vitro, corilagin
exhibits strong inhibitory activity against viruses and
microorganisms (40,41). Staphylococcus aureus,
well-known for its drug resistance, is inhibited by corilagin at a
minimum inhibitory concentration of 25 µg/ml (42). The anti-microorganism activity of
corilagin is expected to solve the problem of bacterial drug
resistance (43).
Thus, corilagin regulates several signal pathways to
participate in biological activities (44). In vivo and in vitro
experiments demonstrate that corilagin prevents herpes simplex
virus-1 encephalitis by inhibiting the Toll-like receptor 2
signaling pathways (45). Corilagin
reduces the release of TNF-α, which is considered to stimulate the
growth and progression of early malignant tumor cells (46,47).
Corilagin attenuates radiation-induced brain injury in mice by
inhibiting the expression of NF-κB and TNF-α (48). Our team demonstrated that corilagin
inhibits the proliferation of glioma cells by blocking the NF-κB
pathway (33). However, the present
study demonstrated the inhibitory effect of NRF2-mediated corilagin
on glioma cells, enriching the anti-tumor mechanism of corilagin,
from the perspective of apoptosis and autophagy. NRF2 mainly
regulates transcriptional activation through antioxidant response
elements (AREs) present in the promoters of NRF2 target genes
(49). Studies (50–52)
have revealed that NRF2 is overexpressed in the KB-derived
drug-resistant cancer cell panel. Silencing NRF2 expression can
enhance the drug sensitivity of chemoresistant cells and NRF2
activator can enable cells to acquire drug resistance (50). In addition, a previous study
demonstrated that disulfiram can selectively inhibit the NRF2 and
NF-κB signaling pathways of leukemia stem cells, restricting their
proliferation and inducing apoptosis (53). Our team previously demonstrated that
corilagin possesses a strong inhibitory effect on glioma stem cells
(33), however, whether glioma stem
cells are more addictive to NRF2 than glioma cells requires further
research. The present study mainly focused on the regulation of
corilagin in glioma cells by inhibiting NRF2 expression. It was
demonstrated that NRF2 was highly expressed in glioma. That glioma
stem cells are addictive to NRF2 (28,29)
supports the conclusion of the present study to some extent.
Nevertheless, the small samples size remains a limitation of the
present study. In theory, the larger the sample size, the more
accurate the research results. When the sample size is not large
enough, it is more difficult to use the limited data to prove the
significance of the NRF2 overexpression with glioma. Therefore,
increasing the samples size needs be addressed in future research.
With enough samples, the detection of NRF2 expression can be
performed to provide more evidence to support the hypothesis of the
present study. Previous studies have investigated the mechanism by
which corilagin regulates microRNA (54,55).
One reported that corilagin reduces the expression of miR-21 and
inhibits schistosomiasis-induced hepatic fibrosis (54). Previous studies (33,42,44–48,54)
have demonstrated that corilagin regulates numerous pathways and
possesses various biological properties, including anti-viral,
anti-bacterial and anti-cancer.
Cell apoptosis is considered to be cell death type
I, with the ability to maintain cell fate and homeostasis (56). Apoptotic procedures are disrupted
during tumorigenesis. The Bcl-2 family of proteins regulate cell
apoptosis (57). The family
includes proapoptotic proteins, such as Bad, Bax and Bid and
antiapoptotic proteins, such as Bcl-2 and Bcl-XL. The balance
between proapoptotic and antiapoptotic proteins determines the
progress of apoptosis (57). The
present study focused on the anti-apoptotic protein Bcl-2 and
demonstrated that corilagin induced cell apoptosis through the
reduction of Bcl-2 expression. Cell apoptosis is closely associated
with autophagy proteins. Autophagy-related (ATG)5 and ATG12, which
are necessary for induction of cell autophagy, contribute to the
induction of apoptosis. ATG5 interacts with Bcl-XL to promote the
release of cytochrome c to regulate cell apoptosis (58,59).
The present study focused on NRF2, considered an apoptosis
inhibitor. The significance of the NRF2-related apoptosis process
is diverse for various physiological processes (25,60,61).
For example, a previous study revealed that exposure of
non-small-cell lung cancer cells to ionizing radiation induces cell
apoptosis and knocking down NRF2 expression boosts cell apoptosis
(25). NRF2 acts as a protective
factor against normal tissue damage and possesses a protective
effect on brain injury by inhibiting mitochondria-related apoptosis
(60). Cisplatin resistance is a
significant obstacle in the treatment of tumors and it has been
demonstrated that NRF2 expression induces cisplatin resistance in
ovarian cancer (61). Several
studies have reported that NRF2 inhibits apoptosis by activating
AREs and that this process is a protective program against cell
death (62–64).
The expression level of NRF2 is inextricably linked
to autophagy intensity. A previous study clarifies the relationship
between NRF2 and autophagy, demonstrating that autophagic flow
disorder leads to p62 accumulation and then detachment of NRF2 from
Keap1, which in turn regulates the ARE pathway (65). The present study identified that
NRF2 expression was reduced by corilagin stimulation. The
ubiquitination pathway is a protein degradation pathway independent
of lysosomal degradation (66). The
26S proteasome is the major protease responsible for protein
degradation (67). Numerous
factors, including human telomerase reverse transcriptase (68) and cyclic adenosine monophosphate
(69), regulate 26S proteasome
activity. Whether corilagin inhibits NRF2 expression by regulating
26S proteasome activity should be further explored. A previous
study reported that corilagin induces autophagy in breast cancer
cells by inhibiting Akt/mTOR/p70S6K and this induced autophagy can
be downregulated by the ROS scavenger N-acetylcysteine (14). This study also showed that the
corilagin-induced autophagy of breast cancer cell may depend on ROS
generation. Another study demonstrated that corilagin also promotes
ROS production and induces autophagy (12). ROS was not detected in the present
study, which was one of its limitations. However, a number of
studies reveal that NRF2 inhibition induces ROS production
(70–72) and these results support the
conclusion of the present study that corilagin induced autophagy
via downregulating NRF2. In addition, to confirm that the action of
corilagin is specific to the reduction of NRF2 expression, it would
be more convincing to administer corilagin to NRF2-overexpressing
U251 cells to verify the suppression of cell death. The
overexpressed vector of NRF2 has not been successfully created yet,
and this another limitation to the present study. It is hoped, once
the vector is successfully constructed, to remedy this defect.
In conclusion, the present study demonstrated the
mechanisms of corilagin in glioma treatment and explored its
relationship with NRF2 expression. It was identified that corilagin
induced apoptosis and autophagy in the U251 glioma cell line. NRF2
was overexpressed in glioma tissues and its reduced expression
contributed to the anti-glioma effect of corilagin. The data from
the present study provided solid evidence for corilagin as a
candidate drug for glioma treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the project of
Health and Family Planning Commission of Shandong province (grant
no. 2017WS513), Supporting Fund for Teachers' research of Jining
Medical University (grant nos. JYFC2019FKJ125, JYFC2018FKJ035 and
JYFC2018FKJ107), the project of Jining Science and Technology
Bureau (grant no. 2016-56-60), Scientific Research Project of
Jining Medical University (grant no. JY2015KJ022), Project of
Scientific Research Program of Affiliated Hospital of Jining
Medical University (grant nos. MP-2018-012, MP-2015-003) and
Scientific Research Project of Jining Medical University (grant no.
JY2015KJ022).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JL, XQ and FJ conceived and designed the study. JL
and XQ conducted the experiments, analyzed the data and drafted the
manuscript. WM collected the clinical tissues and participated in
protein determination. SJ and XZ participated in the data
acquisition and analysis. FJ, XY and DP provided experimental
guidance and participated in data analysis. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Affiliated Hospital of Jining Medical University
(Jining, China) and the approval number was 2019C008. The present
study was performed in accordance with the Declaration of Helsinki
and guidelines of the Ethics Committee of Affiliated Hospital of
Jining Medical University. Each participant (or their legal
representative) provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen R, Smith-Cohn M, Cohen AL and Colman
H: Glioma subclassifications and their clinical significance.
Neurotherapeutics. 14:284–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Delgado-Lopez PD, Corrales-Garcia EM,
Martino J, Lastra-Aras E and Duenas-Polo MT: Diffuse low-grade
glioma: A review on the new molecular classification, natural
history and current management strategies. Clin Transl Oncol.
19:931–944. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Duffau H and Taillandier L: New concepts
in the management of diffuse low-grade glioma: Proposal of a
multistage and individualized therapeutic approach. Neuro Oncol.
17:332–342. 2015.PubMed/NCBI
|
|
4
|
Chen X, Zhang M, Gan H, Wang H, Lee JH,
Fang D, Kitange GJ, He L, Hu Z, Parney IF, et al: A novel enhancer
regulates MGMT expression and promotes temozolomide resistance in
glioblastoma. Nat Commun. 9:29492018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schmidt OT and Lademann R: Corilagin, ein
weiterer kristallisierter Gerbstoff aus Dividivi. X. Mitteilung
über natürliche Gerbstoffe. Justus Liebigs Ann Chem. 571:232–237.
1951. View Article : Google Scholar
|
|
6
|
Kakiuchi N, Hattori M, Namba T, Nishizawa
M, Yamagishi T and Okuda T: Inhibitory effect of tannins on reverse
transcriptase from RNA tumor virus. J Nat Prod. 48:614–621. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qiu F, Liu L, Lin Y, Yang Z and Qiu F:
Corilagin inhibits esophageal squamous cell carcinoma by inducing
DNA damage and down-regulation of RNF8. Anticancer Agents Med Chem.
19:1021–1028. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ding Y, Ren D, Xu H, Liu W, Liu T, Li L,
Li J, Li Y and Wen A: Antioxidant and pro-angiogenic effects of
corilagin in rat cerebral ischemia via Nrf2 activation. Oncotarget.
8:114816–114828. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu FC, Chaudry IH and Yu HP:
Hepatoprotective effects of corilagin following hemorrhagic shock
are through akt-dependent pathway. Shock. 47:346–345. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo S, Fu Y, Xiong S and Lv J: Corilagin
protects the acute lung injury by ameliorating the apoptosis
pathway. Biomed Pharmacother. 95:1743–1748. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li HR, Liu J, Zhang SL, Luo T, Wu F, Dong
JH, Guo YJ and Zhao L: Corilagin ameliorates the extreme
inflammatory status in sepsis through TLR4 signaling pathways. BMC
Complement Altern Med. 17:182017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu J, Zhang G, Tong Y, Yuan J, Li Y and
Song G: Corilagin induces apoptosis, autophagy and ROS generation
in gastric cancer cells in vitro. Int J Mol Med. 43:967–979.
2019.PubMed/NCBI
|
|
13
|
Deng Y, Li X, Li X, Zheng Z, Huang W, Chen
L, Tong Q and Ming Y: Corilagin induces the apoptosis of
hepatocellular carcinoma cells through the mitochondrial apoptotic
and death receptor pathways. Oncol Rep. 39:2545–2552.
2018.PubMed/NCBI
|
|
14
|
Tong Y, Zhang G, Li Y, Xu J, Yuan J, Zhang
B, Hu T and Song G: Corilagin inhibits breast cancer growth via
reactive oxygen species-dependent apoptosis and autophagy. J Cell
Mol Med Jun. 22:3795–3807. 2018. View Article : Google Scholar
|
|
15
|
Li N, Lin Z, Chen W, Zheng Y, Ming Y,
Zheng Z, Huang W, Chen L, Xiao J and Lin H: Corilagin from longan
seed: Identification, quantification, and synergistic cytotoxicity
on SKOv3ip and hey cells with ginsenoside Rh2 and 5-fluorouracil.
Food Chem Toxicol. 119:133–140. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Milani R, Brognara E, Fabbri E, Finotti A,
Borgatti M, Lampronti I, Marzaro G, Chilin A, Lee KK, Kok SH, et
al: Corilagin induces high levels of apoptosis in the
temozolomide-resistant T98G glioma cell line. Oncol Res.
26:1307–1315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Noguchi M, Hirata N, Tanaka T, Suizu F,
Nakajima H and Chiorini JA: Autophagy as a modulator of cell death
machinery. Cell Death Dis. 11:5172020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choi AM, Ryter SW and Levine B: Autophagy
in human health and disease. N Engl J Med. 368:651–662. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baechler BL, Bloemberg D and Quadrilatero
J: Mitophagy regulates mitochondrial network signaling, oxidative
stress, and apoptosis during myoblast differentiation. Autophagy.
15:1606–1619. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang P and Mizushima N: LC3- and
p62-based biochemical methods for the analysis of autophagy
progression in mammalian cells. Methods. 75:13–18. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He F, Ru X and Wen T: NRF2, a
transcription factor for stress response and beyond. Int J Mol Sci.
21:47772020. View Article : Google Scholar
|
|
22
|
Baird L and Yamamoto M: The molecular
mechanisms regulating the KEAP1-NRF2 pathway. Mol Cell Biol.
40:e00099-20. 2020. View Article : Google Scholar
|
|
23
|
Unoki T, Akiyama M and Kumagai Y: Nrf2
activation and its coordination with the protective defense systems
in response to electrophilic stress. Int J Mol Sci. 21:5452020.
View Article : Google Scholar
|
|
24
|
Menegon S, Columbano A and Giordano S: The
dual roles of NRF2 in cancer. Trends Mol Med. 22:578–593. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao Q, Mao A, Yan J, Sun C, Di C, Zhou X,
Li H, Guo R and Zhang H: Downregulation of Nrf2 promotes
radiation-induced apoptosis through Nrf2 mediated Notch signaling
in non-small cell lung cancer cells. Int J Oncol. 48:765–773. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meng QT, Chen R, Chen C, Su K, Li W, Tang
LH, Liu HM, Xue R, Sun Q, Leng Y, et al: Transcription factors Nrf2
and NF-kappaB contribute to inflammation and apoptosis induced by
intestinal ischemia-reperfusion in mice. Int J Mol Med.
40:1731–1740. 2017.PubMed/NCBI
|
|
27
|
Wan ZH, Jiang TY, Shi YY, Pan YF, Lin YK,
Ma YH, Yang C, Feng XF, Huang LF, Kong XN, et al: RPB5-mediating
protein promotes cholangiocarcinoma tumorigenesis and drug
resistance by competing with NRF2 for KEAP1 binding. Hepatology.
71:2005–2022. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kitamura H and Motohashi H: NRF2 addiction
in cancer cells. Cancer Sci. 109:900–911. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ji XJ, Chen SH, Zhu L, Pan H, Zhou Y, Li
W, You WC, Gao CC, Zhu JH, Jiang K, et al: Knockdown of
NF-E2-related factor 2 inhibits the proliferation and growth of
U251MG human glioma cells in a mouse xenograft model. Oncol Rep.
30:157–164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Taguchi K and Yamamoto M: The KEAP1-NRF2
System in Cancer. Front Oncol. 7:852017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang WT, Li GH, Li ZY, Feng S, Liu XQ, Han
GK, Zhang H, Qin XY, Zhang R, Nie QM, et al: Effect of corilagin on
the proliferation and NF-κB in U251 glioblastoma cells and U251
glioblastoma stem-like cells. Evid Based Complement Alternat Med.
2016:14183092016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Basso J, Miranda A, Sousa J, Pais A and
Vitorino C: Repurposing drugs for glioblastoma: From bench to
bedside. Cancer Lett. 428:173–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hau DK, Zhu GY, Leung AK, Wong RS, Cheng
GY, Lai PB, Tong SW, Lau FY, Chan KW, Wong WY, et al: In vivo
anti-tumour activity of corilagin on Hep3B hepatocellular
carcinoma. Phytomedicine. 18:11–15. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pham AT, Malterud KE, Paulsen BS, Diallo D
and Wangensteen H: DPPH radical scavenging and xanthine oxidase
inhibitory activity of Terminalia macroptera leaves. Nat
Prod Commun. 6:1125–1128. 2011.PubMed/NCBI
|
|
37
|
Gaudreault R and Mousseau N: Mitigating
Alzheimer's disease with natural polyphenols: A review. Curr
Alzheimer Res. 16:529–543. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moraes LS, Donza MR, Rodrigues AP, Silva
BJ, Brasil DS, Zoghbi M, Andrade EH, Guilhon GM and Silva EO:
Leishmanicidal activity of (+)-phyllanthidine and the phytochemical
profile of Margaritaria nobilis (Phyllanthaceae). Molecules.
20:22157–22169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li Y, Xu D, Sun A, Ho SL, Poon CY, Chan
HN, Ng OT, Yung KK, Yan H, Li HW, et al: Fluoro-substituted cyanine
for reliable in vivo labelling of amyloid-β oligomers and
neuroprotection against amyloid-β induced toxicity. Chem Sci.
8:8279–8284. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Adesina SK, Idowu O, Ogundaini AO,
Oladimeji H, Olugbade TA, Onawunmi GO and Pais M: Antimicrobial
constituents of the leaves of Acalypha wilkesiana and
Aacalypha hispida. Phytother Res. 14:371–374. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yeo SG, Song JH, Hong EH, Lee BR, Kwon YS,
Chang SY, Kim SH, Lee SW, Park JH and Ko HJ: Antiviral effects of
Phyllanthus urinaria containing corilagin against human
enterovirus 71 and Coxsackievirus A16 in vitro. Arch Pharm Res.
38:193–202. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Burapadaja S and Bunchoo A: Antimicrobial
activity of tannins from Terminalia citrina. Planta Med.
61:365–366. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Teodoro GR, Brighenti FL, Delbem AC,
Delbem ÁC, Khouri S, Gontijo AV, Pascoal AC, Salvador MJ and
Koga-Ito CY: Antifungal activity of extracts and isolated compounds
from Buchenavia tomentosa on Candida albicans and
non-albicans. Future Microbiol. 10:917–927. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li X, Deng Y, Zheng Z, Huang W, Chen L,
Tong Q and Ming Y: Corilagin, a promising medicinal herbal agent.
Biomed Pharmacother. 99:43–50. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Guo YJ, Luo T, Wu F, Liu H, Li HR, Mei YW,
Zhang SL, Tao JY, Dong JH, Fang Y, et al: Corilagin protects
against HSV1 encephalitis through inhibiting the TLR2 signaling
pathways in vivo and in vitro. Mol Neurobiol. 52:1547–1560. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Komori A, Yatsunami J, Suganuma M, Okabe
S, Abe S, Sakai A, Sasaki K and Fujiki H: Tumor necrosis factor
acts as a tumor promoter in BALB/3T3 cell transformation. Cancer
Res. 53:1982–1985. 1993.PubMed/NCBI
|
|
47
|
Okabe S, Suganuma M, Imayoshi Y, Taniguchi
S, Yoshida T and Fujiki H: New TNF-alpha releasing inhibitors,
geraniin and corilagin, in leaves of Acer nikoense, Megusurino-ki.
Biol Pharm Bull. 24:1145–1148. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tong F, Zhang J, Liu L, Gao X, Cai Q, Wei
C, Dong J, Hu Y, Wu G and Dong X: Corilagin attenuates
radiation-induced brain injury in mice. Mol Neurobiol.
53:6982–6996. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu MC, Ji JA, Jiang ZY and You QD: The
Keap1-Nrf2-ARE pathway as a potential preventive and therapeutic
target: An update. Med Res Rev. 36:924–963. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen HH, Chang HH, Chang JY, Tang YC,
Cheng YC, Lin LM, Cheng SY, Huang CH, Sun MW, Chen CT, et al:
Enhanced B-Raf-mediated NRF2 gene transcription and HATs-mediated
NRF2 protein acetylation contributes to ABCC1-mediated
chemoresistance and glutathione-mediated survival in acquired
topoisomerase II poison-resistant cancer cells. Free Radic Biol
Med. 113:505–518. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Beidler DR, Chang JY, Zhou BS and Cheng
YC: Camptothecin resistance involving steps subsequent to the
formation of protein-linked DNA breaks in human
camptothecin-resistant KB cell lines. Cancer Res. 56:345–353.
1996.PubMed/NCBI
|
|
52
|
Ferguson PJ, Fisher MH, Stephenson J, Li
DH, Zhou BS and Cheng YC: Combined modalities of resistance in
etoposide-resistant human KB cell lines. Cancer Res. 48:5956–5964.
1988.PubMed/NCBI
|
|
53
|
Xu B, Wang S, Li R, Chen K, He L, Deng M,
Kannappan V, Zha J, Dong H and Wang W: Disulfiram/copper
selectively eradicates AML leukemia stem cells in vitro and in vivo
by simultaneous induction of ROS-JNK and inhibition of NF-κB and
Nrf2. Cell Death Dis. 8:e27972017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang F, Wang Y, Xue J, Ma Q, Zhang J, Chen
YF, Shang ZZ, Li QQ, Zhang SL and Zhao L: Effect of Corilagin on
the miR-21/smad7/ERK signaling pathway in a schistosomiasis-induced
hepatic fibrosis mouse model. Parasitol Int. 65:308–315. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou X, Xiong J, Lu S, Luo L, Chen ZL,
Yang F, Jin F, Wang Y, Ma Q, Luo YY, et al: Inhibitory effect of
corilagin on miR-21-regulated hepatic fibrosis signaling pathway.
Am J Chin Med. 47:1541–1569. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rami A and Kögel D: Apoptosis meets
autophagy-like cell death in the ischemic penumbra: Two sides of
the same coin? Autophagy. 4:422–426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Siddiqui WA, Ahad A and Ahsan H: The
mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update.
Arch Toxicol. 89:289–317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yousefi S, Perozzo R, Schmid I, Ziemiecki
A, Schaffner T, Scapozza L, Brunner T and Simon HU:
Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis.
Nat Cell Biol. 8:1124–1132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Booth LA, Tavallai S, Hamed HA,
Cruickshanks N and Dent P: The role of cell signalling in the
crosstalk between autophagy and apoptosis. Cell Signal. 26:549–555.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang Z and Guo S: Nrf2/HO-1 mediates the
neuroprotective effect of mangiferin on early brain injury after
subarachnoid hemorrhage by attenuating mitochondria-related
apoptosis and neuroinflammation. Sci Rep. 7:118832017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wu J, Zhang L, Li H, Wu S and Liu Z: Nrf2
induced cisplatin resistance in ovarian cancer by promoting CD99
expression. Biochem Biophys Res Commun. 518:698–705. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu P, Rojo de la Vega M, Sammani S,
Mascarenhas JB, Kerins M, Dodson M, Sun X, Wang T, Ooi A, Garcia
JG, et al: RPA1 binding to NRF2 switches ARE-dependent
transcriptional activation to ARE-NRE-dependent repression. Proc
Natl Acad Sci USA. 115:E10352–E10361. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Buendia I, Michalska P, Navarro E, Gameiro
I, Egea J and León R: Nrf2-ARE pathway: An emerging target against
oxidative stress and neuroinflammation in neurodegenerative
diseases. Pharmacol Ther. 157:84–104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shaw P and Chattopadhyay A: Nrf2-ARE
signaling in cellular protection: Mechanism of action and the
regulatory mechanisms. J Cell Physiol. 235:3119–3130. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jiang T, Harder B, Rojo de la Vega M, Wong
PK, Chapman E and Zhang DD: p62 links autophagy and Nrf2 signaling.
Free Radic Biol Med. 88:199–204. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen RH, Chen YH and Huang TY:
Ubiquitin-mediated regulation of autophagy. J Biomed Sci.
26:802019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Narayanan S, Cai CY, Assaraf YG, Guo HQ,
Cui Q, Wei L, Huang JJ, Ashby CR Jr and Chen ZS: Targeting the
ubiquitin-proteasome pathway to overcome anti-cancer drug
resistance. Drug Resist Updat. 48:1006632020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Im E, Yoon JB, Lee HW and Chung KC: Human
telomerase reverse transcriptase (hTERT) positively regulates 26S
proteasome activity. J Cell Physiol. 232:2083–2093. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lokireddy S, Kukushkin NV and Goldberg AL:
cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11
enhances their activity and the degradation of misfolded proteins.
Proc Natl Acad Sci USA. 112:E7176–E7185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Roh JL, Kim EH, Jang H and Shin D: Nrf2
inhibition reverses the resistance of cisplatin-resistant head and
neck cancer cells to artesunate-induced ferroptosis. Redox Biol.
11:254–262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang Y, Mandal AK, Son YO, Pratheeshkumar
P, Wise JT, Wang L, Zhang Z, Shi X and Chen Z: Roles of ROS, Nrf2,
and autophagy in cadmium-carcinogenesis and its prevention by
sulforaphane. Toxicol Appl Pharmacol. 353:23–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kovac S, Angelova PR, Holmström KM, Zhang
Y, Dinkova-Kostova AT and Abramov AY: Nrf2 regulates ROS production
by mitochondria and NADPH oxidase. Biochim Biophys Acta.
1850:794–801. 2015. View Article : Google Scholar : PubMed/NCBI
|